Metabolome Profiling in the Plasma of Dogs with Idiopathic Dilated Cardiomyopathy: A Multiplatform Mass-Spectrometry-Based Approach
Abstract
:1. Introduction
2. Results
2.1. The Metabolomics Data Set in Dogs with Diagnosed iDCM
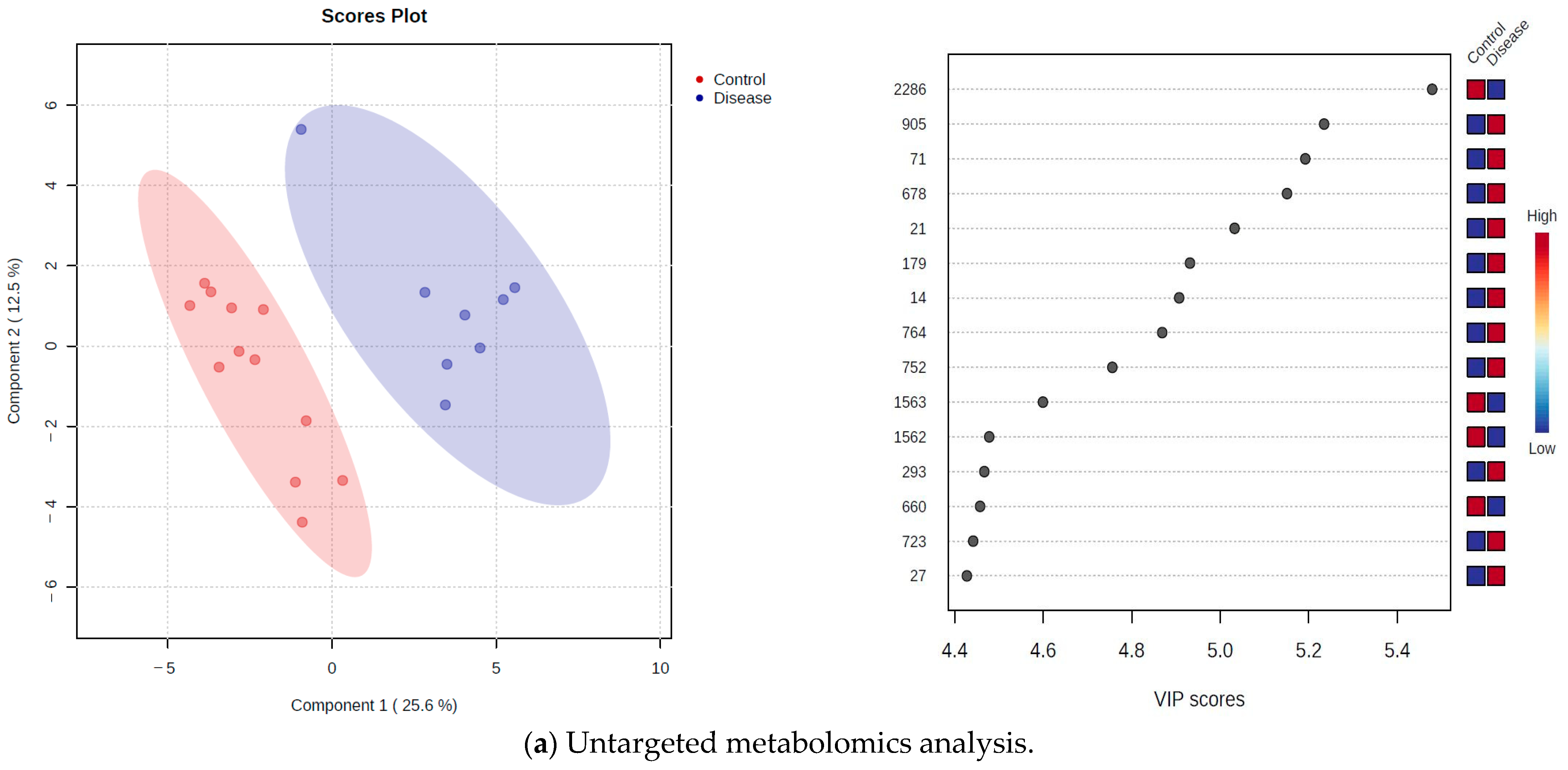
2.2. Untargeted Metabolomics Approach
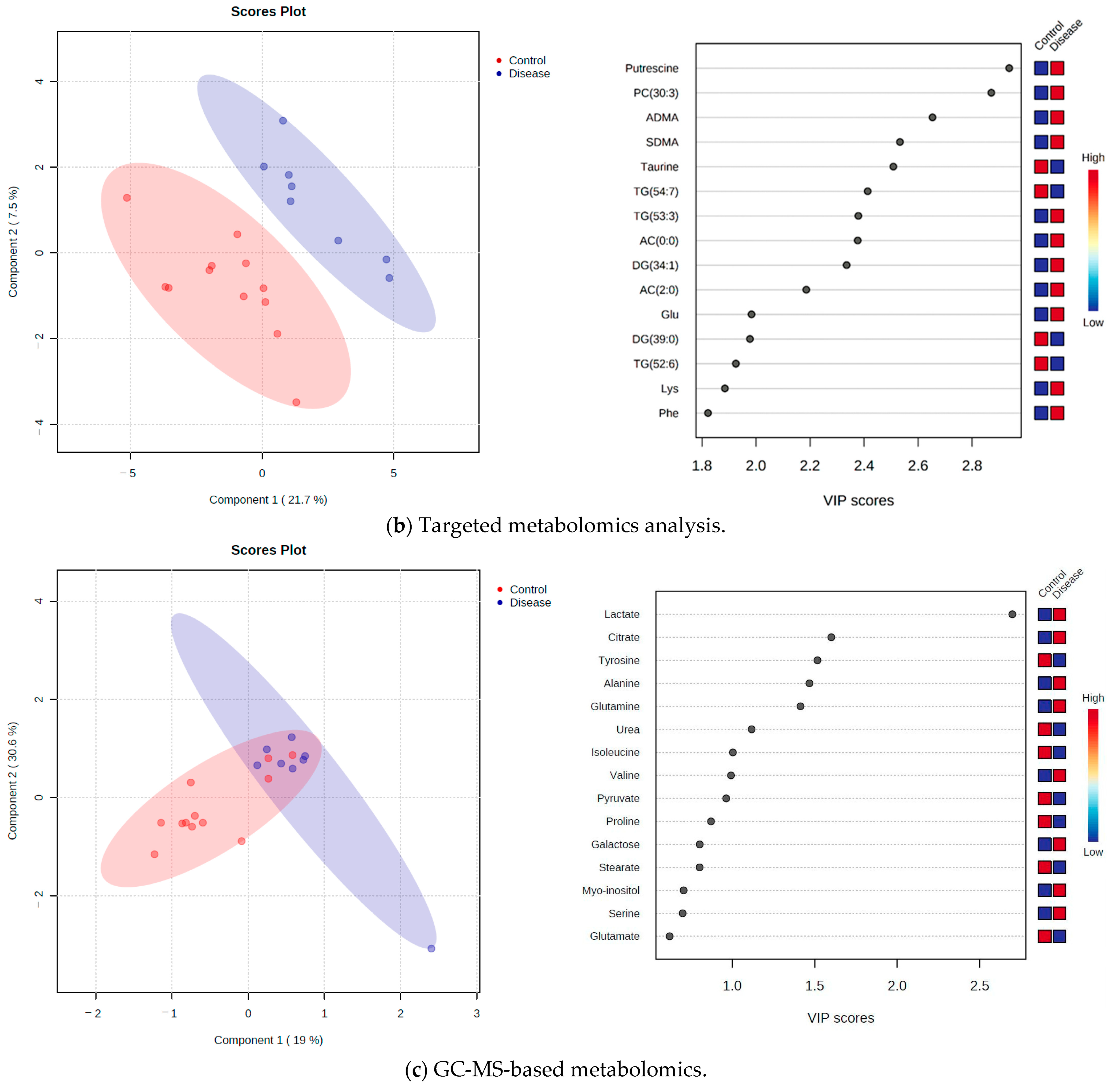
2.3. Targeted Metabolomics Approach
2.4. GC-MS-Based Metabolomics
2.5. Identification of Metabolites Showing Differential Abundance in the Plasma Metabolome of Dogs with iDCM and Healthy Dogs
3. Discussion
4. Materials and Methods
4.1. Experimental Design
4.2. Blood Sample Preparation and Analysis
4.3. Untargeted Metabolomics Approach
4.3.1. Sample Preparation
4.3.2. Metabolite Analysis
4.3.3. Data Processing
4.4. Targeted Metabolomics Approach
4.4.1. Sample Preparation
4.4.2. Metabolite Analysis
4.4.3. Data Processing
4.5. GC-MS-Based Metabolomics Analysis
4.5.1. Sample Preparation
4.5.2. Metabolite Analysis
4.5.3. Data Processing
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wynne, J.; Braunwald, E. The cardiomyopathies and myocarditides. In Heart Disease: A Textbook of Cardiovascular Medicine, 5th ed.; Braunwald, E., Saunders, E.W.B., Eds.; Elsevier: Philadelphia, PA, USA, 1997; p. 1404. [Google Scholar]
- Oyama, M. Cardiomyopathy. In Manual of Canine and Feline Cardiology; Smith, F., Tilley, L., Oyama, M., Sleeper, M., Eds.; Elsevier: St. Louis, MO, USA, 2015; pp. 141–151. [Google Scholar]
- Richardson, P.; McKenna, W.; Bristow, M.; Maisch, B.; Mautner, B.; O’Connell, J.; Olsen, E.; Thiene, G.; Goodwin, J.; Gyarfas, I.; et al. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of Cardiomyopathies. Circulation 1996, 93, 841–842. [Google Scholar] [PubMed]
- O’Grady, M.R.; O’Sullivan, M.L.; Minors, S.L.; Horne, R. Efficacy of benazepril hydrochloride to delay the progression of occult dilated cardiomyopathy in Doberman Pinschers. J. Vet. Intern. Med. 2009, 23, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Summerfield, N.J.; Boswood, A.; O’Grady, M.R.; Gordon, S.G.; Dukes-McEwan, J.; Oyama, M.A.; Smith, S.; Patteson, M.; French, A.T.; Culshaw, G.J.; et al. Efficacy of Pimobendan in the Prevention of Congestive Heart Failure or Sudden Death in Doberman Pinschers with Preclinical Dilated Cardiomyopathy (The PROTECT Study). J. Vet. Intern. Med. 2012, 26, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Wess, G.; Schulze, A.; Butz, V.; Simak, J.; Killich, M.; Keller, L.; Maeurer, J.; Hartmann, K. Prevalence of dilated cardiomyopathy in Doberman Pinschers in various age groups. J. Vet. Intern. Med. 2010, 24, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Schild, D.P.; Ricciardi, S.I.; Hellige, J.G.; Vogel, R.; Arenja, N. Current Pathophysiological and Genetic Aspects of Dilated Cardiomyopathy. In Visions of Cardiomyocyte—Fundamental Concepts of Heart Life and Disease; IntechOpen: London, UK, 2019. [Google Scholar]
- Kordalewska, M.; Markuszewski, M.J. Metabolomics in cardiovascular diseases. J. Pharm. Biomed. Anal. 2015, 113, 121–136. [Google Scholar] [CrossRef]
- Niiranen, T.J.; Vasan, R.S. Epidemiology of cardiovascular disease: Recent novel outlooks on risk factors and clinical approaches. Expert. Rev. Cardiovasc. Ther. 2016, 14, 855–869. [Google Scholar] [CrossRef]
- Shah, S.H.; Sun, J.L.; Stevens, R.D.; Bain, J.R.; Muehlbauer, M.J.; Pieper, K.S.; Haynes, C.; Hauser, E.R.; Kraus, W.E.; Granger, C.B.; et al. Baseline metabolomics profiles predict cardiovascular events in patients at risk for coronary artery disease. Am. Heart J. 2012, 163, 844–850. [Google Scholar] [CrossRef]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; DuGar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.-M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef]
- Diaz, M.L.; Burgess, K.; Burchmore, R.; Gómez, M.A.; Gómez-Ochoa, S.A.; Echeverría, L.E.; Morillo, C.; González, C.I. Metabolomic Profiling of End-Stage Heart Failure Secondary to Chronic Chagas Cardiomyopathy. Int. J. Mol. Sci. 2022, 23, 10456. [Google Scholar] [CrossRef]
- Alexander, D.; Lombardi, R.; Rodriguez, G.; Mitchell, M.M.; Marian, A.J. Metabolomic distinction and insights into the pathogenesis of human primary dilated cardiomyopathy. Eur. J. Clin. Investig. 2011, 41, 527–538. [Google Scholar] [CrossRef]
- West, J.A.; Beqqali, A.; Ament, Z.; Elliott, P.; Pinto, Y.M.; Arbustini, E.; Griffin, J.L. A targeted metabolomics assay for cardiac metabolism and demonstration using a mouse model of dilated cardiomyopathy. Metabolomics 2016, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, S.; Jing, R.; Jin, H.; Hu, Y.; Wang, J.; Gu, M.; Niu, H.; Zhang, S.; Chen, L.; et al. Plasma Metabolomic Profiles Differentiate Patients With Dilated Cardiomyopathy and Ischemic Cardiomyopathy. Front. Cardiovasc. Med. 2020, 7, 597546. [Google Scholar] [CrossRef] [PubMed]
- Ritterhoff, J.; Tian, R. Metabolism in cardiomyopathy: Every substrate matters. Cardiovasc. Res. 2017, 113, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Qian, L.; Cheng, Z.; Chen, C.; Wang, K.; Hu, S.; Zhang, X.; Wu, T. Lactate and Myocadiac Energy Metabolism. Front. Physiol. 2021, 12, 715081. [Google Scholar] [CrossRef]
- Haas, J.; Frese, K.S.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Tappu, R.; Nietsch, R.; Tugrul, O.F.; Wisdom, M.; Dietrich, C.; Amr, A.; et al. Energy Metabolites as Biomarkers in Ischemic and Dilated Cardiomyopathy. Int. J. Mol. Sci. 2021, 22, 1999. [Google Scholar] [CrossRef]
- Cerovic, O.; Golubovic, V.; Spec-Marn, A.; Kremzar, B.; Vidmar, G. Relationship between injury severity and lactate levels in severely injured patients. Intensive Care Med. 2003, 29, 1300–1305. [Google Scholar] [CrossRef]
- Nguyen, H.B.; Rivers, E.P.; Knoblich, B.P.; Jacobsen, G.; Muzzin, A.; Ressler, J.A.; Tomlanovich, M.C. Early lactate clearance is associated with improved outcome in severe sepsis and septic shock. Crit. Care Med. 2004, 32, 1637–1642. [Google Scholar] [CrossRef]
- Khosravani, H.; Shahpori, R.; Stelfox, H.T.; Kirkpatrick, A.W.; Laupland, K.B. Occurrence and adverse effect on outcome of hyperlactatemia in the critically ill. Crit. Care 2009, 13, R90. [Google Scholar] [CrossRef]
- Bremer, J.; Roberts, P.A.; Bouitbir, J.; Bonifacio, A.; Singh, F.; Kaufmann, P.; Urwyler, A.; Krähenbühl, S.; Kong, J.Y.; Rabkin, S.W.; et al. Carnitine- metabolism and functions. Physiol. Rev. 1983, 63, 1420–1480. [Google Scholar] [CrossRef]
- Hoppel, C. The Physiological Role of Carnitine. L-carnitine and Its Role in Medicine: From Function to Therapy; Ferrari, R., DiMauro, S., Sherwood, G., Eds.; Academic Press: London, UK, 1992; pp. 5–17. [Google Scholar]
- Paulson, D.J.; Shug, A.L. Tissue specific depletion of L-carnitine rat heart and skeletal muscle by L-carnitine. Life Sci. 1981, 28, 2931–2938. [Google Scholar] [CrossRef]
- Rebouche, C.J.; Engel, A.G. Carnitine metabolism and deficiency syndromes. Mayo Clin.Proc. 1983, 58, 533–540. [Google Scholar] [PubMed]
- Magoulas, P.L.; El-Hattab, A.W. Systemic primary carnitine deficiency: An overview of clinical manifestations, diagnosis, and management. Orphanet J. Rare Dis. 2012, 7, 68. [Google Scholar] [CrossRef]
- Pierpont, M.E.; Judd, D.; Goldenberg, I.; Ring, W.S.; Olivari, M.T.; Pierpont, G.L. Myocardial carnitine in end-stage congestive heart failure. Am. J. Cardiol. 1989, 64, 56–60. [Google Scholar] [CrossRef] [PubMed]
- El-Aroussy, W.; Rizk, A.; Mayhoub, G.; Aleem, S.A.; El-Tobgy, S.; Mokhtar, M.S. Plasma carnitine levels as a marker of impaired left ventricular functions. Mol. Cell. Biochem. 2000, 213, 37–41. [Google Scholar] [CrossRef]
- Regitz, V.; Shug, A.L.; Fleck, E. Defective myocardial carnitine metabolism in congestive heart failure secondary to dilated cardiomyopathy and to coronary, hypertensive, and valvular heart disease. Am. J. Cardiol. 1990, 65, 755–760. [Google Scholar] [CrossRef]
- Regitz, V.; Bossaller, C.; Strasser, R.; Müller, M.; Shug, A.L.; Fleck, E. Metabolic alterations in end stage and less severe heart failure-myocardial carnitine decrease. J. Clin. Chem. Clin. Biochem. 1990, 28, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Wittles, B.; Spann, J.F., Jr. Defective lipid metabolism in the failing heart. J. Clin. Invest. 1968, 47, 1787–1794. [Google Scholar]
- Kukharenko, A.; Brito, A.; Kozhevnikova, M.V.; Moskaleva, N.; Markin, P.A.; Bochkareva, N.; Korobkova, E.O.; Belenkov, Y.N.; Privalova, E.V.; Larcova, E.V.; et al. Relationship between the plasma acylcarnitine profile and cardiometabolic risk factors in adults diagnosed with cardiovascular diseases. Clin. Chim. Acta. 2020, 507, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.E.; Evans, A.M. Carnitine and acylcarnitines. Clin. Pharmacokinet. 2012, 51, 553–572. [Google Scholar] [CrossRef]
- Strand, E.; Pedersen, E.R.; Svingen, G.F.T.; Olsen, T.; Bjørndal, B.; Karlsson, T.; Dierkes, J.; Njølstad, P.R.; Mellgren, G.; Tell, G.S.; et al. Serum acylcarnitines and risk of cardiovascular death and acute myocardial infarction in patients with stable angina pectoris. J. Am. Heart Assoc. 2017, 6, e003620. [Google Scholar] [CrossRef] [PubMed]
- Zobel, E.H.; Von Scholten, B.J.; Reinhard, H.; Persson, F.; Teerlink, T.; Hansen, T.W.; Parving, H.-H.; Jacobsen, P.K.; Rossing, P. Symmetric and asymmetric dimethylarginine as risk markers of cardiovascular disease, all-cause mortality and deterioration in kidney function in persons with type 2 diabetes and microalbuminuria. Cardiovasc. Diabetol. 2017, 16, 88. [Google Scholar] [CrossRef]
- Vallance, P.; Leone, A.; Calver, A.; Collier, J.; Moncada, S. Accumulation of an endogenous inhibitor of nitric oxide synthesis in chronic renal failure. Lancet 1992, 339, 572–575. [Google Scholar]
- Valkonen, V.P.; Paiva, H.; Salonen, J.T.; Lakka, T.A.; Lehtimäki, T.; Laakso, J.; Laaksonen, R. Risk of acute coronary events and serum concentration of asymmetrical dimethylarginine. Lancet 2001, 358, 2127–2128. [Google Scholar] [CrossRef] [PubMed]
- Boger, R.H.; Bode-Boger, S.M.; Thiele, W.; Junker, W.; Alexander, K.; Frolich, J.C. Biochemical evidence for impaired nitric oxide synthesis in patients with peripheral arterial occlusive disease. Circulation 1997, 95, 2068–2074. [Google Scholar] [CrossRef] [PubMed]
- Mittermayer, F.; Krzyzanowska, K.; Exner, M.; Mlekusch, W.; Amighi, J.; Sabeti, S.; Minar, E.; Muller, M.; Wolzt, M.; Schillinger, M. Asymmetric dimethylarginine predicts major adverse cardiovascular events in patients with advanced peripheral artery disease. Arter. Thromb. Vasc. Biol. 2006, 26, 2536–2540. [Google Scholar] [CrossRef]
- Usui, M.; Matsuoka, H.; Miyazaki, H.; Ueda, S.; Okuda, S.; Imaizumi, T. Increased endogenous nitric oxide synthase inhibitor in patients with congestive heart failure. Life Sci. 1998, 62, 2425–2430. [Google Scholar] [CrossRef] [PubMed]
- Gorenflo, M.; Zheng, C.; Werle, E.; Fiehn, W.; Ulmer, H.E. Plasma levels of asymmetrical dimethyl-L-arginine in patients with congenital heart disease and pulmonary hypertension. J. Cardiovasc. Pharmacol. 2001, 37, 489–492. [Google Scholar] [CrossRef]
- Pettersson, A.; Hedner, T.; Milsom, I. Increased circulating concentrations of asymmetric dimethyl arginine (ADMA), an endogenous inhibitor of nitric oxide synthesis, in preeclampsia. Acta Obstet. Gynecol. Scand. 1998, 77, 808–813. [Google Scholar] [PubMed]
- Yoo, J.H.; Lee, S.C. Elevated levels of plasma homocyst(e)ine and asymmetric dimethylarginine in elderly patients with stroke. Atherosclerosis 2001, 158, 425–430. [Google Scholar] [CrossRef]
- Dimitrow, P.P.; Undas, A.; Bober, M.; Tracz, W.; Dubiel, J.S. Plasma biomarkers of endothelial dysfunction in patients with hypertrophic cardiomyopathy. Pharmacol. Rep. 2007, 59, 715–720. [Google Scholar]
- Ali, O.A.; Chapman, M.; Nguyen, T.H.; Chirkov, Y.Y.; Heresztyn, T.; Mundisugih, J.; Horowitz, J.D. Interactions between inflammatory activation and endothelial dysfunction selectively modulate valve disease progression in patients with bicuspid aortic valve. Heart 2014, 100, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Anderssohn, M.; Rosenberg, M.; Schwedhelm, E.; Zugck, C.; Lutz, M.; Lüneburg, N.; Frey, N.; Böger, R.H. The L-Arginine-asymmetric dimethylarginine ratio is an independent predictor of mortality in dilated cardiomyopathy. J. Card. Fail. 2012, 18, 904–911. [Google Scholar] [CrossRef] [PubMed]
- Tutarel, O.; Denecke, A.; Bode-Böger, S.M.; Martens-Lobenhoffer, J.; Lovric, S.; Bauersachs, J.; Schieffer, B.; Westhoff-Bleck, M.; Kielstein, J.T. Asymmetrical dimethylarginine--more sensitive than NT-proBNP to diagnose heart failure in adults with congenital heart disease. PLoS ONE 2012, 7, e33795. [Google Scholar] [CrossRef]
- Jacobi, J.; Tsao, P.S. Asymmetrical dimethylarginine in renal disease: Limits of variation or variation limits? A systematic review. Am. J. Nephrol. 2008, 28, 224–237. [Google Scholar] [CrossRef]
- Anderssohn, M.; Schwedhelm, E.; Lüneburg, N.; Vasan, R.S.; Böger, R.H. Asymmetric dimethylarginine as a mediator of vascular dysfunction and a marker of cardiovascular disease and mortality: An intriguing interaction with diabetes mellitus. Diab. Vasc. Dis. Res. 2010, 7, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.Y.; Ito, A.; Asagami, T.; Tsao, P.S.; Adimoolam, S.; Kimoto, M.; Tsuji, H.; Reaven, G.M.; Cooke, J.P. Impaired nitric oxide synthase pathway in diabetes mellitus: Role of asymmetric dimethylarginine and dimethylarginine dimethylaminohydrolase. Circulation 2002, 106, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Cengel, A.; Sahinarslan, A.; Biberoğlu, G.; Hasanoğlu, A.; Tavil, Y.; Tulmaç, M.; Ozdemir, M. Asymmetrical dimethylarginine level in atrial fibrillation. Acta Cardiol. 2008, 63, 33–37. [Google Scholar] [CrossRef]
- Nicholls, S.J.; Wang, Z.; Koeth, R.; Levison, B.; DelFraino, B.; Dzavik, V.; Griffith, O.W.; Hathaway, D.; Panza, J.A.; Nissen, S.E.; et al. Metabolic profiling of arginine and nitric oxide pathways predicts hemodynamic abnormalities and mortality in patients with cardiogenic shock after acute myocardial infarction. Circulation 2007, 116, 2315–2324. [Google Scholar] [CrossRef]
- Schnabel, R.; Blankenberg, S.; Lubos, E.; Lackner, K.J.; Rupprecht, H.J.; Espinola-Klein, C.; Jachmann, N.; Post, F.; Peetz, D.; Bickel, C.; et al. Asymmetric dimethylarginine and the risk of cardiovascular events and death in patients with coronary artery disease: Results from the Athero Gene Study. Circ. Res. 2005, 97, e53–e59. [Google Scholar] [CrossRef] [PubMed]
- Böger, R.H.; Sullivan, L.M.; Schwedhelm, E.; Wang, T.J.; Maas, R.; Benjamin, E.J.; Schulze, F.; Xanthakis, V.; Benndorf, R.A.; Vasan, R.S. Plasma asymmetric dimethylarginine and incidence of cardiovascular disease and death in the community. Circulation 2009, 119, 1592–1600. [Google Scholar] [CrossRef]
- Djordjević, B.V.; Pavlović, R.; Ćosić, V.; Deljanin-Ilić, M.; Ristić, T.; Krstić, N.; Jevtović-Stoimenov, T. High clinical accuracy of asymmetric dimethylarginine and symmetric dimethylarginine in patients with ischemic heart disease. Amino Acids 2012, 43, 2293–2300. [Google Scholar] [CrossRef]
- Meinitzer, A.; Kielstein, J.T.; Pilz, S.; Drechsler, C.; Ritz, E.; Boehm, B.O.; Winkelmann, B.R.; März, W. Symmetrical and asymmetrical dimethylarginine as predictors for mortality in patients referred for coronary angiography: The Ludwigshafen Risk and Cardiovascular Health study. Clin. Chem. 2011, 57, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Ampong, I. Metabolic and Metabolomics Insights into Dilated Cardiomyopathy. Ann. Nutr. Metab. 2022, 78, 147–155. [Google Scholar] [CrossRef]
- Ferdinandy, P.; Fazekas, T.; Kadar, E. Effects of orotic acid on ischaemic/reperfused myocardial function and glycogen content in isolated working rat hearts. Pharmacol. Res. 1998, 37, 111–114. [Google Scholar] [CrossRef]
- Das, S.; Kumar, Y.; Sharma, S.; Ray, R.; Arava, S.; Seth, S.; Agarwal, A.; Sharma, G. An Untargeted LC–MS based approach for identification of altered metabolites in blood plasma of rheumatic heart disease patients. Sci. Rep. 2022, 12, 5238. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.F.; Donohoe, J.A.; Rosenfeldt, F.L.; Munsch, C.M. Biochemistry and functional roles of orotic acid for support of the infarcted heart during open heart surgery. In Orotic Acid in Cardiology, Proceedings of the International Symposium on Orotic Acid and Magnesium Orotate, Rudesheim, Germany, 16 November 1991; Williams, J.F., Ed.; Thieme Medical Publishers Inc.: New York, NY, USA, 1992; pp. 1–24. [Google Scholar]
- Iacoviello, M.; Leone, M.; Valeria Antoncecchi, V.; Ciccone, M.M. Evaluation of chronic kidney disease in chronic heart failure: From biomarkers to arterial renal resistances. World J. Clin. Cases 2015, 3, 10–19. [Google Scholar] [CrossRef]
- Bagheri, B.; Radmard, N.; Makrani, A.; Rasouli, M. Serum Creatinine and Occurrence and Severity of Coronary Artery Disease. Med. Arch. 2019, 73, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Dries, D.L.; Exner, D.V.; Domanski, M.J.; Greenberg, B.; Stevenson, L.W. The prognostic implications of renal insufficiency in asymptomatic and symptomatic patients with left ventricular systolic dysfunction. J. Am. Coll. Cardiol. 2000, 35, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Mahon, N.G.; Blackstone, E.H.; Francis, G.S.; Starling, R.C.; Young, J.B.; Lauer, M.S. The prognostic value of estimated creatinine clearance alongside functional capacity in ambulatory patients with chronic congestive heart failure. J. Am. Coll. Cardiol. 2002, 40, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- McClellan, W.M.; Flanders, W.D.; Langston, R.D.; Jurkovitz, C.; Presley, R. Anemia and renal insufficiency are independent risk factors for death among patients with congestive heart failure admitted to community hospitals: A population-based study. J. Am. Soc. Nephrol. 2002, 13, 1928–1936. [Google Scholar] [CrossRef]
- Chávez-Iñiguez, J.; Sánchez-Villaseca, S.; García-Macías, L. Cardiorenal syndrome: Classification, pathophysiology, diagnosis and management. Literature review. Arch. Cardiol. Mex. 2022, 92, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W.P.; Gaber, C.E.; Jacobs, G.J.; Kaplan, P.M.; Lombard, C.W.; Sydney Moise, N.; Moses, B.L. Recommendations for Standards in Transthoracic Two-Dimensional Echocardiography in the Dog and Cat. J. Vet. Intern. Med. 1993, 7, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.; Couto, C.G. Small Animal Internal Medicine, 5th ed.; Elsevier Mosby: St. Louis, MO, USA, 2013; p. 1504. [Google Scholar]
- Cornell, C.C.; Kittleson, M.D.; Della Torre, P.; Häggström, J.; Lombard, C.W.; Pedersen, H.D.; Vollmar, A.; Wey, A. Allometric Scaling of M-Mode Cardiac Measurements in Normal Adult Dogs. J. Vet. Intern. Med. 2004, 18, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Bonagura, J.D.; Visser, L.C. Echocardiographic assessment of dilated cardiomyopathy in dogs. J. Vet. Cardiol. 2022, 40, 15–50. [Google Scholar] [CrossRef] [PubMed]
- Dukes-McEwan, J.; Borgarelli, M.; Tidholm, A.; Vollmar, A.C.; Häggström, J. Proposed guidelines for the diagnosis of canine idiopathic dilated cardiomyopathy. J. Vet. Cardiol. 2003, 5, 7–19. [Google Scholar] [CrossRef]
- Gloaguen, Y.; Morton, F.; Daly, R.; Gurden, R.; Rogers, S.; Wandy, J.; Wilson, D.; Barrett, M.; Burgess, K. PiMP my metabolome: An integrated, web-based tool for LC-MS metabolomics data. Bioinformatics 2017, 33, 4007–4009. [Google Scholar] [CrossRef]
- Smith, A.C.; Want, J.E.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Scheltema, R.A.; Jankevics, A.; Jansen, C.R.; Swertz, A.M.; Breitling, R. PeakML/mzMatch: A File Format, Java Library, R Library, and Tool-Chain for Mass Spectrometry Data Analysis. Anal. Chem. 2011, 83, 2786–2793. [Google Scholar] [CrossRef]
- Prince, T.J.; Marcotte, M.E. Chromatographic Alignment of ESI-LC-MS Proteomics Data Sets by Ordered Bijective Interpolated Warping. Anal. Chem. 2006, 78, 6140–6152. [Google Scholar] [CrossRef]
- Holman, J.D.; Tabb, D.L.; Mallick, P. Employing ProteoWizard to Convert Raw Mass Spectrometry Data. Curr. Protoc. Bioinform. 2014, 46, 13–24. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
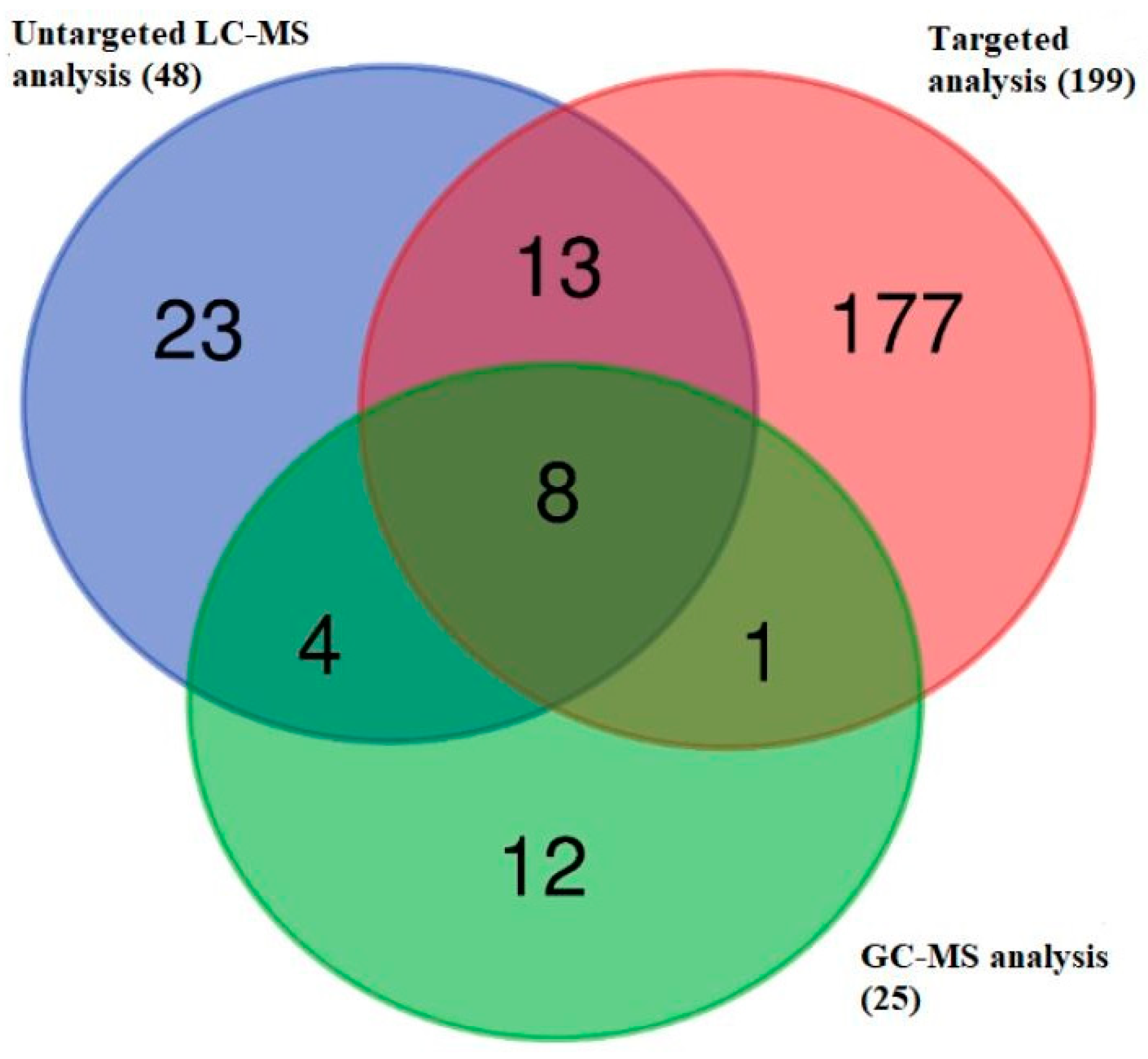
| Metabolites | Peak ID | Mass | RT(s) | p-Value (FDR) | Log2 (FC) |
|---|---|---|---|---|---|
| Cystine | 1021 | 241.0309 | 730.41 | 0.043 | −0.84 |
| 4-Hydroxyproline | 180 | 132.0656 | 684.95 | 0.045 | −0.69 |
| Creatinine | 4 | 114.0662 | 525.25 | 0.030 | 0.45 |
| 3-Hydroxybutanoate | 2002 | 103.0403 | 514.83 | 0.030 | 0.71 |
| Orotate | 2559 | 155.0103 | 555.73 | 0.001 | 0.94 |
| Lactate | 1367 | 89.0246 | 528.32 | 0.025 | 1.00 |
| Carnitine | 3 | 162.1124 | 636.28 | 0.023 | 1.70 |
| cis-Aconitate | 2003 | 173.0097 | 800.83 | 0.0065 | 1.72 |
| O-Acetyl-L-carnitine | 21 | 204.123 | 553.67 | 0.010 | 2.20 |
| 3-Metylhistidine | 27 | 170.0924 | 616.4 | 0.034 | 2.37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubić, I.; Weidt, S.; Burchmore, R.; Kovačević, A.; Kuleš, J.; Eckersall, P.D.; Torti, M.; Jović, I.; Kovačić, M.; Gotić, J.; et al. Metabolome Profiling in the Plasma of Dogs with Idiopathic Dilated Cardiomyopathy: A Multiplatform Mass-Spectrometry-Based Approach. Int. J. Mol. Sci. 2023, 24, 15182. https://doi.org/10.3390/ijms242015182
Rubić I, Weidt S, Burchmore R, Kovačević A, Kuleš J, Eckersall PD, Torti M, Jović I, Kovačić M, Gotić J, et al. Metabolome Profiling in the Plasma of Dogs with Idiopathic Dilated Cardiomyopathy: A Multiplatform Mass-Spectrometry-Based Approach. International Journal of Molecular Sciences. 2023; 24(20):15182. https://doi.org/10.3390/ijms242015182
Chicago/Turabian StyleRubić, Ivana, Stefan Weidt, Richard Burchmore, Alan Kovačević, Josipa Kuleš, Peter David Eckersall, Marin Torti, Ines Jović, Mislav Kovačić, Jelena Gotić, and et al. 2023. "Metabolome Profiling in the Plasma of Dogs with Idiopathic Dilated Cardiomyopathy: A Multiplatform Mass-Spectrometry-Based Approach" International Journal of Molecular Sciences 24, no. 20: 15182. https://doi.org/10.3390/ijms242015182








