Gut Microbiota Patterns in Patients with Non-Alcoholic Fatty Liver Disease: A Comprehensive Assessment Using Three Analysis Methods
Abstract
1. Introduction
2. Results
2.1. MALDI-TOF-MS
2.2. Real-Time qPCR Analysis
2.2.1. Data Preprocessing
2.2.2. Statistical Testing using the Mann–Whitney Test
2.2.3. Machine Learning Model
2.2.4. Correlations within Case/Control Groups
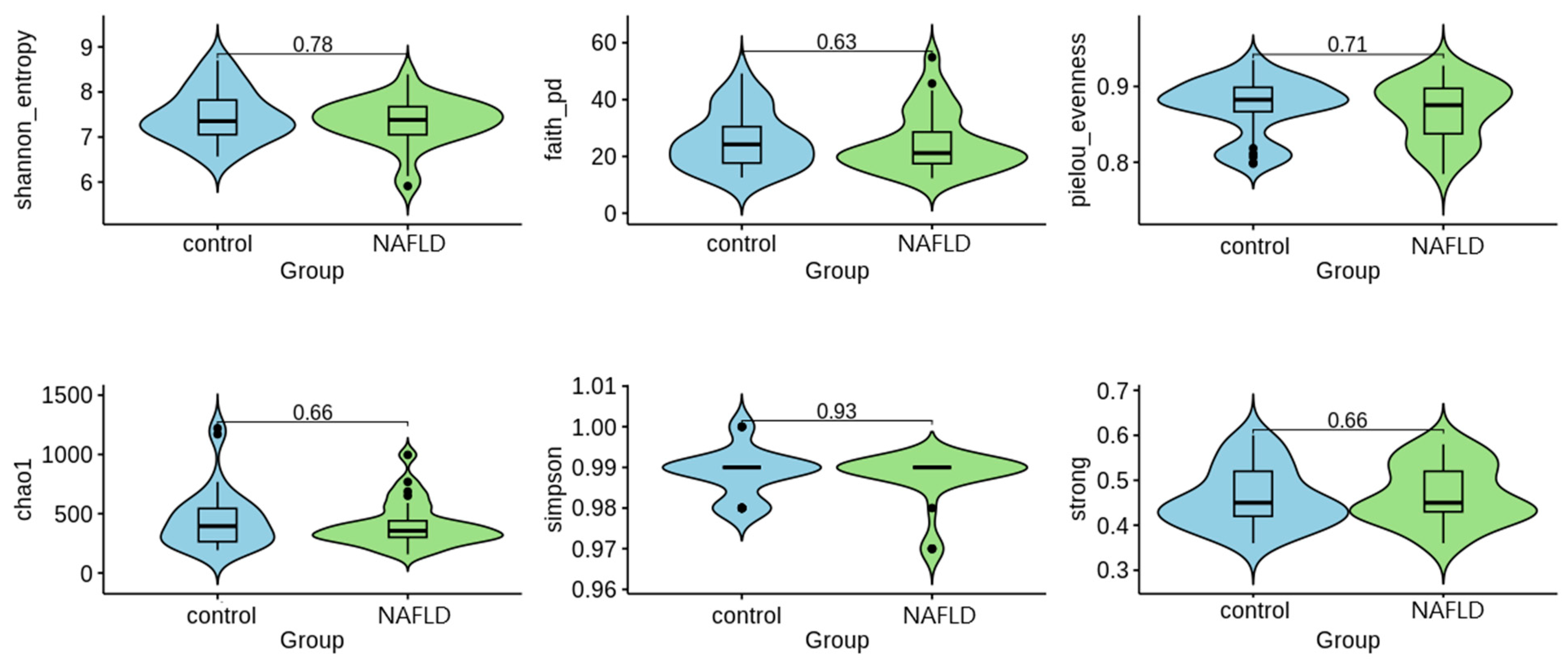
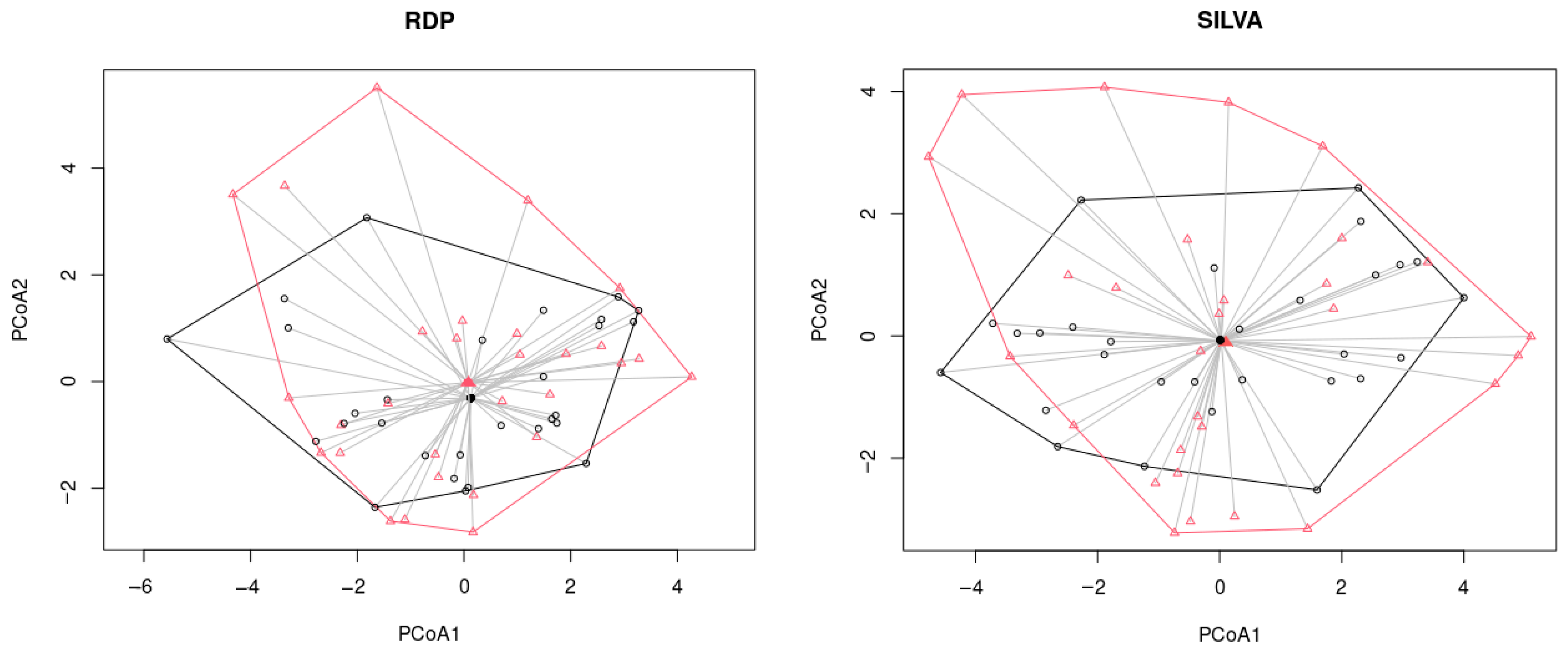
2.3. NGS
2.4. Comparative Analysis of Several Assessment Methods
3. Discussion
3.1. Single Method Results
3.2. Cross-Method Comparative Analysis
3.3. Bacterial Correlations
4. Materials and Methods
4.1. Cohort Description
4.2. Microbial Identification using MALDI-TOF-MS
4.3. Real-Time qPCR Quantification of Bacterial DNA
4.3.1. Study Group
4.3.2. PCR Reaction
4.4. Preparation of DNA Libraries and Sequencing
4.5. Metagenome Data Processing and Analysis
4.6. Statistical Analysis
4.6.1. Software
4.6.2. Data Conversion
- -
- all zero elements were retained,
- -
- non-zero elements were converted using a formula (Formula (1)):
4.6.3. Data Filtering
4.6.4. Exploratory and Statistical Data Analysis
Mann–Whitney Test
Principal Coordinate Analysis
Permutational Multivariate Analysis of Variance
Latent Dirichlet Distribution Analysis
Penalized Regression
Simulated Data Synthesis
Machine Learning Model
Correlation Scores
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Younossi, Z.M.; Marchesini, G.; Pinto-Cortez, H.; Petta, S. Epidemiology of Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis: Implications for Liver Transplantation. Transplantation 2019, 103, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Dhamija, E.; Paul, S.; Kedia, S. Non-alcoholic fatty liver disease associated with hepatocellular carcinoma: An increasing concern. Indian J. Med. Res. 2019, 149, 9. [Google Scholar] [CrossRef] [PubMed]
- Sogabe, M.; Okahisa, T.; Kurihara, T.; Takehara, M.; Kagemoto, K.; Okazaki, J.; Kida, Y.; Hirao, A.; Tanaka, H.; Tomonari, T.; et al. Differences among patients with and without nonalcoholic fatty liver disease having elevated alanine aminotransferase levels at various stages of metabolic syndrome. PLoS ONE 2020, 15, e0238388. [Google Scholar] [CrossRef] [PubMed]
- Hazlehurst, J.M.; Woods, C.; Marjot, T.; Cobbold, J.F.; Tomlinson, J.W. Non-alcoholic fatty liver disease and diabetes. Metabolism 2016, 65, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Jaruvongvanich, V.; Wirunsawanya, K.; Sanguankeo, A.; Upala, S. Nonalcoholic fatty liver disease is associated with coronary artery calcification: A systematic review and meta-analysis. Dig. Liver Dis. 2016, 48, 1410–1417. [Google Scholar] [CrossRef] [PubMed]
- Colognesi, M.; Gabbia, D.; De Martin, S. Depression and Cognitive Impairment—Extrahepatic Manifestations of NAFLD and NASH. Biomedicines 2020, 8, 229. [Google Scholar] [CrossRef]
- Mantovani, A.; Petracca, G.; Beatrice, G.; Csermely, A.; Tilg, H.; Byrne, C.D.; Targher, G. Non-alcoholic fatty liver disease and increased risk of incident extrahepatic cancers: A meta-analysis of observational cohort studies. Gut 2022, 71, 778–788. [Google Scholar] [CrossRef]
- Campo, L.; Eiseler, S.; Apfel, T.; Pyrsopoulos, N. Fatty Liver Disease and Gut Microbiota: A Comprehensive Update. J. Clin. Transl. Hepatol. 2019, 7, 56–60. [Google Scholar] [CrossRef]
- Hrncir, T.; Hrncirova, L.; Kverka, M.; Hromadka, R.; Machova, V.; Trckova, E.; Kostovcikova, K.; Kralickova, P.; Krejsek, J.; Tlaskalova-Hogenova, H. Gut Microbiota and NAFLD: Pathogenetic Mechanisms, Microbiota Signatures, and Therapeutic Interventions. Microorganisms 2021, 9, 957. [Google Scholar] [CrossRef]
- Aron-Wisnewsky, J.; Vigliotti, C.; Witjes, J.; Le, P.; Holleboom, A.G.; Verheij, J.; Nieuwdorp, M.; Clément, K. Gut microbiota and human NAFLD: Disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 279–297. [Google Scholar] [CrossRef]
- Jiang, X.; Zheng, J.; Zhang, S.; Wang, B.; Wu, C.; Guo, X. Advances in the Involvement of Gut Microbiota in Pathophysiology of NAFLD. Front. Med. 2020, 7, 361. [Google Scholar] [CrossRef] [PubMed]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [PubMed]
- Mbaye, B.; Borentain, P.; Magdy Wasfy, R.; Alou, M.T.; Armstrong, N.; Mottola, G.; Meddeb, L.; Ranque, S.; Gérolami, R.; Million, M.; et al. Endogenous Ethanol and Triglyceride Production by Gut Pichia kudriavzevii, Candida albicans and Candida glabrata Yeasts in Non-Alcoholic Steatohepatitis. Cells 2022, 11, 3390. [Google Scholar] [CrossRef] [PubMed]
- Layden, B.T.; Angueira, A.R.; Brodsky, M.; Durai, V.; Lowe, W.L. Short chain fatty acids and their receptors: New metabolic targets. Transl. Res. 2013, 161, 131–140. [Google Scholar] [CrossRef]
- Scheppach, W.; Bartram, P.; Richter, A.; Richter, F.; Liepold, H.; Dusel, G.; Hofstetter, G.; Rüthlein, J.; Kasper, H. Effect of Short-Chain Fatty Acids on the Human Colonic Mucosa in Vitro. J. Parenter. Enter. Nutr. 1992, 16, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Pingitore, A.; Chambers, E.S.; Hill, T.; Maldonado, I.R.; Liu, B.; Bewick, G.; Morrison, D.J.; Preston, T.; Wallis, G.A.; Tedford, C.; et al. The diet-derived short chain fatty acid propionate improves beta-cell function in humans and stimulates insulin secretion from human islets in vitro. Diabetes Obes. Metab. 2017, 19, 257–265. [Google Scholar] [CrossRef]
- Meng, Q.; Duan, X.; Wang, C.; Liu, Z.; Sun, P.; Huo, X.; Sun, H.; Peng, J.; Liu, K. Alisol B 23-acetate protects against non-alcoholic steatohepatitis in mice via farnesoid X receptor activation. Acta Pharmacol. Sin. 2017, 38, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Zhai, Q.; Chen, W. Investigations of Bacteroides spp. towards next-generation probiotics. Food Res. Int. 2019, 116, 637–644. [Google Scholar] [CrossRef]
- Gauffin Cano, P.; Santacruz, A.; Moya, Á.; Sanz, Y. Bacteroides uniformis CECT 7771 Ameliorates Metabolic and Immunological Dysfunction in Mice with High-Fat-Diet Induced Obesity. PLoS ONE 2012, 7, e41079. [Google Scholar] [CrossRef]
- Safari, Z.; Gérard, P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD). Cell. Mol. Life Sci. 2019, 76, 1541–1558. [Google Scholar] [CrossRef]
- Mabwi, H.A.; Hitayezu, E.; Mauliasari, I.R.; Mwaikono, K.S.; Yoon, H.S.; Komba, E.V.G.; Pan, C.-H.; Cha, K.H. Simulation of the mucosal environment in the re-construction of the synthetic gut microbial ecosystem. J. Microbiol. Methods 2021, 191, 106351. [Google Scholar] [CrossRef] [PubMed]
- Mayengbam, S.; Chleilat, F.; Reimer, R.A. Dietary Vitamin B6 Deficiency Impairs Gut Microbiota and Host and Microbial Metabolites in Rats. Biomedicines 2020, 8, 469. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Zou, Y.; Dai, Y.; Luo, G.; Zhang, X.; Xiao, L. Characterization a Novel Butyric Acid-Producing Bacterium Collinsella aerofaciens Subsp. Shenzhenensis Subsp. Nov. Microorganisms 2019, 7, 78. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Wang, X.; Ren, X.; Liu, Y. Changes of intestinal bacterial microbiota in coronary heart disease complicated with nonalcoholic fatty liver disease. BMC Genom. 2019, 20, 862. [Google Scholar] [CrossRef] [PubMed]
- Astbury, S.; Atallah, E.; Vijay, A.; Aithal, G.P.; Grove, J.I.; Valdes, A.M. Lower gut microbiome diversity and higher abundance of proinflammatory genus Collinsella are associated with biopsy-proven nonalcoholic steatohepatitis. Gut Microbes 2020, 11, 569–580. [Google Scholar] [CrossRef]
- Sui, G.; Jia, L.; Quan, D.; Zhao, N.; Yang, G. Activation of the gut microbiota-kynurenine-liver axis contributes to the development of nonalcoholic hepatic steatosis in nondiabetic adults. Aging 2021, 13, 21309–21324. [Google Scholar] [CrossRef]
- Demir, M.; Lang, S.; Martin, A.; Farowski, F.; Wisplinghoff, H.; Vehreschild, M.J.G.T.; Krawczyk, M.; Nowag, A.; Scholz, C.J.; Kretzschmar, A.; et al. Phenotyping non-alcoholic fatty liver disease by the gut microbiota: Ready for prime time? J. Gastroenterol. Hepatol. 2020, 35, 1969–1977. [Google Scholar] [CrossRef]
- Raman, M.; Ahmed, I.; Gillevet, P.M.; Probert, C.S.; Ratcliffe, N.M.; Smith, S.; Greenwood, R.; Sikaroodi, M.; Lam, V.; Crotty, P.; et al. Fecal Microbiome and Volatile Organic Compound Metabolome in Obese Humans With Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2013, 11, 868–875.e3. [Google Scholar] [CrossRef]
- Vallianou, N.; Christodoulatos, G.S.; Karampela, I.; Tsilingiris, D.; Magkos, F.; Stratigou, T.; Kounatidis, D.; Dalamaga, M. Understanding the Role of the Gut Microbiome and Microbial Metabolites in Non-Alcoholic Fatty Liver Disease: Current Evidence and Perspectives. Biomolecules 2021, 12, 56. [Google Scholar] [CrossRef]
- Da Silva, H.E.; Teterina, A.; Comelli, E.M.; Taibi, A.; Arendt, B.M.; Fischer, S.E.; Lou, W.; Allard, J.P. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci. Rep. 2018, 8, 1466. [Google Scholar] [CrossRef]
- Jiao, N.; Loomba, R.; Yang, Z.-H.; Wu, D.; Fang, S.; Bettencourt, R.; Lan, P.; Zhu, R.; Zhu, L. Alterations in bile acid metabolizing gut microbiota and specific bile acid genes as a precision medicine to subclassify NAFLD. Physiol. Genom. 2021, 53, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Bashir, A.; Miskeen, A.Y.; Hazari, Y.M.; Asrafuzzaman, S.; Fazili, K.M. Fusobacterium nucleatum, inflammation, and immunity: The fire within human gut. Tumor Biol. 2016, 37, 2805–2810. [Google Scholar] [CrossRef] [PubMed]
- Michels, N.; Zouiouich, S.; Vanderbauwhede, B.; Vanacker, J.; Indave Ruiz, B.I.; Huybrechts, I. Human microbiome and metabolic health: An overview of systematic reviews. Obes. Rev. 2022, 23, e13409. [Google Scholar] [CrossRef] [PubMed]
- Satapathy, S.K.; Banerjee, P.; Pierre, J.F.; Higgins, D.; Dutta, S.; Heda, R.; Khan, S.D.; Mupparaju, V.K.; Mas, V.; Nair, S.; et al. Characterization of Gut Microbiome in Liver Transplant Recipients With Nonalcoholic Steatohepatitis. Transplant. Direct 2020, 6, e625. [Google Scholar] [CrossRef]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the Environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef]
- Yun, Y.; Kim, H.-N.; Lee, E.; Ryu, S.; Chang, Y.; Shin, H.; Kim, H.-L.; Kim, T.H.; Yoo, K.; Kim, H.Y. Fecal and blood microbiota profiles and presence of nonalcoholic fatty liver disease in obese versus lean subjects. PLoS ONE 2019, 14, e0213692. [Google Scholar] [CrossRef]
- Price, L.B.; Liu, C.M.; Melendez, J.H.; Frankel, Y.M.; Engelthaler, D.; Aziz, M.; Bowers, J.; Rattray, R.; Ravel, J.; Kingsley, C.; et al. Community Analysis of Chronic Wound Bacteria Using 16S rRNA Gene-Based Pyrosequencing: Impact of Diabetes and Antibiotics on Chronic Wound Microbiota. PLoS ONE 2009, 4, e6462. [Google Scholar] [CrossRef]
- Liu, Y.; Méric, G.; Havulinna, A.S.; Teo, S.M.; Åberg, F.; Ruuskanen, M.; Sanders, J.; Zhu, Q.; Tripathi, A.; Verspoor, K.; et al. Early prediction of incident liver disease using conventional risk factors and gut-microbiome-augmented gradient boosting. Cell Metab. 2022, 34, 719–730.e4. [Google Scholar] [CrossRef]
- Lee, G.; You, H.J.; Bajaj, J.S.; Joo, S.K.; Yu, J.; Park, S.; Kang, H.; Park, J.H.; Kim, J.H.; Lee, D.H.; et al. Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat. Commun. 2020, 11, 4982. [Google Scholar] [CrossRef]
- Wang, B.; Jiang, X.; Cao, M.; Ge, J.; Bao, Q.; Tang, L.; Chen, Y.; Li, L. Altered Fecal Microbiota Correlates with Liver Biochemistry in Nonobese Patients with Non-alcoholic Fatty Liver Disease. Sci. Rep. 2016, 6, 32002. [Google Scholar] [CrossRef]
- Sobhonslidsuk, A.; Chanprasertyothin, S.; Pongrujikorn, T.; Kaewduang, P.; Promson, K.; Petraksa, S.; Ongphiphadhanakul, B. The Association of Gut Microbiota with Nonalcoholic Steatohepatitis in Thais. Biomed Res. Int. 2018, 2018, 9340316. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; Lee, J.H.; Cho, M.S.; Kim, H.; Chun, J.; Lee, J.H.; Yoon, Y.; Kang, W. Characterization of Gut Microbiome in Korean Patients with Metabolic Associated Fatty Liver Disease. Nutrients 2021, 13, 1013. [Google Scholar] [CrossRef] [PubMed]
- Rau, M.; Rehman, A.; Dittrich, M.; Groen, A.K.; Hermanns, H.M.; Seyfried, F.; Beyersdorf, N.; Dandekar, T.; Rosenstiel, P.; Geier, A. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United Eur. Gastroenterol. J. 2018, 6, 1496–1507. [Google Scholar] [CrossRef]
- Jiang, W.; Wu, N.; Wang, X.; Chi, Y.; Zhang, Y.; Qiu, X.; Hu, Y.; Li, J.; Liu, Y. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci. Rep. 2015, 5, 8096. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Heuman, D.M.; Hylemon, P.B.; Sanyal, A.J.; White, M.B.; Monteith, P.; Noble, N.A.; Unser, A.B.; Daita, K.; Fisher, A.R.; et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 2014, 60, 940–947. [Google Scholar] [CrossRef]
- Zeng, Q.; Li, D.; He, Y.; Li, Y.; Yang, Z.; Zhao, X.; Liu, Y.; Wang, Y.; Sun, J.; Feng, X.; et al. Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci. Rep. 2019, 9, 13424. [Google Scholar] [CrossRef] [PubMed]
- Palmas, V.; Pisanu, S.; Madau, V.; Casula, E.; Deledda, A.; Cusano, R.; Uva, P.; Vascellari, S.; Loviselli, A.; Manzin, A.; et al. Gut microbiota markers associated with obesity and overweight in Italian adults. Sci. Rep. 2021, 11, 5532. [Google Scholar] [CrossRef] [PubMed]
- Kwan, S.; Jiao, J.; Joon, A.; Wei, P.; Petty, L.E.; Below, J.E.; Daniel, C.R.; Wu, X.; Zhang, J.; Jenq, R.R.; et al. Gut microbiome features associated with liver fibrosis in Hispanics, a population at high risk for fatty liver disease. Hepatology 2022, 75, 955–967. [Google Scholar] [CrossRef] [PubMed]
- Ottosson, F.; Brunkwall, L.; Ericson, U.; Nilsson, P.M.; Almgren, P.; Fernandez, C.; Melander, O.; Orho-Melander, M. Connection between BMI-Related Plasma Metabolite Profile and Gut Microbiota. J. Clin. Endocrinol. Metab. 2018, 103, 1491–1501. [Google Scholar] [CrossRef]
- Grigor’eva, I.N. Gallstone Disease, Obesity and the Firmicutes/Bacteroidetes Ratio as a Possible Biomarker of Gut Dysbiosis. J. Pers. Med. 2020, 11, 13. [Google Scholar] [CrossRef]
- Bai, J.; Wan, Z.; Zhang, Y.; Wang, T.; Xue, Y.; Peng, Q. Composition and diversity of gut microbiota in diabetic retinopathy. Front. Microbiol. 2022, 13, 926926. [Google Scholar] [CrossRef] [PubMed]
- Jaagura, M.; Viiard, E.; Karu-Lavits, K.; Adamberg, K. Low-carbohydrate high-fat weight reduction diet induces changes in human gut microbiota. Microbiologyopen 2021, 10, e1194. [Google Scholar] [CrossRef] [PubMed]
- Companys, J.; Gosalbes, M.J.; Pla-Pagà, L.; Calderón-Pérez, L.; Llauradó, E.; Pedret, A.; Valls, R.M.; Jiménez-Hernández, N.; Sandoval-Ramirez, B.A.; del Bas, J.M.; et al. Gut Microbiota Profile and Its Association with Clinical Variables and Dietary Intake in Overweight/Obese and Lean Subjects: A Cross-Sectional Study. Nutrients 2021, 13, 2032. [Google Scholar] [CrossRef] [PubMed]
- Burakova, I.; Smirnova, Y.; Gryaznova, M.; Syromyatnikov, M.; Chizhkov, P.; Popov, E.; Popov, V. The Effect of Short-Term Consumption of Lactic Acid Bacteria on the Gut Microbiota in Obese People. Nutrients 2022, 14, 3384. [Google Scholar] [CrossRef]
- Yeo, W.-Z.; Lim, S.-P.; Say, Y.-H. Salivary cariogenic bacteria counts are associated with obesity in student women at a Malaysian university. Asia Pac. J. Clin. Nutr. 2018, 27, 99–106. [Google Scholar] [CrossRef]
- Barkeling, B.; Linné, Y.; Lindroos, A.; Birkhed, D.; Rooth, P.; Rössner, S. Intake of sweet foods and counts of cariogenic microorganisms in relation to body mass index and psychometric variables in women. Int. J. Obes. 2002, 26, 1239–1244. [Google Scholar] [CrossRef][Green Version]
- Letchumanan, G.; Abdullah, N.; Marlini, M.; Baharom, N.; Lawley, B.; Omar, M.R.; Mohideen, F.B.S.; Addnan, F.H.; Nur Fariha, M.M.; Ismail, Z.; et al. Gut Microbiota Composition in Prediabetes and Newly Diagnosed Type 2 Diabetes: A Systematic Review of Observational Studies. Front. Cell. Infect. Microbiol. 2022, 12, 985. [Google Scholar] [CrossRef]
- Wu, X.; Park, S. Fecal Bacterial Community and Metagenome Function in Asians with Type 2 Diabetes, According to Enterotypes. Biomedicines 2022, 10, 2998. [Google Scholar] [CrossRef]
- Chumponsuk, T.; Gruneck, L.; Gentekaki, E.; Jitprasertwong, P.; Kullawong, N.; Nakayama, J.; Popluechai, S. The salivary microbiota of Thai adults with metabolic disorders and association with diet. Arch. Oral Biol. 2021, 122, 105036. [Google Scholar] [CrossRef]
- Huh, J.-W.; Roh, T.-Y. Opportunistic detection of Fusobacterium nucleatum as a marker for the early gut microbial dysbiosis. BMC Microbiol. 2020, 20, 208. [Google Scholar] [CrossRef]
- Elbere, I.; Silamikelis, I.; Dindune, I.I.; Kalnina, I.; Ustinova, M.; Zaharenko, L.; Silamikele, L.; Rovite, V.; Gudra, D.; Konrade, I.; et al. Baseline gut microbiome composition predicts metformin therapy short-term efficacy in newly diagnosed type 2 diabetes patients. PLoS ONE 2020, 15, e0241338. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ajami, N.J.; El-Serag, H.B.; Hair, C.; Graham, D.Y.; White, D.L.; Chen, L.; Wang, Z.; Plew, S.; Kramer, J.; et al. Dietary quality and the colonic mucosa–associated gut microbiome in humans. Am. J. Clin. Nutr. 2019, 110, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Limón, P.; Mena-Vázquez, N.; Moreno-Indias, I.; Manrique-Arija, S.; Lisbona-Montañez, J.M.; Cano-García, L.; Tinahones, F.J.; Fernández-Nebro, A. Collinsella is associated with cumulative inflammatory burden in an established rheumatoid arthritis cohort. Biomed. Pharmacother. 2022, 153, 113518. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Naito, Y.; Kashiwagi, S.; Uchiyama, K.; Mizushima, K.; Kamada, K.; Ishikawa, T.; Inoue, R.; Okuda, K.; Tsujimoto, Y.; et al. Changes in the Gut Microbiota are Associated with Hypertension, Hyperlipidemia, and Type 2 Diabetes Mellitus in Japanese Subjects. Nutrients 2020, 12, 2996. [Google Scholar] [CrossRef] [PubMed]
- Medawar, E.; Haange, S.-B.; Rolle-Kampczyk, U.; Engelmann, B.; Dietrich, A.; Thieleking, R.; Wiegank, C.; Fries, C.; Horstmann, A.; Villringer, A.; et al. Gut microbiota link dietary fiber intake and short-chain fatty acid metabolism with eating behavior. Transl. Psychiatry 2021, 11, 500. [Google Scholar] [CrossRef]
- Guo, X.; Dai, S.; Lou, J.; Ma, X.; Hu, X.; Tu, L.; Cui, J.; Lu, H.; Jiang, T.; Xu, J. Distribution characteristics of oral microbiota and its relationship with intestinal microbiota in patients with type 2 diabetes mellitus. Front. Endocrinol. 2023, 14, 1119201. [Google Scholar] [CrossRef]
- Quaglio, A.E.V.; Grillo, T.G.; De Oliveira, E.C.S.; Di Stasi, L.C.; Sassaki, L.Y. Gut microbiota, inflammatory bowel disease and colorectal cancer. World J. Gastroenterol. 2022, 28, 4053–4060. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.E.; Imai, K.; King, G.; Stuart, E.A. MatchIt: Nonparametric Preprocessing for Parametric Causal Inference. J. Stat. Softw. 2011, 42, 1–28. [Google Scholar] [CrossRef]
- Hittmeir, M.; Mayer, R.; Ekelhart, A. Utility and Privacy Assessment of Synthetic Microbiome Data. In Data and Applications Security and Privacy XXXVI; DBSec 2022. Lecture Notes in Computer Science; Sural, S., Lu, H., Eds.; Springer: Cham, Switzerland, 2022; Volume 13383, pp. 15–27. [Google Scholar] [CrossRef]
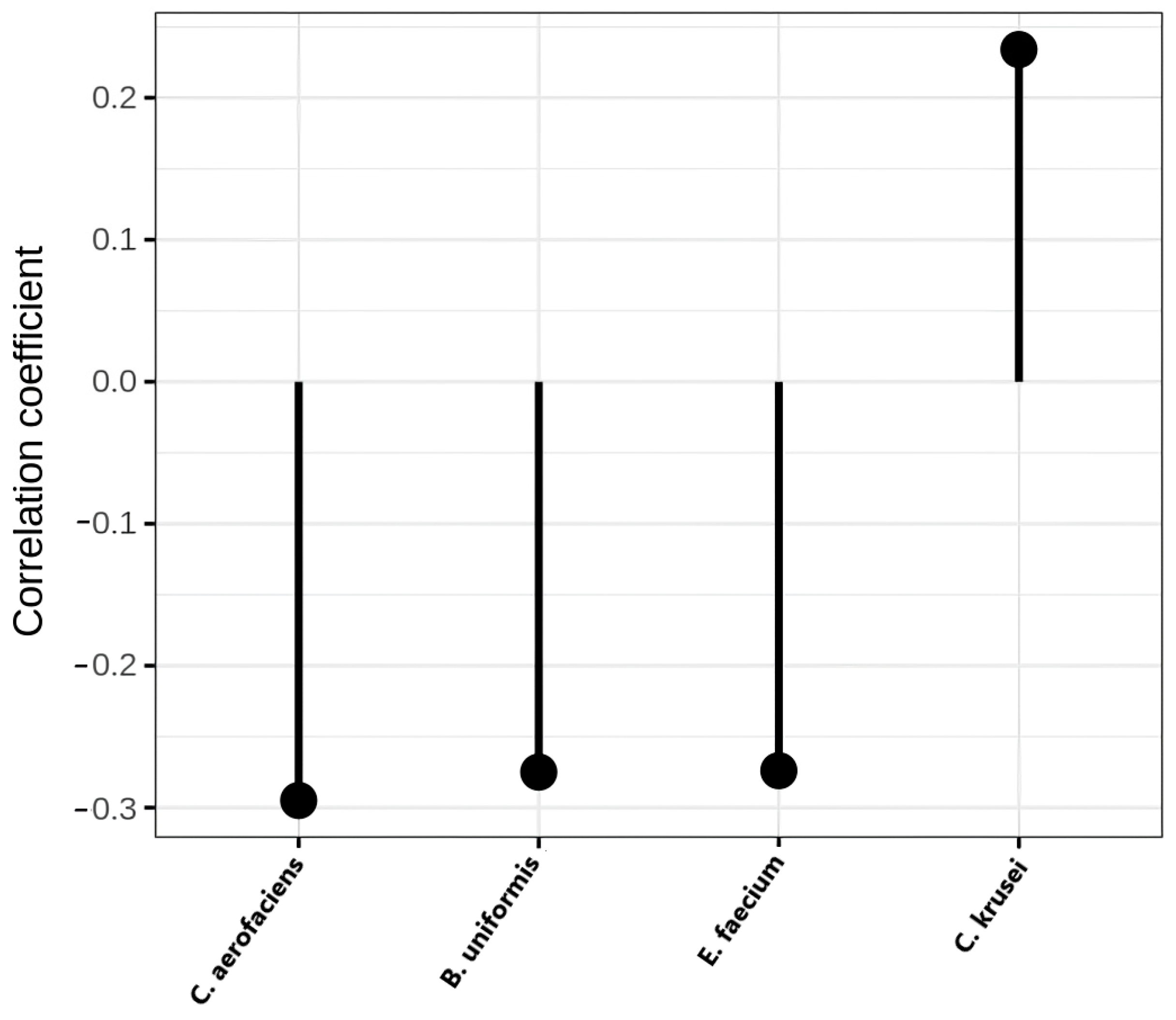





| List of Bacterial Species | Adjusted p-Value | Abundance |
|---|---|---|
| Bacteroides uniformis | 0.021 | Control |
| Enterococcus faecium | 0.018 | Control |
| Collinsella aerofaciens | 0.011 | Control |
| Candida krusei | 0.045 | NAFLD |
| Control | NAFLD | |
|---|---|---|
| Bacteroides sp.: Oscillibacter sp. | −0.45 | −0.52 |
| Blautia sp.: Clostridium symbiosum | 0.67 | 0.48 |
| Blautia sp.: Dorea sp. | 0.39 | 0.72 |
| Blautia sp.: Streptococcus sp. | 0.41 | 0.53 |
| Clostridium symbiosum: Dorea sp. | 0.31 | 0.63 |
| Fusobcaterium nucleatum: Veillonella sp. | 0.51 | 0.4 |
| Control | NAFLD | |
|---|---|---|
| Age: Coprococcus sp. | −0.3 | −0.59 |
| BMI: Enterococcus faecalis | 0.61 | 0.41 |
| BMI: Lactobacillaceae | 0.43 | 0.5 |
| Akkermansia muciniphila: Fusobacterium nucleatum | −0.5 | −0.39 |
| Bacteroides sp.: Odoribacter sp. | 0.77 | 0.53 |
| Bacteroides sp.: Parabacteroides sp. | 0.6 | 0.62 |
| Bifidobacterium sp.: Blautia sp. | 0.38 | 0.56 |
| Bifidobacterium sp.: Clostridium symbiosum | 0.72 | 0.3 |
| Bifidobacterium sp.: Coprococcus sp. | 0.6 | 0.52 |
| Bifidobacterium sp.: Roseburia sp. | 0.67 | 0.26 |
| Bifidobacterium sp.: Ruminococcus sp. | 0.5 | 0.37 |
| Blautia sp.: Roseburia sp. | 0.63 | 0.43 |
| Blautia sp. Collinsella sp. | 0.52 | 0.61 |
| Blautia sp.: Clostridium symbiosum | 0.51 | 0.47 |
| Blautia sp.: Ruminococcus sp. | 0.47 | 0.5 |
| Blautia sp.: Dorea sp. | 0.8 | 0.64 |
| Clostridium symbiosum: Faecalibacterium prausnitzii | 0.68 | 0.34 |
| Clostridium symbiosum: Fusobacterium nucleatum | −0.59 | −0.25 |
| Clostridium symbiosum: Oscillibacter sp. | 0.51 | 0.37 |
| Clostridium symbiosum: Roseburia sp. | 0.64 | 0.63 |
| Clostridium symbiosum: Ruminococcus sp. | 0.63 | 0.29 |
| Collinsella sp.: Streptococcus sp. | 0.7 | 0.42 |
| Coprococcus sp.: Clostridium symbiosum | 0.58 | 0.49 |
| Coprococcus sp.: Roseburia sp. | 0.63 | 0.42 |
| Dorea sp.: Clostridium symbiosum | 0.58 | 0.64 |
| Dorea sp.: Collinsella sp. | 0.5 | 0.76 |
| Dorea sp.: Coprococcus sp. | 0.5 | 0.59 |
| Dorea sp.: Odoribacter sp. | −0.29 | −0.5 |
| Dorea sp.: Roseburia sp. | 0.81 | 0.52 |
| Desulfovibrio sp.: Enterococcus faecalis | 0.52 | 0.45 |
| Enterococcus faecalis: Streptococcus sp. | 0.33 | 0.69 |
| Faecalibacterium prausnitzii: Roseburia sp. | 0.39 | 0.65 |
| Lactobacillaceae: Ruminococcus sp. | 0.3 | 0.77 |
| Lactobacillaceae: Streptococcus sp. | 0.71 | 0.54 |
| Odoribacter sp.: Parabacteroides sp. | 0.75 | 0.8 |
| Roseburia sp.: Streptococcus sp. | 0.39 | 0.58 |
| Ruminococcus sp.: Roseburia sp. | 0.52 | 0.5 |
| Ruminococcus sp.: Streptococcus sp. | 0.34 | 0.71 |
| Domain | Phylum | Class | Order | Family | Adjusted p-Value | Abundance |
|---|---|---|---|---|---|---|
| Bacteria | Firmicutes | Clostridia | Clostridiales | Gracilibacteraceae | 0.017 | NAFLD |
| Bacteria | Firmicutes | Negativicutes | Acidaminococcales | Acidaminococcaceae | 0.034 | control |
| Bacteria | Bacteroidetes | Chitinophagia | Chitinophagales | Chitinophagaceae | 0.038 | NAFLD |
| Bacteria | Planctomycetes | Planctomycetacia | Pirellulales | Pirellulaceae | 0.044 | NAFLD |
| Bacteria | Firmicutes | Erysipelotrichia | Erysipelotrichales | Erysipelatoclostridiaceae | 0.046 | NAFLD |
| Bacteria | Bacteroidetes | Bacteroidia | Bacteroidales | Muribaculaceae | 0.048 | NAFLD |
| Bacteria | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | 0.048 | NAFLD |
| Domain | Phylum | Class | Order | Family | Adjusted p-Value | Abundance |
|---|---|---|---|---|---|---|
| Bacteria | Firmicutes | Negativicutes | Acidaminococcales | Acidaminococcaceae | 0.035 | control |
| NGS 16S | qPCR | Culture | |
|---|---|---|---|
| Collinsella sp. | Enrichment in control (discovered in 38–48% of samples, content less than 0.5%) | Statistically significant taxon according to the Mann–Whitney method (PF group) | Enrichment in control (discovered in 38–48% of samples, content less than 0.5%) |
| Fusobacterium nucleatum | Not presented—discovered in 2 patients (content less than 0.05%) | Statistically significant taxon according to the Mann–Whitney method (FS group) An important taxon in XGBoost analysis: PF group—#7 FS group—#2 Enrichment in NAFLD | Not presented |
| Acidaminococcaceae | An important taxon in the latent Dirichlet distribution analysis (SILVA) An important taxon in Linear Discriminant Analysis (SILVA, RDP) Statistically significant taxon according to the Mann–Whitney method (SILVA, RDP) Enrichment in control | Not analyzed | Not presented—discovered only in 1 patient |
| Bifidobacteriaceae | An important taxon in the latent Dirichlet distribution analysis (SILVA) Enrichment in control | An important taxon in XGBoost analysis Bifidobacterium sp. (#4, PF group) Enrichment in NAFLD | Important taxa in XGBoost analysis: Bifidobacterium adolescentis (#1) Bifidobacterium longum (#2) An important taxon in Principal Component Analysis: Bifidobacterium adolescentis Enrichment in control |
| Enterococcaceae | An important taxon in Penalty Regression analysis (SILVA) An important taxon in Linear Discriminant Analysis (SILVA, RDP) Enrichment in control (Discovered in 17–21% of samples, content less than 0.5%) E. faecalis—discovered only in 3 patients (content less than 0.1%) | Statistically significant taxon according to the Mann–Whitney method (PF2020 group): E. faecalis Enrichment in NAFLD | Statistically significant taxon according to the Mann–Whitney method Enterococcus faecium An important taxon in XGBoost analysis: Enterococcus faecium (#5) Enrichment in control An important taxon in XGBoost analysis: Enterococcus faecalis (#4) Enrichment in control |
| Lachnospiraceae | An important taxon in the latent Dirichlet distribution analysis (SILVA, RDP) Enrichment in NAFLD | Important taxa in XGBoost analysis (PF group): Dorea sp. (#8) Roseburia sp. (#9) Statistically significant taxon according to the Mann–Whitney method Dorea sp. (FS group) All family representatives analyzed (Blautia sp., Dorea sp., Roseburia sp.) are enriched in NAFLD | Not presented—discovered only in 1 patient |
| Lactobacillaceae | An important taxon in penalty regression analysis (RDP) Enrichment in control | Statistically significant taxon according to the Mann–Whitney method (PF20 group) Enrichment in NAFLD | An important taxon in XGBoost analysis: Lactobacillus plantarum (#3) Lactobacillus delbrueckii (#6) An important taxon in Principal Component Analysis: Leuconostoc lactis Enrichment in control |
| Muribaculaceae | An important taxon in the latent Dirichlet distribution analysis (RDP) Statistically significant taxon according to the Mann–Whitney method (RDP) Enrichment in NAFLD | Not analyzed | Not presented |
| Oscillospiraceae | An important taxon in the latent Dirichlet distribution analysis (SILVA) Enrichment in control | Important taxa in XGBoost analysis (PF group): Ruminococcus sp. (#1) Faecalibacterium prausnitzii (#7) All family representatives analyzed (Faecalibacterium prausnitzii, Oscillibacter sp., Ruminococcus sp.) are enriched in control | Not presented |
| Ruminococcaceae | An important taxon in the latent Dirichlet distribution analysis (SILVA, RDP) An important taxon in penalty regression analysis (RDP) Enrichment in control (RDP), in NAFLD (SILVA) | An important taxon in XGBoost analysis: Ruminococcus sp. (#1, group PF) Enrichment in control | Not presented—discovered only in 1 patient |
| Veillonellaceae | An important taxon in the latent Dirichlet distribution analysis (RDP) Family—Enrichment in NAFLD Veillonella sp.—slight enrichment in control (content less than 0.5%) | Statistically significant taxon according to the Mann–Whitney method (group PF21) Veillonella sp. Enrichment in control | Not presented—discovered only in 3 patients |
| PF Control | FS Control | PF NAFLD | FS NAFLD | References | |
|---|---|---|---|---|---|
| Bacteroides sp.: Parabacteroides sp. | 0.35 | 0.6 | 0.46 | 0.62 | [46,47,48] |
| Blautia sp.: Clostridium symbiosum | 0.67 | 0.51 | 0.48 | 0.47 | - |
| Blautia sp.: Dorea sp. | 0.39 | 0.8 | 0.72 | 0.64 | [29,46,49,50] |
| Dorea sp.: Clostridium symbiosum | 0.31 | 0.58 | 0.63 | 0.64 | - |
| Dorea sp.: Collinsella sp. | 0.43 | 0.5 | 0.32 | 0.76 | [51,52,53] |
| Lactobacillaceae: Streptococcus sp. | 0.43 | 0.71 | 0.4 | 0.54 | [54,55,56] |
| Blautia sp: Collinsella sp. | 0.34 | 0.52 | - | ||
| Collinsella sp.: Streptococcus sp. | 0.34 | 0.7 | [57,58] | ||
| Fusobacterium nucleatum: Veillonella sp. | 0.51 | 0.49 | [59,60] | ||
| Odoribacter sp.: Parabacteroides sp. | 0.39 | 0.75 | [61,62] | ||
| Bifidobacterium sp.: Collinsella sp. | 0.35 | 0.78 | [24,58,63,64] | ||
| Blautia sp.: Streptococcus sp. | 0.53 | 0.52 | [54,65,66] |
| Control | NAFLD | |
|---|---|---|
| qPCR E. faecalis—Culture E. faecalis | 0.41 | 0.33 |
| qPCR Enterobacteriaceae—Culture Enterobacteriaceae | 0.38 | 0.48 |
| qPCR Enterobacteriaceae—qPCR E. faecalis | 0.43 | 0.08 |
| Culture Enterobacteriaceae—Culture E. faecalis | 0.35 | 0.32 |
| Parameter | NAFLD Patients (n = 155) | Controls (n = 44) |
|---|---|---|
| Sex: female, n (%) | 91 (58.7) | 34 (77.3) |
| Age, years, median (IR) | 52 (44–60) | 47 (36–51.8) |
| BMI, kg/m2, median (IR) | 30.5 (28.1–33.8) | 23.4 (21.5–26.4) |
| Normal weight (BMI < 25 kg/m2), n (%) | 11 (7.1) | 26 (59.1) |
| Overweight (25 ≤ BMI < 30 kg/m2), n (%) | 56 (36.1) | 17 (38.6) |
| Obesity (BMI ≥ 30 kg/m2), n (%) | 88 (56.8) | 1 (2.3) |
| Waist circumference, cm, median (IR) | 101 (93–109) | 79.5 (70.4–89.8) |
| Hip circumference, cm, median (IR) | 111.5 (105–120) | 99 (94.8–103.5) |
| Epicardial fat, mm, median (IR) | 11 (8–35) | 7.5 (4.3–9.8) |
| Platelet count, 109/L, median (IR) | 236 (202–278) | 233 (205–255) |
| FLI, median (IR) | 74 (60.8–91.3) | 11 (5.5–32.5) |
| ALT, IU/L, median (IR) | 24 (17–37) | 14 (10–24) |
| AST, IU/L, median (IR) | 21 (17–26) | 18 (15.5–23) |
| GGT, IU/L, median (IR) | 30 (20.3–49.8) | 17 (13–30) |
| Total bilirubin, mg/dL, median (IR) | 13 (10–18) | 11.6 (8.8–14.1) |
| Glucose, mmol/L, median (IR) | 5.7 (5.3–6.3) | 5.3 (5.03–5.8) |
| Total cholesterol, mg/dL, median (IR) | 5.4 (4.6–6.3) | 5.3 (4.5–6.5) |
| Triglycerides, mg/dL, median (IR) | 1.44 (1.03–2.04) | 0.92 (0.68–1.3) |
| Fibrinogen, g/L, median (IR) | 3.8 (3.4–4.2) | 3.5 (3.2–4.0) |
| CRP, mg/L, median (IR) | 1.8 (0.9–3.3) | 0.88 (0.43–1.7) |
| Creatinine, μmol/L, median (IR) | 75 (69–87) | 70 (63.3–79.8) |
| Uric acid, μmol/L, median (IR) | 5.8 (4.9–7.0) | 4.6 (4.0–5.9) |
| Insulin, μIU/mL, median (IR) | 11.7 (8.2–15.2) | 6.3 (4.9–8.1) |
| HOMA-IR > 2.7, n (%) | 86 (55.4) | 7 (15.9) |
| TyG, median (IR) | 4.7 (4.6–4.9) | 4.6 (4.4–4.8) |
| Fibrotest, median (IR) | 0.16 (0.10–0.22) | 0.12 (0.08–0.21) |
| Control Group (n = 38) | NAFLD Group (n = 38) | |
|---|---|---|
| Age, years Average ± SD [min; max] | 46 ± 8.6 [34; 65] | 50.3 ± 8.5 [34; 67] |
| BMI, kg/m2 Average ± SD [min; max] | 26.8 ± 3.1 [23.3; 33.9] | 27.3 ± 2.4 [23.6; 33.6] |
| Sex, male, n (%) | 46 | 50 |
| Group PF20 | Group FS20 | Group PF21 | Group PF21+ | |
|---|---|---|---|---|
| Collection year | 2020 | 2020 | 2021 | 2021 |
| Extraction method | PF | FS | PF | PF |
| Samples | NAFLD and Control | NAFLD and Control | NAFLD and Control | NAFLD |
| Peculiarities | - | - | - | Higher BMI, mostly women (see Table 12) |
| Control Group | NAFLD Group | ||||||
|---|---|---|---|---|---|---|---|
| Subgroup # | 1 (n = 23) | 2 (n = 14) | 3 (n = 15) | 1 (n = 19) | 2 (n = 14) | 3 (n = 19) | 4 |
| Age, years Average ± SD [min; max] | 46.2 ± 8.1 [32; 64] | 47.2 ± 11 [31; 64] | 45.3 ± 9.7 [30; 64] | 52.9 ± 7.6 [35; 65] | 51.8 ± 9.1 [36; 65] | 47.7 ± 8.7 [34; 62] | 50.3 ± 10.1 [27; 70] |
| BMI, kg/m2 Average ± SD [min; max] | 27.7 ± 3.2 [22.7; 33.2] | 24.6 ± 3.9 [19; 34] | 25.3 ± 2.1 [21.5; 28.8] | 28 ± 2.7 [22.5; 33.2] | 27.3 ± 3.4 [21; 32] | 26.7 ± 2 [23.5; 30.1] | 34.2 ± 4.1 [29.5; 46.1] |
| Sex, male, n (%) | 87 | 71.4 | 53.3 | 42.1 | 71.4 | 57.9 | 13.5 |
| Control Group (n = 29) | NAFLD Group (n = 29) | |
|---|---|---|
| Age, years Average ± SD [min; max] | 45.6 ± 8.1 [31; 64] | 51 ± 8.2 [34; 65] |
| BMI, kg/m2 Average ± SD [min; max] | 27.3 ± 2.9 [22.9; 33.2] | 27.5 ± 2.5 [22.5; 33.2] |
| Sex, male, n (%) | 79 | 48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korobeinikova, A.V.; Zlobovskaya, O.A.; Sheptulina, A.F.; Ashniev, G.A.; Bobrova, M.M.; Yafarova, A.A.; Akasheva, D.U.; Kabieva, S.S.; Bakoev, S.Y.; Zagaynova, A.V.; et al. Gut Microbiota Patterns in Patients with Non-Alcoholic Fatty Liver Disease: A Comprehensive Assessment Using Three Analysis Methods. Int. J. Mol. Sci. 2023, 24, 15272. https://doi.org/10.3390/ijms242015272
Korobeinikova AV, Zlobovskaya OA, Sheptulina AF, Ashniev GA, Bobrova MM, Yafarova AA, Akasheva DU, Kabieva SS, Bakoev SY, Zagaynova AV, et al. Gut Microbiota Patterns in Patients with Non-Alcoholic Fatty Liver Disease: A Comprehensive Assessment Using Three Analysis Methods. International Journal of Molecular Sciences. 2023; 24(20):15272. https://doi.org/10.3390/ijms242015272
Chicago/Turabian StyleKorobeinikova, Anna V., Olga A. Zlobovskaya, Anna F. Sheptulina, German A. Ashniev, Maria M. Bobrova, Adel A. Yafarova, Dariga U. Akasheva, Shuanat Sh. Kabieva, Siroj Yu. Bakoev, Anjelica V. Zagaynova, and et al. 2023. "Gut Microbiota Patterns in Patients with Non-Alcoholic Fatty Liver Disease: A Comprehensive Assessment Using Three Analysis Methods" International Journal of Molecular Sciences 24, no. 20: 15272. https://doi.org/10.3390/ijms242015272
APA StyleKorobeinikova, A. V., Zlobovskaya, O. A., Sheptulina, A. F., Ashniev, G. A., Bobrova, M. M., Yafarova, A. A., Akasheva, D. U., Kabieva, S. S., Bakoev, S. Y., Zagaynova, A. V., Lukashina, M. V., Abramov, I. A., Pokrovskaya, M. S., Doludin, Y. V., Tolkacheva, L. R., Kurnosov, A. S., Zyatenkova, E. V., Lavrenova, E. A., Efimova, I. A., ... Yudin, S. M. (2023). Gut Microbiota Patterns in Patients with Non-Alcoholic Fatty Liver Disease: A Comprehensive Assessment Using Three Analysis Methods. International Journal of Molecular Sciences, 24(20), 15272. https://doi.org/10.3390/ijms242015272







