The Functional Characteristics and Soluble Expression of Saffron CsCCD2
Abstract
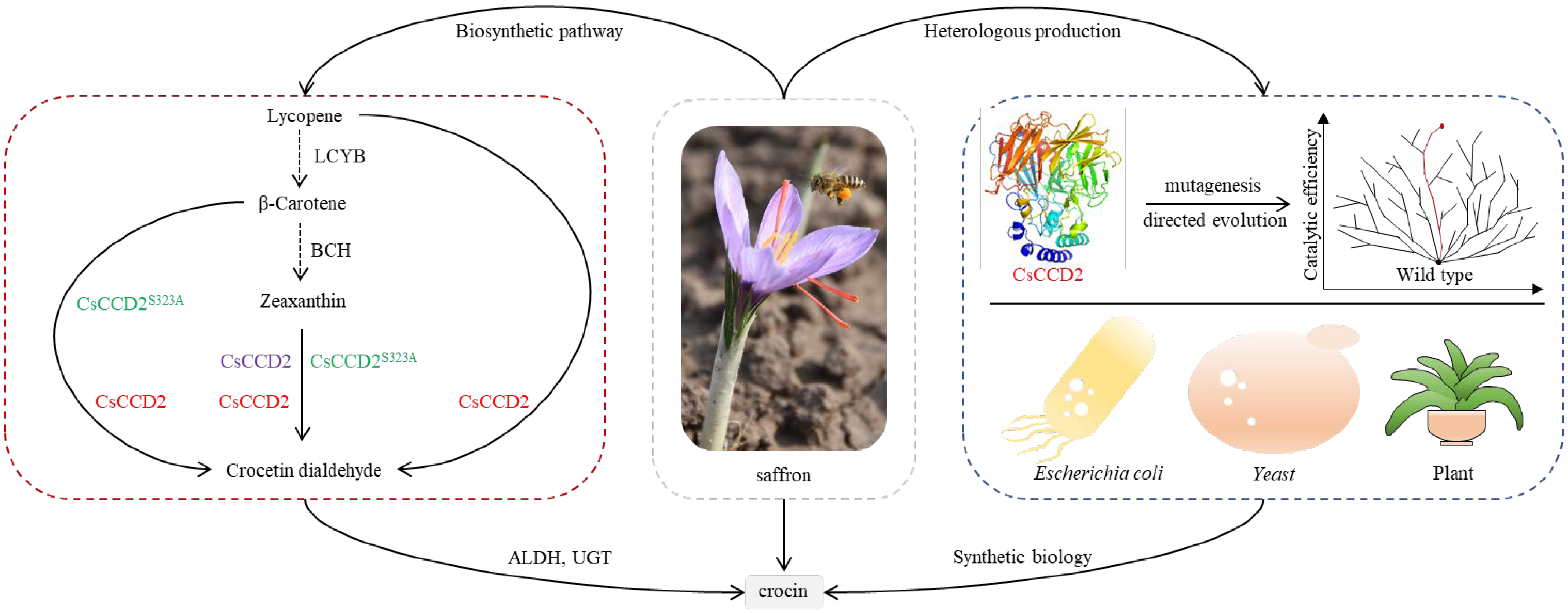
:1. Introduction
2. Results
2.1. Functional Characteristics of CsCCD2
2.2. Optimization of CsCCD2 Protein Expression
2.3. The Catalytic Activity Study of CsCCD2-Fusion Protein
2.4. Transforming the Catalytic Activity of CsCCD2 Using Tailored Truncation
3. Discussion
4. Materials and Methods
4.1. Chemicals and Strains
4.2. Plasmid Construction and Culture Conditions
4.3. The Function Assay of CsCCD2 in Bacteria
4.4. UPLC and LC-MS/MS Analysis of the Product
4.5. Optimization of CsCCD2 Protein Expression Pattern
4.6. The Bioinformatic Analysis of CsCCD2, TRX-CsCCD2, and DAN1-CsCCD2
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, Z.; Pu, X.; Gao, R.; Demurtas, O.C.; Fleck, S.J.; Richter, M.; He, C.; Ji, A.; Sun, W.; Kong, J.; et al. Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. 2020, 18, 63. [Google Scholar] [CrossRef] [PubMed]
- Khorasanchi, Z.; Shafiee, M.; Kermanshahi, F.; Khazaei, M.; Ryzhikov, M.; Parizadeh, M.R.; Kermanshahi, B.; Ferns, G.A.; Avan, A.; Hassanian, S.M. Crocus sativus a natural food coloring and flavoring has potent anti-tumor properties. Phytomedicine 2018, 43, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Rahaiee, S.; Hashemi, M.; Shojaosadati, S.A.; Moini, S.; Razavi, S.H. Nanoparticles based on crocin loaded chitosan-alginate biopolymers: Antioxidant activities, bioavailability and anticancer properties. Int. J. Biol. Macromol. 2017, 99, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Albalawi, G.A.; Albalawi, M.Z.; Alsubaie, K.T.; Albalawi, A.Z.; Elewa, M.A.F.; Hashem, K.S.; Al-Gayyar, M.M.H. Curative effects of crocin in ulcerative colitis via modulating apoptosis and inflammation. Int. Immunopharmacol. 2023, 118, 110138. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Albarral, J.A.; Ramirez, A.I.; de Hoz, R.; Lopez-Villarin, N.; Salobrar-Garcia, E.; Lopez-Cuenca, I.; Licastro, E.; Inarejos-Garcia, A.M.; Almodovar, P.; Pinazo-Duran, M.D.; et al. Neuroprotective and anti-inflammatory effects of a hydrophilic saffron extract in a model of glaucoma. Int. J. Mol. Sci. 2019, 20, 4110. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, G.; Nabavi, S.M.; Sancarlo, D.; Greco, A.; Pieretti, S. Crocus sativus L. (Saffron) in Alzheimer’s disease treatment: Bioactive effects on cognitive impairment. Curr. Neuropharmacol. 2021, 19, 1606–1616. [Google Scholar] [CrossRef] [PubMed]
- El Midaoui, A.; Ghzaiel, I.; Vervandier-Fasseur, D.; Ksila, M.; Zarrouk, A.; Nury, T.; Khallouki, F.; El Hessni, A.; Ibrahimi, S.O.; Latruffe, N.; et al. Saffron (Crocus sativus L.): A source of nutrients for health and for the treatment of neuropsychiatric and age-related diseases. Nutrients 2022, 14, 597. [Google Scholar] [CrossRef] [PubMed]
- Frusciante, S.; Diretto, G.; Bruno, M.; Ferrante, P.; Pietrella, M.; Prado-Cabrero, A.; Rubio-Moraga, A.; Beyer, P.; Gomez-Gomez, L.; Al-Babili, S.; et al. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 12246–12251. [Google Scholar] [CrossRef]
- Ahrazem, O.; Rubio-Moraga, A.; Nebauer, S.G.; Molina, R.V.; Gomez-Gomez, L. Saffron: Its phytochemistry, developmental processes, and biotechnological prospects. J. Agric. Food Chem. 2015, 63, 8751–8764. [Google Scholar] [CrossRef]
- Luo, Y.; Li, B.Z.; Liu, D.; Zhang, L.; Chen, Y.; Jia, B.; Zeng, B.X.; Zhao, H.; Yuan, Y.J. Engineered biosynthesis of natural products in heterologous hosts. Chem. Soc. Rev. 2015, 44, 5265–5290. [Google Scholar] [CrossRef]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Ro, D.K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 2006, 440, 940–943. [Google Scholar] [CrossRef] [PubMed]
- Lau, W.; Sattely, E.S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 2015, 349, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Reiter, M.A.; d’Espaux, L.; Wong, J.; Denby, C.M.; Lechner, A.; Zhang, Y.; Grzybowski, A.T.; Harth, S.; Lin, W.; et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 2019, 567, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, F.; Suire, C.; Mutterer, J.; Camara, B. Oxidative remodeling of chromoplast carotenoids: Identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in crocus secondary metabolite biogenesis. Plant Cell 2003, 15, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Pu, X.; He, C.; Yang, Y.; Wang, W.; Hu, K.; Xu, Z.; Song, J. In Vivo production of five crocins in the engineered Escherichia coli. ACS Synth. Biol. 2020, 9, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Ji, A.; Jia, J.; Xu, Z.; Li, Y.; Bi, W.; Ren, F.; He, C.; Liu, J.; Hu, K.; Song, J. Transcriptome-guided mining of genes involved in crocin biosynthesis. Front. Plant Sci. 2017, 8, 518. [Google Scholar] [CrossRef]
- Walter, M.H.; Strack, D. Carotenoids and their cleavage products: Biosynthesis and functions. Nat. Prod. Rep. 2011, 28, 663–692. [Google Scholar] [CrossRef]
- Pu, X.; Li, Z.; Tian, Y.; Gao, R.; Hao, L.; Hu, Y.; He, C.; Sun, W.; Xu, M.; Peters, R.J.; et al. The honeysuckle genome provides insight into the molecular mechanism of carotenoid metabolism underlying dynamic flower coloration. New Phytol. 2020, 227, 930–943. [Google Scholar] [CrossRef]
- Ohmiya, A.; Kishimoto, S.; Aida, R.; Yoshioka, S.; Sumitomo, K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006, 142, 1193–1201. [Google Scholar] [CrossRef]
- Brandi, F.; Bar, E.; Mourgues, F.; Horváth, G.; Turcsi, E.; Giuliano, G.; Liverani, A.; Tartarini, S.; Lewinsohn, E.; Rosati, C. Study of ‘Redhaven’ peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism. BMC Plant Biol. 2011, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Buah, S.; Mlalazi, B.; Khanna, H.; Dale, J.L.; Mortimer, C.L. The Quest for golden bananas: Investigating carotenoid regulation in a Fe’i group musa cultivar. J. Agric. Food Chem. 2016, 64, 3176–3185. [Google Scholar] [CrossRef] [PubMed]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Yao, M.D.; Wang, Y.; Liu, J.; Feng, L.; Wang, Z.M.; Li, X.Y.; Xiao, W.H.; Yuan, Y.J. CsCCD2 access tunnel design for a broader substrate profile in crocetin production. J. Agric. Food Chem. 2021, 69, 11626–11636. [Google Scholar] [CrossRef] [PubMed]
- Daruwalla, A.; Zhang, J.; Lee, H.J.; Khadka, N.; Farquhar, E.R.; Shi, W.; von Lintig, J.; Kiser, P.D. Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase. Proc. Natl. Acad. Sci. USA 2020, 117, 19914–19925. [Google Scholar] [CrossRef]
- Misawa, N.; Satomi, Y.; Kondo, K.; Yokoyama, A.; Kajiwara, S.; Saito, T.; Ohtani, T.; Miki, W. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J. Bacteriol. 1995, 177, 6575–6584. [Google Scholar] [CrossRef] [PubMed]
- Trautmann, D.; Beyer, P.; Al-Babili, S. The ORF slr0091 of Synechocystis sp. PCC6803 encodes a high-light induced aldehyde dehydrogenase converting apocarotenals and alkanals. FEBS J. 2013, 280, 3685–3696. [Google Scholar] [CrossRef]
- Liu, T.; Yu, S.; Xu, Z.; Tan, J.; Wang, B.; Liu, Y.G.; Zhu, Q. Prospects and progress on crocin biosynthetic pathway and metabolic engineering. Comput. Struct. Biotechnol. J. 2020, 18, 3278–3286. [Google Scholar] [CrossRef]
- Kloer, D.P.; Ruch, S.; Al-Babili, S.; Beyer, P.; Schulz, G.E. The structure of a retinal-forming carotenoid oxygenase. Science 2005, 308, 267–269. [Google Scholar] [CrossRef]
- Messing, S.A.; Gabelli, S.B.; Echeverria, I.; Vogel, J.T.; Guan, J.C.; Tan, B.C.; Klee, H.J.; McCarty, D.R.; Amzel, L.M. Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 2010, 22, 2970–2980. [Google Scholar] [CrossRef]
- Esposito, D.; Chatterjee, D.K. Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotechnol. 2006, 17, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Waugh, D.S. Making the most of affinity tags. Trends Biotechnol. 2005, 23, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Eicholt, L.A.; Aubel, M.; Berk, K.; Bornberg-Bauer, E.; Lange, A. Heterologous expression of naturally evolved putative de novo proteins with chaperones. Protein Sci. 2022, 31, e4371. [Google Scholar] [CrossRef]
- Smith, D.B.; Johnson, K.S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 1988, 67, 31–40. [Google Scholar] [CrossRef]
- di Guan, C.; Li, P.; Riggs, P.D.; Inouye, H. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene 1988, 67, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Butt, T.R.; Edavettal, S.C.; Hall, J.P.; Mattern, M.R. SUMO fusion technology for difficult-to-express proteins. Protein Expres. Purif. 2005, 43, 1–9. [Google Scholar] [CrossRef]





| Plasmids | Relevant Properties or Genetic Marker a | Source or Reference |
|---|---|---|
| pACCAR25ΔcrtX | pACYC184 plus crtE, crtI, crtB, crtY, and crtZ from E. uredovora, CmR | [26] |
| pACCAR16Δcrt | pACYC184 plus crtE, crtI, crtB, and crtY from E. uredovora, CmR | [26] |
| pACCRT-EIB | pACYC184 plus crtE, crtI, and crtB from E. uredovora, CmR | [26] |
| pTHIO-DAN1 | pBR322 ori and pUC ori, AmpR | [27] |
| DAN1-CsCCD2 | pTHIO-DAN1 plus CsCCD2 from saffron | [8] |
| DAN1-CsCCD2-T | pTHIO-DAN1 plus truncated CsCCD2 | This study |
| pET32a | pBR322 ori and f1 ori, AmpR | Novagen |
| TRX-CsCCD2 | pET32a plus CsCCD2 from saffron | This study |
| pET41a | pBR322 ori and f1 ori, KanR | Novagen |
| GST-CsCCD2 | pET41a plus CsCCD2 from saffron | This study |
| pET28a | pBR322 ori and f1 ori, KanR | Novagen |
| pET28a-SUMO | pET28a plus SUMO•Tag | This study |
| pET28a-MBP | pET28a plus MBP•Tag | This study |
| SUMO-CsCCD2 | pET28a-SUMO•Tag plus CsCCD2 from saffron | This study |
| MBP-CsCCD2 | pET28a-MBP•Tag plus CsCCD2 from saffron | This study |
| Primers | Sequence (5′–3′) | Restriction Endonuclease (Underlined) |
|---|---|---|
| pET41a-GST-F | CATGCCATGGGCGAAAACCTGTACTTTCAAGGCATGGCAAATAAGGAGGAG | NcoI |
| pET41a-GST-R | CCCAAGCTTTCATGTCTCTGCTTGGTGCTTCTG | HindIII |
| pET28a-SUMO-F | CCCAAGCTTCCGAAAACCTGTACTTTCAAGGCATGGCAAATAAGGAGGAG | HindIII |
| pET28a-SUMO-R | AAGGAAAAAAGCGGCCGCTCATGTCTCTGCTTGGTGCTTCTGAAGTTC | NotI |
| pET28a-MBP-F | CTAGCTAGCGAAAACCTGTACTTTCAAGGCCATATGATGGCAAATAAGGAGGAGGCAG | NheI |
| pET28a-MBP-R | CCCAAGCTTTCATGTCTCTGCTTGGTGCTTCTG | HindIII |
| CCD2-T-F | CCAAGCAGAGACATGAGTTTAAACGGTCTCCAGCTT | - |
| CCD2-T-R | ACTCATGTCTCTGCTTGGTGCTTCTGAAGTTCT | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, S.; Zhou, Z.; Sun, L.; Sun, J.; Shen, C.; Gao, R.; Song, J.; Pu, X. The Functional Characteristics and Soluble Expression of Saffron CsCCD2. Int. J. Mol. Sci. 2023, 24, 15090. https://doi.org/10.3390/ijms242015090
Wang Y, Li S, Zhou Z, Sun L, Sun J, Shen C, Gao R, Song J, Pu X. The Functional Characteristics and Soluble Expression of Saffron CsCCD2. International Journal of Molecular Sciences. 2023; 24(20):15090. https://doi.org/10.3390/ijms242015090
Chicago/Turabian StyleWang, Ying, Siqi Li, Ze Zhou, Lifen Sun, Jing Sun, Chuanpu Shen, Ranran Gao, Jingyuan Song, and Xiangdong Pu. 2023. "The Functional Characteristics and Soluble Expression of Saffron CsCCD2" International Journal of Molecular Sciences 24, no. 20: 15090. https://doi.org/10.3390/ijms242015090
APA StyleWang, Y., Li, S., Zhou, Z., Sun, L., Sun, J., Shen, C., Gao, R., Song, J., & Pu, X. (2023). The Functional Characteristics and Soluble Expression of Saffron CsCCD2. International Journal of Molecular Sciences, 24(20), 15090. https://doi.org/10.3390/ijms242015090





