Impact of Sex and Genetic Variation in Relevant Pharmacogenes on the Pharmacokinetics and Safety of Valsartan, Olmesartan and Hydrochlorothiazide
Abstract
:1. Introduction
2. Results
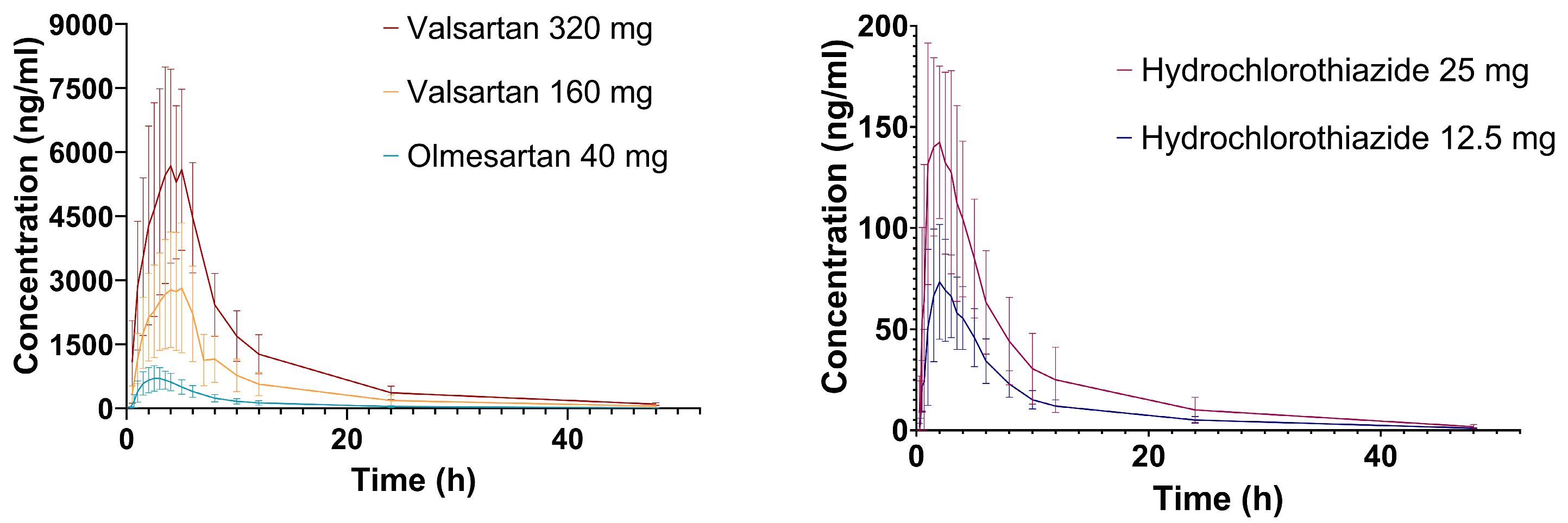
2.1. Hydrochlorothiazide
2.2. Valsartan
2.3. Olmesartan
2.4. Blood Pressure and Heart Rate
2.5. Adverse Drug Reactions
3. Discussion
- LIMITATIONS
4. Material and Methods
4.1. Study Design and Procedures
4.2. Pharmacokinetics, Safety and Pharmacodynamics
4.3. Genotyping and Phenotyping
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FDA (Food and Drug Administration). Highlights of Prescribing Information. Diovan (Valsartan) Tablets. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2011/021283s033lbl.pdf (accessed on 27 July 2023).
- FDA (Food and Drug Administration). Highlights of Prescribing Information. Benicar (Olmesartan Medox-Omil) Tablets. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/021286s023lbl.pdf (accessed on 31 July 2023).
- FDA MICROZIDE® (Hydrochlorothiazide, USP) Capsules 12.5 Mg. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/020504s026lbl.pdf (accessed on 4 August 2023).
- Jordan, J.; Kurschat, C.; Reuter, H. Arterial Hypertension. Dtsch. Ärzteblatt. Int. 2018, 115, 557. [Google Scholar] [CrossRef] [PubMed]
- Zubiaur, P.; Mejía-Abril, G.; Navares-Gómez, M.; Villapalos-García, G.; Soria-Chacartegui, P.; Saiz-Rodríguez, M.; Ochoa, D.; Abad-Santos, F. PriME-PGx: La Princesa University Hospital Multidisciplinary Initiative for the Implementation of Pharmacogenetics. J. Clin. Med. 2021, 10, 3772. [Google Scholar] [CrossRef] [PubMed]
- Soria-Chacartegui, P.; Zubiaur, P.; Ochoa, D.; Villapalos-García, G.; Román, M.; Matas, M.; Figueiredo-Tor, L.; Mejía-Abril, G.; Calleja, S.; de Miguel, A.; et al. Genetic Variation in CYP2D6 and SLC22A1 Affects Amlodipine Pharmacokinetics and Safety. Pharmaceutics 2023, 15, 404. [Google Scholar] [CrossRef]
- Guerrero-García, C.; Rubio-Guerra, F. Combination therapy in the treatment of hypertension. Drugs Context 2018, 7, 212531. [Google Scholar] [CrossRef] [PubMed]
- Cabaleiro, T.; Román, M.; Ochoa, D.; Talegón, M.; Prieto-Pérez, R.; Wojnicz, A.; López-Rodríguez, R.; Novalbos, J.; Abad-Santos, F. Evaluation of the Relationship between Sex, Polymorphisms in CYP2C8 and CYP2C9, and Pharmacokinetics of Angiotensin Receptor Blockers. Drug Metab. Dispos. 2013, 41, 224–229. [Google Scholar] [CrossRef]
- Laeis, P.; Püchler, K.; Kirch, W. The pharmacokinetic and metabolic profile of olmesartan medoxomil limits the risk of clinically relevant drug interaction. J. Hypertens. 2001, 19, S21–S32. [Google Scholar] [CrossRef]
- Rohatagi, S.; Lee, J.; Shenouda, M.; Haworth, S.; Bathala, M.S.; Allison, M.; Rubets, I.; Heyrman, R.; Noveck, R.; Salazar, D.E. Pharmacokinetics of Amlodipine and Olmesartan After Administration of Amlodipine Besylate and Olmesartan Medoxomil in Separate Dosage Forms and as a Fixed-Dose Combination. J. Clin. Pharmacol. 2008, 48, 1309–1322. [Google Scholar] [CrossRef]
- Oh, M.; Shin, J.-G.; Ahn, S.; Kim, B.H.; Kim, J.Y.; Ghim, J.-L. Pharmacokinetic comparison of a fixed-dose combination versus concomitant administration of amlodipine, olmesartan, and rosuvastatin in healthy adult subjects. Drug Des. Dev. Ther. 2019, 13, 991–997. [Google Scholar] [CrossRef]
- CBG-MEB Public Assessment Report: Olmesartan Medoxomil/Amlodipine Rafarm 20 Mg/5 Mg, 40 Mg/5 Mg, 40 Mg/10 Mg, Film-Coated Tablets. Available online: https://www.geneesmiddeleninformatiebank.nl/pars/h125192.pdf (accessed on 10 August 2023).
- Chu, T. Gender Differences in Pharmacokinetics. US Pharm. 2014, 39, 40–43. [Google Scholar]
- Jensen, C.E.D.M.; Dos Santos, R.A.S.; Denadai, A.M.L.; Santos, C.F.F.; Braga, A.N.G.; Sinisterra, R.D. Pharmaceutical Composition of Valsartan: β-Cyclodextrin: Physico–Chemical Characterization and Anti-Hypertensive Evaluation. Molecules 2010, 15, 4067–4084. [Google Scholar] [CrossRef]
- Bredella, M.A. Sex Differences in Body Composition. Adv. Exp. Med. Biol. 2017, 1043, 9–27. [Google Scholar] [CrossRef]
- Soldin, O.P.; Chung, S.H.; Mattison, D.R. Sex Differences in Drug Disposition. J. Biomed. Biotechnol. 2011, 2011, 187103. [Google Scholar] [CrossRef] [PubMed]
- Soldin, O.P.; Mattison, D.R. Sex Differences in Pharmacokinetics and Pharmacodynamics. Clin. Pharmacokinet. 2009, 48, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Kalibala, J.; Pechère-Bertschi, A.; Desmeules, J. Gender Differences in Cardiovascular Pharmacotherapy—The Example of Hypertension: A Mini Review. Front. Pharmacol. 2020, 11, 564. [Google Scholar] [CrossRef]
- SingleCare Team Understanding Blood Pressure Levels. Available online: https://www.singlecare.com/blog/blood-pressure-levels/ (accessed on 27 July 2023).
- Fan, X.; Han, Y.; Sun, K.; Wang, Y.; Xin, Y.; Bai, Y.; Li, W.; Yang, T.; Song, X.; Wang, H.; et al. Sex Differences in Blood Pressure Response to Antihypertensive Therapy in Chinese Patients with Hypertension. Ann. Pharmacother. 2008, 42, 1772–1781. [Google Scholar] [CrossRef] [PubMed]
- Di Rienzo, M.; Parati, G.; Radaelli, A.; Castiglioni, P. Baroreflex contribution to blood pressure and heart rate oscillations: Time scales, time-variant characteristics and nonlinearities. Philos. Trans. A Math. Phys. Eng. Sci. 2009, 367, 1301–1318. [Google Scholar] [CrossRef] [PubMed]
- Oparil, S.; Williams, D.; Chrysant, S.G.; Marbury, T.C.; Neutel, J. Comparative Efficacy of Olmesartan, Losartan, Valsartan, and Irbesartan in the Control of Essential Hypertension. J. Clin. Hypertens. 2001, 3, 283–318. [Google Scholar] [CrossRef]
- Shin, H.-B.; Jung, E.H.; Kang, P.; Lim, C.W.; Oh, K.-Y.; Cho, C.-K.; Lee, Y.J.; Choi, C.-I.; Jang, C.-G.; Lee, S.-Y.; et al. ABCB1 c.2677G>T/c.3435C>T diplotype increases the early-phase oral absorption of losartan. Arch. Pharmacal Res. 2020, 43, 1187–1196. [Google Scholar] [CrossRef]
- PharmGKB Candesartan Pathway, Pharmacokinetics. Available online: https://www.pharmgkb.org/pathway/PA166182962 (accessed on 28 July 2023).
- Linnet, K.; Ejsing, T.B. A review on the impact of P-glycoprotein on the penetration of drugs into the brain. Focus on psychotropic drugs. Eur. Neuropsychopharmacol. 2008, 18, 157–169. [Google Scholar] [CrossRef]
- Goswami, S.; Gong, L.; Giacomini, K.; Altman, R.B.; Klein, T.E. PharmGKB summary: Very Important Pharmacogene In-formation for SLC22A1. Pharmacogenet. Genom. 2014, 24, 324–328. [Google Scholar] [CrossRef]
- DrugBank Hydrochlorothiazide. Available online: https://go.drugbank.com/drugs/DB00999 (accessed on 7 August 2023).
- Hypha Discovery Differential Pharmacological Activities of Valsartan and Its Main Human Metabolite 4-Hydroxyvaleryl-Valsartan. Available online: https://www.hyphadiscovery.com/wp-content/uploads/2021/06/Hypha-Valsartan-metabolism.pdf (accessed on 9 August 2023).
- Nakashima, A.; Kawashita, H.; Masuda, N.; Saxer, C.; Niina, M.; Nagae, Y.; Iwasaki, K. Identification of cytochrome P450 forms involved in the 4-hydroxylation of valsartan, a potent and specific angiotensin II receptor antagonist, in human liver microsomes. Xenobiotica 2005, 35, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Arcas, M.R.; García-Jiménez, E.; Martínez-Martínez, F.; Conesa-Zamora, P. Papel del citocromo P450 en la farmacocinética y en la farmacogenética de los fármacos antihipertensivos. Farm. Hosp. 2011, 35, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Hanna, I.; Alexander, N.; Crouthamel, M.H.; Davis, J.; Natrillo, A.; Tran, P.; Vapurcuyan, A.; Zhu, B. Transport properties of valsartan, sacubitril and its active metabolite (LBQ657) as determinants of disposition. Xenobiotica 2018, 48, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Chung, J.-E.; Yee, J.; Lee, K.-E.; Park, K.; Gwak, H.-S. Effects of SLCO1B1 and SLCO1B3 Genetic Polymorphisms on Valsartan Pharmacokinetics in Healthy Korean Volunteers. J. Pers. Med. 2021, 11, 862. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Gong, L.; Lee, S.; Wagner, J.B.; Zhou, X.; Sangkuhl, K.; Adams, S.M.; Straka, R.J.; Empey, P.E.; Boone, E.C.; et al. PharmVar GeneFocus: SLCO1B1. Clin. Pharmacol. Ther. 2022, 113, 782–793. [Google Scholar] [CrossRef]
- Suwannakul, S.; Ieiri, I.; Kimura, M.; Kawabata, K.; Kusuhara, H.; Hirota, T.; Irie, S.; Sugiyama, Y.; Higuchi, S. Pharmacokinetic interaction between pravastatin and olmesartan in relation to SLCO1B1 polymorphism. J. Hum. Genet. 2008, 53, 899–904. [Google Scholar] [CrossRef]
- Cooper-DeHoff, R.M.; Niemi, M.; Ramsey, L.B.; Luzum, J.A.; Tarkiainen, E.K.; Straka, R.J.; Gong, L.; Tuteja, S.; Wilke, R.A.; Wadelius, M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and Statin-Associated Musculoskeletal Symptoms. Clin. Pharmacol. Ther. 2022, 111, 1007–1021. [Google Scholar] [CrossRef]
- Vijayananthan, A.; Nawawi, O. The importance of Good Clinical Practice guidelines and its role in clinical trials. Biomed. Imaging Interv. J. 2008, 4, e5. [Google Scholar] [CrossRef]
- World Medical Association World Medical Association Declaration of Helsinki Ethical Principles for Medical Research Involving Human Subjects. Available online: https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/ (accessed on 3 August 2023).
- Aguirre, C.; García, M. Causality assessment in adverse drug reaction reports. Algorithm of the Spanish Pharmacovigilance System. Med. Clínica 2016, 147, 461–464. [Google Scholar] [CrossRef]
- Desta, Z.; Gammal, R.S.; Gong, L.; Whirl-Carrillo, M.; Gaur, A.H.; Sukasem, C.; Hockings, J.; Myers, A.; Swart, M.; Tyndale, R.F.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2B6 and Efavirenz-Containing Antiretroviral Therapy. Clin. Pharmacol. Ther. 2019, 106, 726–733. [Google Scholar] [CrossRef]
- Scott, S.A.; Sangkuhl, K.; Stein, C.M.; Hulot, J.-S.; Mega, J.L.; Roden, D.M.; Klein, T.E.; Sabatine, M.S.; Johnson, J.A.; Shuldiner, A.R. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C19 Genotype and Clopidogrel Therapy: 2013 Update. Clin. Pharmacol. Ther. 2013, 94, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Theken, K.N.; Lee, C.R.; Gong, L.; Caudle, K.E.; Formea, C.M.; Gaedigk, A.; Klein, T.E.; Agúndez, J.A.G.; Grosser, T. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clin. Pharmacol. Ther. 2020, 108, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Sangkuhl, K.; Guchelaar, H.-J.; Schwab, M.; Province, M.; Whirl-Carrillo, M.; Symmans, W.F.; McLeod, H.L.; Ratain, M.J.; Zembutsu, H.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin. Pharmacol. Ther. 2018, 103, 770–777. [Google Scholar] [CrossRef]
- Birdwell, K.; Decker, B.; Barbarino, J.; Peterson, J.; Stein, C.; Sadee, W.; Wang, D.; Vinks, A.; He, Y.; Swen, J.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin. Pharmacol. Ther. 2015, 98, 19–24. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Johnson, S.G.; Caudle, K.E.; Haidar, C.E.; Voora, D.; Wilke, R.A.; Maxwell, W.D.; McLeod, H.L.; Krauss, R.M.; Roden, D.M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1 and Simvastatin-Induced Myopathy: 2014 Update. Clin. Pharmacol. Ther. 2014, 96, 423–428. [Google Scholar] [CrossRef]
- Gammal, R.; Court, M.; Haidar, C.; Iwuchukwu, O.; Gaur, A.; Alvarellos, M.; Guillemette, C.; Lennox, J.; Whirl-Carrillo, M.; Brummel, S.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for UGT1A1 and Atazanavir Prescribing. Clin. Pharmacol. Ther. 2016, 99, 363–369. [Google Scholar] [CrossRef] [PubMed]
- DPWG Annotation of DPWG Guideline for Quetiapine and CYP3A4. 2021. Available online: https://www.pharmgkb.org/guidelineAnnotation/PA166265421 (accessed on 4 August 2023).
- Campodónico, D.M.; Zubiaur, P.; Soria-Chacartegui, P.; Casajús, A.; Villapalos-García, G.; Navares-Gómez, M.; Gómez-Fernández, A.; Parra-Garcés, R.; Mejía-Abril, G.; Román, M.; et al. CYP2C8 *3 and *4 define CYP2C8 phenotype: An approach with the substrate cinitapride. Clin. Transl. Sci. 2022, 15, 2613–2624. [Google Scholar] [CrossRef]

| Variable | n | Age (Years) | Height (m) | Weight (kg) | BMI (kg/m2) |
|---|---|---|---|---|---|
| Sex | |||||
| Men | 37 | 24.57 (4.39) | 1.78 (0.08) | 78.77 (10.38) | 24.89 (3.01) |
| Women | 27 | 31.15 (10.69) * | 1.62 (0.05) * | 60.56 (7.57) * | 23.21 (2.89) * |
| Biogeographic origin | |||||
| European | 43 | 26.16 (7.35) | 1.72 (0.10) | 71.02 (11.97) | 23.84 (3.08) |
| Other # | 21 | 29.76 (9.72) | 1.68 (0.12) | 71.21 (15.04) | 24.89 (2.95) |
| CT | |||||
| 1 | 6 | 22.17 (2.48) | 1.73 (0.12) | 65.83 (7.16) | 22.11 (2.73) |
| 2 | 4 | 30.00 (8.76) | 1.70 (0.16) | 77.93 (14.58) | 26.85 (2.45) |
| 3 | 8 | 27.25 (7.72) | 1.68 (0.10) | 63.50 (13.41) | 22.23 (2.68) |
| 4 | 8 | 26.13 (10.33) | 1.68 (0.10) | 64.96 (12.34) | 22.93 (2.34) |
| 5 | 12 | 30.58 (11.70) | 1.73 (0.10) | 76.54 (12.17) | 25.65 (2.93) |
| 6 | 10 | 28.40 (9.35) | 1.72 (0.09) | 73.50 (12.83) | 24.86 (2.98) |
| 7 | 16 | 26.19 (4.18) | 1.72 (0.12) | 72.60 (13.29) | 24.36 (3.06) |
| Total | 64 | 27.34 (8.30) | 1.71 (0.11) | 71.09 (12.94) | 24.18 (3.05) |
| Variable | n | AUC∞/DW (ng*h*kg/mL*mg) | Cmax/DW (ng*kg/mL*mg) | tmax (h) | t1/2 (h) |
|---|---|---|---|---|---|
| Sex | |||||
| Men | 35 | 3213.59 (584.71) | 465.31 (109.02) | 1.89 (0.61) | 9.70 (1.39) |
| Women | 23 | 3508.15 (911.76) | 491.54 (104.02) | 2.04 (0.84) | 8.66 (1.30) * |
| SLC22A1 rs34059508 | |||||
| G/G | 55 | 3296.92 (731.93) | 474.71 (107.74) | 1.90 (0.67) | 9.29 (1.46) |
| G/A | 2 | 4344.26 (190.17) * | 553.98 (48.15) | 2.50 (0.71) | 9.78 (0.81) |
| Variable | n | AUC∞/DW (ng*h*kg/mL*mg) | Cmax/DW (ng*kg/mL*mg) | tmax (h) | t1/2 (h) |
|---|---|---|---|---|---|
| Sex | |||||
| Men | 11 | 13,374.37 (3839.37) | 1490.04 (382.23) | 3.37 (0.78) | 8.80 (1.50) |
| Women | 15 | 9658.52 (4290.87) * | 1303.44 (509.40) | 3.61 (1.03) | 8.64 (2.75) |
| ABCB1 rs1128503 | |||||
| T/T | 3 | 13,196.33 (7954.48) | 1526.29 (916.55) | 5.00 (0.25) * | 9.06 (2.82) |
| T/C | 10 | 12,529.81 (3854.25) | 1449.58 (317.14) | 3.52 (0.58) | 9.89 (2.51) |
| C/C | 12 | 9812.22 (4005.06) | 1286.24 (478.30) | 3.02 (0.80) | 7.74 (1.62) |
| ABCB1 rs1045642 | |||||
| T/T | 5 | 12,816.62 (5724.14) | 1554.04 (672.27) | 4.50 (0.88) * | 8.56 (2.18) |
| T/C | 9 | 11,536.06 (4803.69) | 1322.30 (381.24) | 3.13 (0.57) | 9.96 (2.59) |
| C/C | 11 | 10,429.64 (3912.43) | 1348.96 (464.47) | 3.25 (0.84) | 7.86 (1.79) |
| ABCB1 rs2032582 | |||||
| T/T | 4 | 12,627.06 (6593.84) | 1481.92 (753.60) | 4.50 (1.02) * | 8.95 (2.32) |
| T/G | 10 | 11,455.73 (4429.94) | 1414.61 (367.05) | 3.39 (0.81) | 9.16 (2.59) |
| G/A | 2 | 11,798.75 (6567.18) | 1634.43 (628.50) | 3.88 (0.35) | 6.50 (1.24) |
| G/G | 9 | 10,987.88 (3298.50) | 1321.33 (410.01) | 3.23 (0.86) | 8.72 (2.16) |
| CYP2D6 | |||||
| UM | 2 | 11,533.20 (393.31) | 1609.28 (269.46) | 4.44 (0.44) | 6.84 (1.63) |
| NM | 11 | 12,329.25 (5352.08) | 1462.70 (503.56) | 3.65 (0.99) | 8.70 (2.19) |
| IM | 9 | 10,298.34 (4685.46) | 1271.91 (531.72) | 3.09 (0.85) | 8.23 (1.50) |
| PM | 2 | 10,170.64 (1857.41) | 1202.63 (316.61) | 3.32 (0.09) | 13.43 (2.23) * |
| CYP3A5 | |||||
| IM | 4 | 11,497.08 (4442.61) | 1439.20 (550.88) | 4.44 (0.71) * | 7.35 (1.34) |
| PM | 21 | 11,268.83 (4635.61) | 1369.18 (468.67) | 3.27 (0.82) | 9.03 (2.37) |
| n | AUC∞/DW (ng*h*kg/mL*mg) | Cmax/DW (ng*kg/mL*mg) | tmax (h) | t1/2 (h) | |
|---|---|---|---|---|---|
| Sex | |||||
| Men | 25 | 10,539.83 (3009.88) | 1406.74 (454.05) | 2.72 (1.16) | 9.24 (1.44) |
| Women | 12 | 11,494.44 (3398.77) | 1445.91 (344.42) | 2.58 (0.76) | 8.56 (1.08) |
| CT | |||||
| 5 | 12 | 10,960.25 (3810.09) | 1457.24 (538.37) | 1.96 (0.69) * | 9.36 (1.58) |
| 6 | 10 | 12,067.51 (3220.87) | 1596.56 (363.70) | 3.20 (0.92) | 8.85 (1.54) |
| 7 | 15 | 9948.73 (2262.70) | 1271.12 (295.86) | 2.90 (1.07) | 8.86 (1.06) |
| CT | Eudra-CT | Test Formulation | Volunteers # |
|---|---|---|---|
| 1 | 2013-004147-23 | Amlodipine/Valsartan 10/160 mg | 6 |
| 2 | 2017-000547-40 | Amlodipine/Valsartan/Hydrochlorothiazide 10/160/25 mg | 4 |
| 3 | 2017-001716-10 | Amlodipine/Valsartan/Hydrochlorothiazide 10/160/12.5 mg | 8 |
| 4 | 2017-001757-14 | Amlodipine/Valsartan/Hydrochlorothiazide 10/320/25 mg | 8 |
| 5 | 2017-005024-25 | Amlodipine/Olmesartan/Hydrochlorothiazide 10/40/25 mg | 12 |
| 6 | 2018-001378-11 | Amlodipine/Olmesartan/Hydrochlorothiazide 10/40/12.5 mg | 10 |
| 7 | 2018-002075-18 | Amlodipine/Olmesartan/Hydrochlorothiazide 10/40/12.5 mg | 16 |
| Gene | Allele | Genetic Variants Present in the Allele | Frequency (European Population) $ | Frequency (Latin-American Population) $ |
|---|---|---|---|---|
| ABCB1 | N/A | rs1045642 (T>C) | T: 51.8%, C: 48.2% | T: 42.8%, C: 57.2% |
| N/A | rs2032582 (T>G/A) | T: 41.0%, G: 57.3%, A: 1.8% | T: 36.9%, G: 57.2%, A: 5.9% | |
| N/A | rs1128503 (T>C) | T: 41.6%, C: 58.4% | T: 40.3%, C: 59.7% | |
| ABCC2 | N/A | rs2273697 (G>A) | G: 79.6%, A: 20.4% | G: 84.1%, A: 15.9% |
| ABCG2 | N/A | rs2231142 (C>A) | C: 89.6%, A: 10.4% | C: 77.6%, A: 22.4% |
| SLC22A1 | N/A | rs12208357 (C>T) | C: 93.7%, T:6.3% | C: 98.0%, T: 2.0% |
| N/A | rs34059508 (G>A) | G:98.0%, A: 2.0% | G: 98.1%, A: 1.9% | |
| N/A | rs72552763 (GAT>delGAT) | GAT: 81.6%, delGAT: 18.4% | GAT: 71.2%, delGAT: 28.8% | |
| SLCO1B1 | *5 | rs4149056 (T>C) | <0.01% | 2.0% |
| *15 | rs4149056 (T>C), rs2306283 (A>G) | 15.0% | 24.0% | |
| *37 | rs2306283 (A>G) | 25.3% | 39.0% | |
| UGT1A1 # | *80 | rs887829 (C>T) | 31.4% | 38.3% |
| CYP2D6 | *3 | rs35742686 (A>delA) | 1.6% | 0.7% |
| *4 | rs3892097 (G>A), rs1065852(C>T) | 18.5% | 12.0% | |
| *6 | rs5030655 (T>delT) | 1.1% | 0.5% | |
| *7 | rs5030867 (A>C) | <0.01% | <0.01% | |
| *8 | rs5030865 (G>T) | <0.01% | <0.01% | |
| *9 | rs5030656 (TCT>delTCT) | 2.8% | 1.6% | |
| *10 | rs1065852 (C>T) | 1.6% | 2.6% | |
| *14 | rs5030865 (G>A) | <0.01% | <0.01% | |
| *17 | rs28371706 (C>T) | 0.4% | 2.3% | |
| *41 | rs28371725 (G>A) | 9.2% | 5.1% | |
| CYP2A6 | *9 | rs28399433 (T>G) | 7.1% | 7.1% |
| CYP2B6 | *4 | rs2279343 (A>G) | 40.9% | 10.6% |
| *5 | rs3211371 (C>T) | 11.6% | 3.8% | |
| *6 | rs2279343(A>G), rs3745274 (G>T) | 23.3% | 21.2% | |
| *7 | rs2279343 (A>G), rs3745274 (G>T), rs3211371 (C>T) | 2.5% | 0.8% | |
| *9 | rs3745274 (G>T) | 1.5% | 7.3% | |
| CYP2C8 | *2 | rs11572103 (A>T) | A: 99.6%, T: 0.4% | A: 98.8%, T: 1.2% |
| *3 | rs10509681 (A>G) | A: 88.2%, G: 11.8% | A: 90.1%, G: 9.9% | |
| *4 | rs1058930 (C>G) | C: 94.2%, G:5.8% | C: 98.1%, G: 1.9% | |
| CYP2C9 | *2 | rs1799853 (C>T) | 12.7% | 7.6% |
| *3 | rs1057910 (A>C) | 7.6% | 4.0% | |
| CYP2C19 | *2 | rs4244285 (G>A) | 14.7% | 7.2% |
| *3 | rs4986893 (G>A) | 0.2% | <0.01% | |
| *4 | rs28399504 (A>G) | 0.2% | <0.01% | |
| *17 | rs12248560 (C>T) | 21.5% | 16.7% | |
| CYP3A4 | *2 | rs55785340 (T>C) | T: >99.9% C: <0.01% | T: >99.9% C: <0.01% |
| *6 | rs46464389 (delA>A) | delA: >99.9%, A: <0.01% | delA: >99.9%, A: <0.01% | |
| *20 | rs67666821(delA>A) | 0.09% | 0.09% | |
| *22 | rs35599367 (C>T) | 3.7% | 2.6% | |
| CYP3A5 | *3 | rs776746 (A>G) | 92.4% | 76.5% |
| *6 | rs10264272 (G>A) | <0.01% | 0.2% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soria-Chacartegui, P.; Zubiaur, P.; Ochoa, D.; Navares-Gómez, M.; Abbes, H.; Villapalos-García, G.; de Miguel, A.; González-Iglesias, E.; Rodríguez-Lopez, A.; Mejía-Abril, G.; et al. Impact of Sex and Genetic Variation in Relevant Pharmacogenes on the Pharmacokinetics and Safety of Valsartan, Olmesartan and Hydrochlorothiazide. Int. J. Mol. Sci. 2023, 24, 15265. https://doi.org/10.3390/ijms242015265
Soria-Chacartegui P, Zubiaur P, Ochoa D, Navares-Gómez M, Abbes H, Villapalos-García G, de Miguel A, González-Iglesias E, Rodríguez-Lopez A, Mejía-Abril G, et al. Impact of Sex and Genetic Variation in Relevant Pharmacogenes on the Pharmacokinetics and Safety of Valsartan, Olmesartan and Hydrochlorothiazide. International Journal of Molecular Sciences. 2023; 24(20):15265. https://doi.org/10.3390/ijms242015265
Chicago/Turabian StyleSoria-Chacartegui, Paula, Pablo Zubiaur, Dolores Ochoa, Marcos Navares-Gómez, Houwaida Abbes, Gonzalo Villapalos-García, Alejandro de Miguel, Eva González-Iglesias, Andrea Rodríguez-Lopez, Gina Mejía-Abril, and et al. 2023. "Impact of Sex and Genetic Variation in Relevant Pharmacogenes on the Pharmacokinetics and Safety of Valsartan, Olmesartan and Hydrochlorothiazide" International Journal of Molecular Sciences 24, no. 20: 15265. https://doi.org/10.3390/ijms242015265
APA StyleSoria-Chacartegui, P., Zubiaur, P., Ochoa, D., Navares-Gómez, M., Abbes, H., Villapalos-García, G., de Miguel, A., González-Iglesias, E., Rodríguez-Lopez, A., Mejía-Abril, G., Martín-Vilchez, S., Luquero-Bueno, S., Román, M., & Abad-Santos, F. (2023). Impact of Sex and Genetic Variation in Relevant Pharmacogenes on the Pharmacokinetics and Safety of Valsartan, Olmesartan and Hydrochlorothiazide. International Journal of Molecular Sciences, 24(20), 15265. https://doi.org/10.3390/ijms242015265









