Flubendazole Enhances the Inhibitory Effect of Paclitaxel via HIF1α/PI3K/AKT Signaling Pathways in Breast Cancer
Abstract
:1. Introduction
2. Results
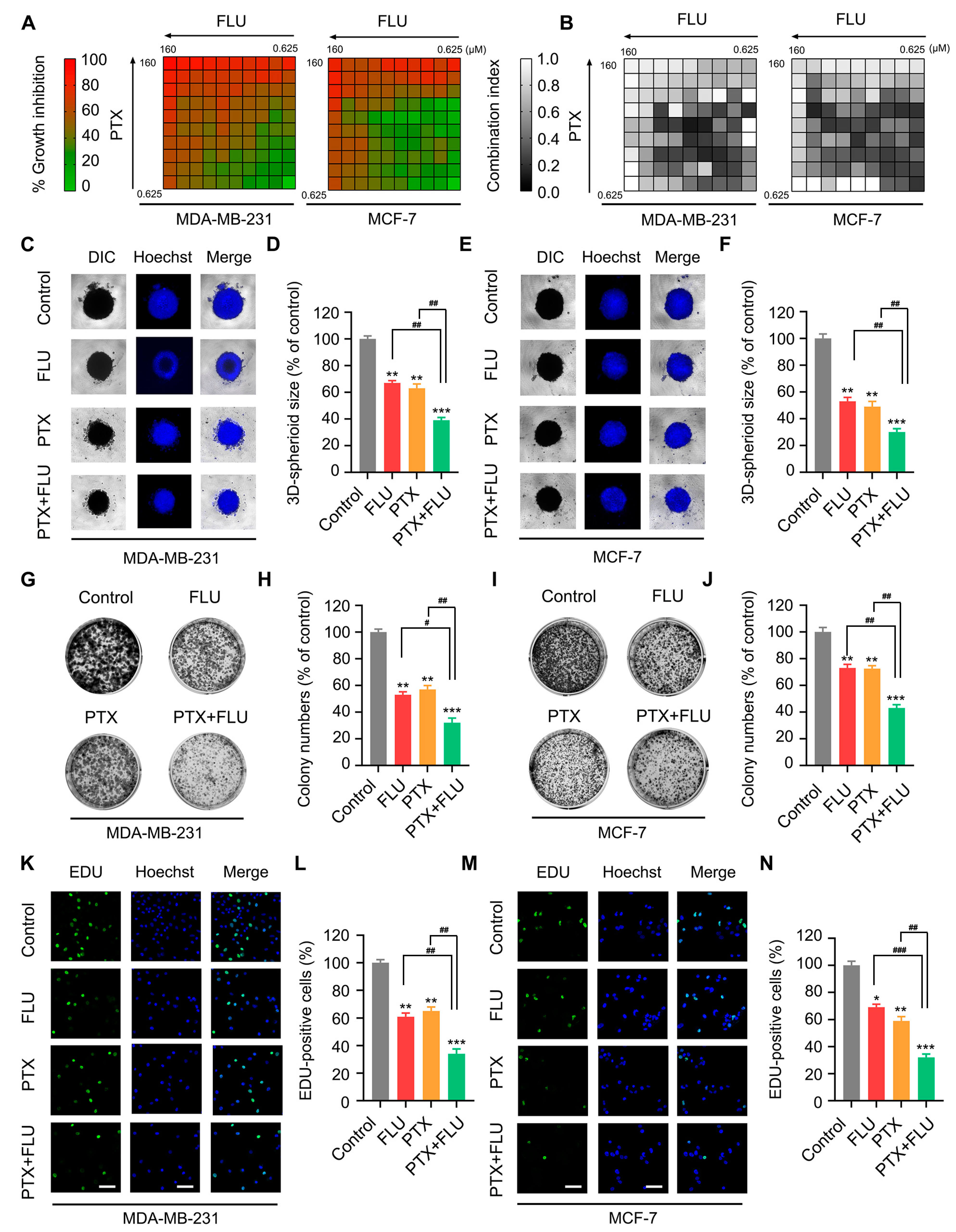
2.1. Flubendazole Enhances the Inhibitory Effect of Paclitaxel in BC Cells
2.2. Combination of Flubendazole and Paclitaxel Leads to Aberrant Mitosis and Induces Apoptosis in BC Cells
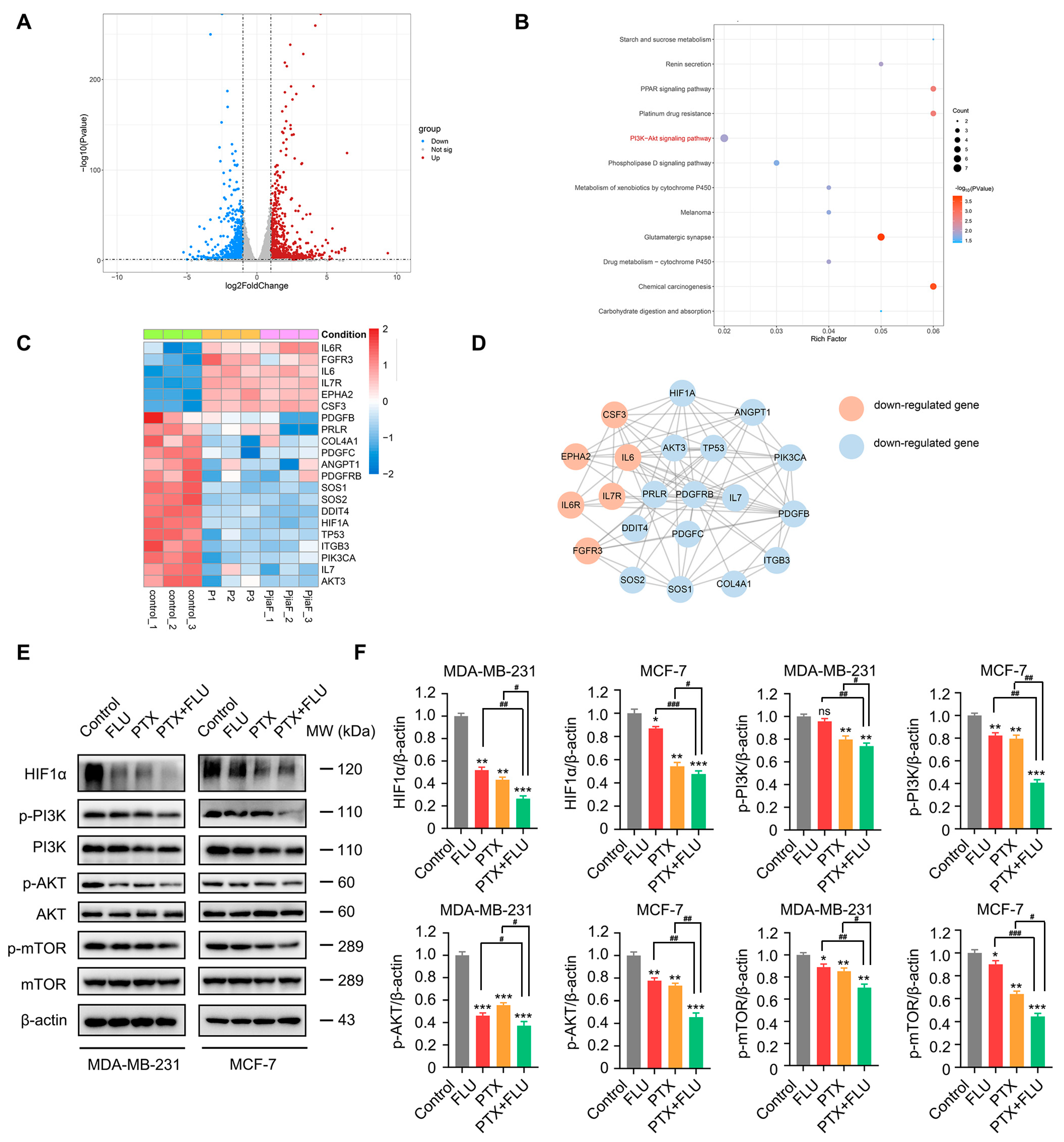
2.3. Combination of Flubendazole and Paclitaxel Regulates HIF1α/PI3K/AKT Signaling Pathways
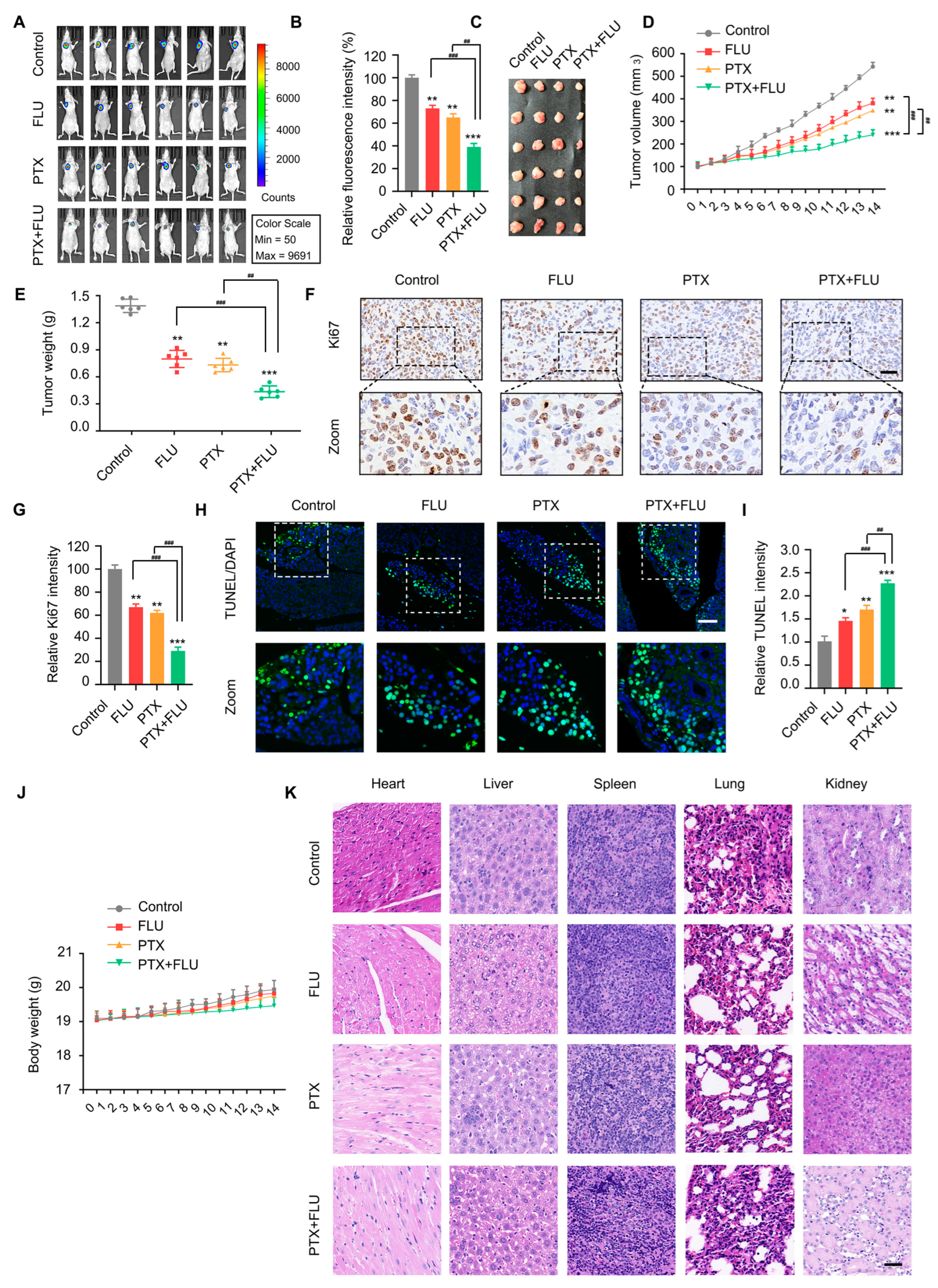
2.4. Flubendazole Combined with Paclitaxel Suppresses Tumor Growth and Metastasis In Vivo
2.5. Flubendazole Inhibits the Proliferation and Migration of MCF-7/PTX Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Cell Viability Assay
4.3. Combined Drug Effect Analysis
4.4. Colony Formation Assay
4.5. Three-Dimensional Cell Culture
4.6. Immunofluorescence
4.7. Western Blotting
4.8. Annexin-V/PI Dual Staining
4.9. Scratch Assay
4.10. Transwell Migration Assay
4.11. Cell Cycle Distribution
4.12. Animal Experiments
4.13. Immunohistochemistry
4.14. TUNEL Assays
4.15. H&E Staining
4.16. Transcriptome Analysis
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Pondé, N.F.; Zardavas, D.; Piccart, M. Progress in adjuvant systemic therapy for breast cancer. Nat. Rev. Clin. Oncol. 2019, 16, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Johnston, S.R. The role of chemotherapy and targeted agents in patients with metastatic breast cancer. Eur. J. Cancer 2011, 47 (Suppl. S3), S38–S47. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, M.O.; Prota, A.E. Microtubule-Targeting Agents: Strategies To Hijack the Cytoskeleton. Trends Cell Biol. 2018, 28, 776–792. [Google Scholar] [CrossRef]
- Staff, N.P.; Fehrenbacher, J.C.; Caillaud, M.; Damaj, M.I.; Segal, R.A.; Rieger, S. Pathogenesis of paclitaxel-induced peripheral neuropathy: A current review of in vitro and in vivo findings using rodent and human model systems. Exp. Neurol. 2020, 324, 113121. [Google Scholar] [CrossRef]
- Abu Samaan, T.M.; Samec, M.; Liskova, A.; Kubatka, P.; Büsselberg, D. Paclitaxel’s Mechanistic and Clinical Effects on Breast Cancer. Biomolecules 2019, 9, 789. [Google Scholar] [CrossRef]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef]
- Fu, L.; Jin, W.; Zhang, J.; Zhu, L.; Lu, J.; Zhen, Y.; Zhang, L.; Ouyang, L.; Liu, B.; Yu, H. Repurposing non-oncology small-molecule drugs to improve cancer therapy: Current situation and future directions. Acta Pharm. Sin. B 2022, 12, 532–557. [Google Scholar] [CrossRef]
- Kralova, V.; Hanušová, V.; Caltová, K.; Špaček, P.; Hochmalová, M.; Skálová, L.; Rudolf, E. Flubendazole and mebendazole impair migration and epithelial to mesenchymal transition in oral cell lines. Chem. Biol. Interact. 2018, 293, 124–132. [Google Scholar] [CrossRef]
- Lin, S.; Yang, L.; Yao, Y.; Xu, L.; Xiang, Y.; Zhao, H.; Wang, L.; Zuo, Z.; Huang, X.; Zhao, C. Flubendazole demonstrates valid antitumor effects by inhibiting STAT3 and activating autophagy. J. Exp. Clin. Cancer Res. 2019, 38, 293. [Google Scholar] [CrossRef]
- Kim, Y.J.; Sung, D.; Oh, E.; Cho, Y.; Cho, T.M.; Farrand, L.; Seo, J.H.; Kim, J.Y. Flubendazole overcomes trastuzumab resistance by targeting cancer stem-like properties and HER2 signaling in HER2-positive breast cancer. Cancer Lett. 2018, 412, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Kim, Y.J.; An, H.; Sung, D.; Cho, T.M.; Farrand, L.; Jang, S.; Seo, J.H.; Kim, J.Y. Flubendazole elicits anti-metastatic effects in triple-negative breast cancer via STAT3 inhibition. Int. J. Cancer 2018, 143, 1978–1993. [Google Scholar] [CrossRef]
- Zhen, Y.; Zhao, R.; Wang, M.; Jiang, X.; Gao, F.; Fu, L.; Zhang, L.; Zhou, X.L. Flubendazole elicits anti-cancer effects via targeting EVA1A-modulated autophagy and apoptosis in Triple-negative Breast Cancer. Theranostics 2020, 10, 8080–8097. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Yuan, Z.; Zhang, J.; Chen, Y.; Fu, Y.; Liu, Y.; Fu, L.; Zhang, L.; Zhou, X.L. Flubendazole induces mitochondrial dysfunction and DRP1-mediated mitophagy by targeting EVA1A in breast cancer. Cell Death Dis. 2022, 13, 375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Guo, M.; Li, J.; Zheng, Y.; Zhang, S.; Xie, T.; Liu, B. Systems biology-based discovery of a potential Atg4B agonist (Flubendazole) that induces autophagy in breast cancer. Mol. Biosyst. 2015, 11, 2860–2866. [Google Scholar] [CrossRef] [PubMed]
- Rushworth, L.K.; Hewit, K.; Munnings-Tomes, S.; Somani, S.; James, D.; Shanks, E.; Dufès, C.; Straube, A.; Patel, R.; Leung, H.Y. Repurposing screen identifies mebendazole as a clinical candidate to synergise with docetaxel for prostate cancer treatment. Br. J. Cancer 2020, 122, 517–527. [Google Scholar] [CrossRef]
- Chen, J.; Tian, W.; He, H.; Chen, F.; Huang, J.; Wang, X.; Chen, Z. Downregulation of miR-200c-3p contributes to the resistance of breast cancer cells to paclitaxel by targeting SOX2. Oncol. Rep. 2018, 40, 3821–3829. [Google Scholar] [CrossRef]
- Molinaro, A.; Becattini, B.; Mazzoli, A.; Bleve, A.; Radici, L.; Maxvall, I.; Sopasakis, V.R.; Molinaro, A.; Bäckhed, F.; Solinas, G. Insulin-Driven PI3K-AKT Signaling in the Hepatocyte Is Mediated by Redundant PI3Kα and PI3Kβ Activities and Is Promoted by RAS. Cell Metab. 2019, 29, 1400–1409.e5. [Google Scholar] [CrossRef]
- Liao, M.; Zhang, J.; Wang, G.; Wang, L.; Liu, J.; Ouyang, L.; Liu, B. Small-Molecule Drug Discovery in Triple Negative Breast Cancer: Current Situation and Future Directions. J. Med. Chem. 2021, 64, 2382–2418. [Google Scholar] [CrossRef]
- Harbeck, N. Neoadjuvant and adjuvant treatment of patients with HER2-positive early breast cancer. Breast 2022, 62 (Suppl. S1), S12–S16. [Google Scholar] [CrossRef]
- Lopes, D.; Seabra, A.L.; Orr, B.; Maiato, H. α-Tubulin detyrosination links the suppression of MCAK activity with taxol cytotoxicity. J. Cell Biol. 2023, 222, e202205092. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Liao, M.; Qin, R.; Zhu, S.; Peng, C.; Fu, L.; Chen, Y.; Han, B. Regulated cell death (RCD) in cancer: Key pathways and targeted therapies. Signal Transduct. Target Ther. 2022, 7, 286. [Google Scholar] [CrossRef] [PubMed]
- Škubník, J.; Svobodová Pavlíčková, V.; Ruml, T.; Rimpelová, S. Autophagy in cancer resistance to paclitaxel: Development of combination strategies. Biomed. Pharmacother. 2023, 161, 114458. [Google Scholar] [CrossRef] [PubMed]
- Brandão, M.; Pondé, N.F.; Poggio, F.; Kotecki, N.; Salis, M.; Lambertini, M.; de Azambuja, E. Combination therapies for the treatment of HER2-positive breast cancer: Current and future prospects. Expert Rev. Anticancer Ther. 2018, 18, 629–649. [Google Scholar] [CrossRef] [PubMed]
- Gayvert, K.M.; Aly, O.; Platt, J.; Bosenberg, M.W.; Stern, D.F.; Elemento, O. A Computational Approach for Identifying Synergistic Drug Combinations. PLoS Comput. Biol. 2017, 13, e1005308. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Zhen, Y.; Chen, S.; Li, Z.; Fu, L. Small-molecule inhibitor of Fam20C in combination with paclitaxel suppresses tumor growth by LIF-JAK2/STAT3-modulated apoptosis in triple-negative breast cancer. J. Taiwan Inst. Chem. Eng. 2023, 143, 104673. [Google Scholar] [CrossRef]
- Kang, C.; Syed, Y.Y. Atezolizumab (in Combination with Nab-Paclitaxel): A Review in Advanced Triple-Negative Breast Cancer. Drugs 2020, 80, 601–607. [Google Scholar] [CrossRef]
- Jotte, R.; Cappuzzo, F.; Vynnychenko, I.; Stroyakovskiy, D.; Rodríguez-Abreu, D.; Hussein, M.; Soo, R.; Conter, H.J.; Kozuki, T.; Huang, K.C.; et al. Atezolizumab in Combination with Carboplatin and Nab-Paclitaxel in Advanced Squamous NSCLC (IMpower131): Results from a Randomized Phase III Trial. J. Thorac. Oncol. 2020, 15, 1351–1360. [Google Scholar] [CrossRef]
- Song, Y.; Bugada, L.; Li, R.; Hu, H.; Zhang, L.; Li, C.; Yuan, H.; Rajanayake, K.K.; Truchan, N.A.; Wen, F.; et al. Albumin nanoparticle containing a PI3Kγ inhibitor and paclitaxel in combination with α-PD1 induces tumor remission of breast cancer in mice. Sci. Transl. Med. 2022, 14, eabl3649. [Google Scholar] [CrossRef]
- Hou, Z.J.; Luo, X.; Zhang, W.; Peng, F.; Cui, B.; Wu, S.J.; Zheng, F.M.; Xu, J.; Xu, L.Z.; Long, Z.J.; et al. Flubendazole, FDA-approved anthelmintic, targets breast cancer stem-like cells. Oncotarget 2015, 6, 6326–6340. [Google Scholar] [CrossRef]
- Mackenzie, C.D.; Geary, T.G. Flubendazole: A candidate macrofilaricide for lymphatic filariasis and onchocerciasis field programs. Expert Rev. Anti-Infect. Ther. 2011, 9, 497–501. [Google Scholar] [CrossRef]
- Čáňová, K.; Rozkydalová, L.; Rudolf, E. Anthelmintic Flubendazole and Its Potential Use in Anticancer Therapy. Acta Medica 2017, 60, 5–11. [Google Scholar] [CrossRef]
- Chen, C.; Ding, Y.; Liu, H.; Sun, M.; Wang, H.; Wu, D. Flubendazole Plays an Important Anti-Tumor Role in Different Types of Cancers. Int. J. Mol. Sci. 2022, 23, 519. [Google Scholar] [CrossRef]
- Spagnuolo, P.A.; Hu, J.; Hurren, R.; Wang, X.; Gronda, M.; Sukhai, M.A.; Di Meo, A.; Boss, J.; Ashali, I.; Beheshti Zavareh, R.; et al. The antihelmintic flubendazole inhibits microtubule function through a mechanism distinct from Vinca alkaloids and displays preclinical activity in leukemia and myeloma. Blood 2010, 115, 4824–4833. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, M.; Agha, B.; Rothweiler, F.; Löschmann, N.; Voges, Y.; Mittelbronn, M.; Starzetz, T.; Harter, P.N.; Abhari, B.A.; Fulda, S.; et al. Identification of flubendazole as potential anti-neuroblastoma compound in a large cell line screen. Sci. Rep. 2015, 5, 8202. [Google Scholar] [CrossRef] [PubMed]
- Raisová Stuchlíková, L.; Králová, V.; Lněničková, K.; Zárybnický, T.; Matoušková, P.; Hanušová, V.; Ambrož, M.; Šubrt, Z.; Skálová, L. The metabolism of flubendazole in human liver and cancer cell lines. Drug Test. Anal. 2018, 10, 1139–1146. [Google Scholar] [CrossRef]
- Králová, V.; Hanušová, V.; Rudolf, E.; Čáňová, K.; Skálová, L. Flubendazole induces mitotic catastrophe and senescence in colon cancer cells in vitro. J. Pharm. Pharmacol. 2016, 68, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Son, D.S.; Lee, E.S.; Adunyah, S.E. The Antitumor Potentials of Benzimidazole Anthelmintics as Repurposing Drugs. Immune Netw. 2020, 20, e29. [Google Scholar] [CrossRef]
- Lee, H.J.; Han, H.J. Role of Microtubule-Associated Factors in HIF1α Nuclear Translocation. Adv. Exp. Med. Biol. 2020, 1232, 271–276. [Google Scholar]
- Khattab, M.; Al-Karmalawy, A.A. Computational repurposing of benzimidazole anthelmintic drugs as potential colchicine binding site inhibitors. Future Med. Chem. 2021, 13, 1623–1638. [Google Scholar] [CrossRef]
- Ermler, S.; Scholze, M.; Kortenkamp, A. Genotoxic mixtures and dissimilar action: Concepts for prediction and assessment. Arch. Toxicol. 2014, 88, 799–814. [Google Scholar] [CrossRef]
- Miyagi, M.Y.S.; de Oliveira Faria, R.; de Souza, G.B.; Lameu, C.; Tagami, T.; Ozeki, T.; Bezzon, V.D.N.; Yukuyama, M.N.; Bou-Chacra, N.A.; de Araujo, G.L.B. Optimizing adjuvant inhaled chemotherapy: Synergistic enhancement in paclitaxel cytotoxicity by flubendazole nanocrystals in a cycle model approach. Int. J. Pharm. 2023, 644, 123324. [Google Scholar] [CrossRef] [PubMed]
- Jabbarzadeh Kaboli, P.; Salimian, F.; Aghapour, S.; Xiang, S.; Zhao, Q.; Li, M.; Wu, X.; Du, F.; Zhao, Y.; Shen, J.; et al. Akt-targeted therapy as a promising strategy to overcome drug resistance in breast cancer—A comprehensive review from chemotherapy to immunotherapy. Pharmacol. Res. 2020, 156, 104806. [Google Scholar] [CrossRef] [PubMed]
- Cocco, S.; Leone, A.; Roca, M.S.; Lombardi, R.; Piezzo, M.; Caputo, R.; Ciardiello, C.; Costantini, S.; Bruzzese, F.; Sisalli, M.J.; et al. Inhibition of autophagy by chloroquine prevents resistance to PI3K/AKT inhibitors and potentiates their antitumor effect in combination with paclitaxel in triple negative breast cancer models. J. Transl. Med. 2022, 20, 290. [Google Scholar] [CrossRef] [PubMed]
- Kierans, S.J.; Taylor, C.T. Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, K. HIF-1 at the crossroads of hypoxia, inflammation, and cancer. Int. J. Cancer 2016, 138, 1058–1066. [Google Scholar] [CrossRef]
- Mabjeesh, N.J.; Escuin, D.; LaVallee, T.M.; Pribluda, V.S.; Swartz, G.M.; Johnson, M.S.; Willard, M.T.; Zhong, H.; Simons, J.W.; Giannakakou, P. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell 2003, 3, 363–375. [Google Scholar] [CrossRef]
- Ye, J.; Wu, J.; Liu, B. Therapeutic strategies of dual-target small molecules to overcome drug resistance in cancer therapy. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188866. [Google Scholar] [CrossRef]
- Chi, Y.; Xue, J.; Huang, S.; Xiu, B.; Su, Y.; Wang, W.; Guo, R.; Wang, L.; Li, L.; Shao, Z.; et al. CapG promotes resistance to paclitaxel in breast cancer through transactivation of PIK3R1/P50. Theranostics 2019, 9, 6840–6855. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, K.; Feng, X.; Wang, J.; Chu, Y.; Jia, C.; He, Q.; Chen, C. PRMT3 promotes tumorigenesis by methylating and stabilizing HIF1α in colorectal cancer. Cell Death Dis. 2021, 12, 1066. [Google Scholar] [CrossRef]
- Masoud, G.N.; Li, W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 2015, 5, 378–389. [Google Scholar] [CrossRef]
- Icard, P.; Simula, L.; Fournel, L.; Leroy, K.; Lupo, A.; Damotte, D.; Charpentier, M.C.; Durdux, C.; Loi, M.; Schussler, O.; et al. The strategic roles of four enzymes in the interconnection between metabolism and oncogene activation in non-small cell lung cancer: Therapeutic implications. Drug Resist. Updates 2022, 63, 100852. [Google Scholar] [CrossRef] [PubMed]
- Khachigian, L.M. Emerging insights on functions of the anthelmintic flubendazole as a repurposed anticancer agent. Cancer Lett. 2021, 522, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Cai, X.; Tang, Y.; Jiang, C.; Zhou, F.; Yang, L.; Liu, Z.; Wang, L.; Zhao, H.; Zhao, C.; et al. Flubendazole Elicits Antitumor Effects by Inhibiting STAT3 and Activating Autophagy in Non-small Cell Lung Cancer. Front. Cell Dev. Biol. 2021, 9, 680600. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, K.; Rudolf, E. An analysis of mitotic catastrophe induced cell responses in melanoma cells exposed to flubendazole. Toxicol In Vitro 2020, 68, 104930. [Google Scholar] [CrossRef]
- Chen, P.; Li, X.; Zhang, R.; Liu, S.; Xiang, Y.; Zhang, M.; Chen, X.; Pan, T.; Yan, L.; Feng, J.; et al. Combinative treatment of β-elemene and cetuximab is sensitive to KRAS mutant colorectal cancer cells by inducing ferroptosis and inhibiting epithelial-mesenchymal transformation. Theranostics 2020, 10, 5107–5119. [Google Scholar] [CrossRef]
- Chen, Y.M.; Xu, W.; Liu, Y.; Zhang, J.H.; Yang, Y.Y.; Wang, Z.W.; Sun, D.J.; Li, H.; Liu, B.; Chen, L.X. Anomanolide C suppresses tumor progression and metastasis by ubiquitinating GPX4-driven autophagy-dependent ferroptosis in triple negative breast cancer. Int. J. Biol. Sci. 2023, 19, 2531–2550. [Google Scholar] [CrossRef]
- Zhou, X.; Jin, W.; Chen, Y.; Zhu, L.; Mo, A.; Xie, Q. Identification of potential druggable targets of cell cycle with small-molecule inhibitors in oral squamous cell carcinoma. Pharmacogenet. Genom. 2022, 32, 125–137. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Liao, M.; Li, Z.; Ye, J.; Wu, L.; Mou, Y.; Fu, L.; Zhen, Y. Flubendazole Enhances the Inhibitory Effect of Paclitaxel via HIF1α/PI3K/AKT Signaling Pathways in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 15121. https://doi.org/10.3390/ijms242015121
Zhou Y, Liao M, Li Z, Ye J, Wu L, Mou Y, Fu L, Zhen Y. Flubendazole Enhances the Inhibitory Effect of Paclitaxel via HIF1α/PI3K/AKT Signaling Pathways in Breast Cancer. International Journal of Molecular Sciences. 2023; 24(20):15121. https://doi.org/10.3390/ijms242015121
Chicago/Turabian StyleZhou, Yuxin, Minru Liao, Zixiang Li, Jing Ye, Lifeng Wu, Yi Mou, Leilei Fu, and Yongqi Zhen. 2023. "Flubendazole Enhances the Inhibitory Effect of Paclitaxel via HIF1α/PI3K/AKT Signaling Pathways in Breast Cancer" International Journal of Molecular Sciences 24, no. 20: 15121. https://doi.org/10.3390/ijms242015121
APA StyleZhou, Y., Liao, M., Li, Z., Ye, J., Wu, L., Mou, Y., Fu, L., & Zhen, Y. (2023). Flubendazole Enhances the Inhibitory Effect of Paclitaxel via HIF1α/PI3K/AKT Signaling Pathways in Breast Cancer. International Journal of Molecular Sciences, 24(20), 15121. https://doi.org/10.3390/ijms242015121




