MicroRNAs as Potential Biomarkers of Post-Traumatic Epileptogenesis: A Systematic Review
Abstract
1. Introduction
- (1)
- Immediate—arising in the first 24 h after a traumatic event;
- (2)
- Early—from 24 h to 1 week after the traumatic event;
- (3)
- Late—occurring more than 1 week after the traumatic event [3].
2. Methods
2.1. Inclusion Criteria
Study Material
2.2. Exclusion Criteria
3. Results and Discussion
3.1. Circulating miRNAs in Epilepsy
| Experimental Design | Sample | Sampling Time | MiRNAs | Significance | Year | Link | |
|---|---|---|---|---|---|---|---|
| AUC | p-Value | ||||||
| TLE/HV | CSF | miR-219↓ | <0.001 | 2014 | [20] | ||
| DR E/HV | Serum | miR-194-5p↑ miR-301a-3p↑ miR-30b-5p↑ miR-342-5p↑ miR-4446-3p↑ | 0.740 0.893 0.684 0.721 0.703 | 2015 | [30] | ||
| DR E/HV | Serum | miR-4521↑ | 0.718 | 2016 | [31] | ||
| DR E/HV | Plasma | miR-153↓ | 0.001 | 2016 | [32] | ||
| DR E/HV | Plasma, brain tissue | miR-129-2-3p↑ | 0.778 | 0.0008 | [33] | ||
| E/HV | Serum | miR-106b↑ miR-146a↑ miR-301a↑ miR-194-5p↓ | 0.786 0.774 0.686 0.696 | 2016 | [24] | ||
| E/HV | Serum | miR-378↑ miR-30a↑ (associated with the frequency of seizures) miR-106b↑ miR-15a↑ | <0.010 | 2016 | [25] | ||
| Seizure mTLE/mTLE | Serum | 30 min after seizure | miR-143↑ miR-145↑ miR-532↑ miR-365a↑ | <0.050 | 2016 | [15] | |
| DR E/HV | Plasma | miR-323a-5p↑ | 0.014 | 2017 | [34] | ||
| mTLE+HS/HV | Plasma | miR-3613-5p↑ miR-4668-5p↓ miR-8071↓ miR-197-5p↓ miR-4322↓ miR-6781-5p↓ | 0.844 0.789 0.932 0.802 0.714 0.781 | 2017 | [35] | ||
| DR-mTLE/mTLE/HV | Plasma | miR-134↓ | 0.671 | 2017 | [36] | ||
| TLE/SE/HV | CSF | TLE/SE: miR-19b-3p↑ miR-21-5p↑ miR-204-5p↑ miR-223-3p↑ miR-451a↑ miR-886- 3p↓ TLE/HV: miR-19b-3p↑ miR-21-5p↑ miR-204-5p↑ miR-223-3p↑ miR-451a↑ miR-886-3p↑ | 0.780 0.800 0.910 0.730 0.830 0.800 | 2017 | [22] | ||
| E/HV | Plasma | miR-153↓ | <0.010 | 2018 | [37] | ||
| E/HV | Serum | miR-155↑ | <0.010 | 2018 | [27] | ||
| E/E after seizure/HV | Plasma | 24 h | TLE/TLE after seizure: miR-27a-3p↓ miR-328-3p↑ miR-335-5p↓ TLE/HV: miR-27a-3p↓ miR-328-3p↑ miR-335-5p↓ | <0.010 <0.050 <0.050 <0.010 <0.010 <0.010 | 2018 | [14] | |
| DR E/mTLE/HV | Plasma | DR E/HV: miR-145-5p↓ mTLE/HV: miR-145-5p↓ | 0.632 0.829 | 2019 | [18] | ||
| mTLE+HS/HV | Serum | miR-145↑ miR-181c↑ miR199a↑ miR-1183↑ | 0.005 0.030 0.010 0.001 | 2019 | [19] | ||
| DR E/E | Serum | Seizures during 12 months/Without seizures during 12 months | miR-146a-5p↑ miR-134-5p↑ | 0.640 0.617 | <0.001 | 2020 | [38] |
| ChE/HV | Plasma | miR-106b↑ miR-146a↑ | 0.885 0.763 | 2019 | [26] | ||
| GGE/HV | Serum | miR-146a↑ miR-155↑ miR-132↓ | Average: 0.850 | 2020 | [28] | ||
| IE/HV | Plasma | miR-125a↓ miR-181a↓ | 0.815 0.704 | 0.001 0.001 | 2020 | [39] | |
| mTLE+HS(Engel I, Engel III-IV)/HV | Serum | E I/HV: miR-328-3p↑ miR-654-3p↑ E III-IV /HV: miR-328-3p↑ miR-654-3p↑ E I/E III-IV: miR-328-3p↑ miR-654-3p↑ | 0.001 0.004 <0.001 0.89 <0.001 0.190 | [40] | |||
| UEE/HV | Serum, CSF | Serum: miR-1275↑ CSF: miR-1275↓ | 2020 | [21] | |||
| TLE/HV | Serum | miR-142↑ miR-146a↑ miR-223↑ | <0.001 0.020 <0.001 | 2021 | [41] | ||
| Ch TLE/HV | Plasma | miR-194-5p↓ | 0.896 | 2021 | [42] | ||
| TLE/HV | Plasma | miR-146a↑ miR-132↑ | 0.808 0.791 | 2022 | [43] | ||
| ChE/HV | Plasma (sEVs) | miR-584-5p↑ miR-342-5p↑ miR-150-5p↑ | 0.846 0.835 0.826 | 2022 | [44] | ||
| ChE/HV | Serum | miR-155 | 0.813 | 2022 | [29] | ||
| ChE/HV | Serum | miR-324-5p↑ miR-146a-5p↓ miR-138-5p↓ miR-187-3p↓ | 2022 | [45] | |||
| E/HV | Serum | miR-378↓ miR-575↓ | <0.001 <0.001 | 2022 | [46] | ||
| mTLE+HS/HV | Serum | miR-629-3p↑ miR-1202↑ miR-1225-5p↑ | 2022 | [47] | |||
3.2. Circulating miRNAs inTraumatic Brain Injury
| Experimental Design | Sample | Sampling Time | MiRNAs | AUC | p-Value | Year | Link |
|---|---|---|---|---|---|---|---|
| sTBI/HV | Plasma | 24 h after trauma | miR-16↓ miR-92a↓ miR-765↑ | 0.890 0.820 0.860 | 2010 | [61] | |
| sTBI/OT | miR-16↓ miR-92a↓ miR-765↑ | 0.820 0.830 0.790 | |||||
| sTBI/HV | miR-16↓ miR-26a↑ miR-92a↓ miR-638↑ miR-765↑ | 0.820 0.730 0.780 0.510 0.680 | |||||
| TBI of varying severity/HV | Serum | 24 h after trauma–21 days after trauma | miR-93↑ miR-191↑ miR-499↑ | 1.000 0.727 0.801 | 2016 | [57] | |
| sTBI (coma)/HV | CSF | Coma during two weeks after trauma | miR-141↑ miR-572↑ miR-181a-star↑ miR-27b-star↑ miR-483-5p↑ miR-30b↑ miR-431-star↑ miR-193b-star↑ miR-499-3p↑ miR-1297↓ miR-33b↓ miR-933↓ miR-449b↓ | 0.005 0.002 0.001 0.012 0.003 0.030 0.006 0.021 0.002 0.013 0.016 0.009 0.042 | 2016 | [62] | |
| TBI/HV | Serum | 24 h after trauma | miR-155↓ | 0.726 | 2019 | [63] | |
| TBI of varying severit/OT/HV | Serum | 24–48 h after trauma | miR-151-5p↑ miR-195↑ miR-20a↑ miR-328↑ miR362-3p↑ miR-30d↑ miR-451↑ miR-486↑ miR-505*↑ miR-92a↑ | 0.660 0.810 0.780 0.730 0.790 0.750 0.820 0.810 0.820 0.860 | 2016 | [56] | |
| TBI+ hypopituitarism /HV | Serum | 1, 7, 28 days after trauma | miR-3907↑ miR126-3p↓ | < 0.050 | 2016 | [58] | |
| лЧМT/ЗД | Serum | 24 h–15 days after trauma | miR-425-5p↓ miR-21↑ miR-502↓ miR-335↑ | 1.000 0.961 1.000 0.990 | 2017 | [59] | |
| TBI of varying severity/HV | Saliva, CSF | miR-182-5p↓ miR-221-3p↓ mir-26b-5p↓ miR-320c↓ miR-29c-3p↑ miR-30e-5p↑ | Average 0.852 | 2018 | [64] | ||
| TBI of varying severity/HV | Plasma | miR-6867-5p ↑ miR-3665↑ miR-328-5p↑ miR-762↑ miR-3195↑ miR-4669↑ miR-2861↑ | 0.854 0.877 0.888 0.916 0.899 0.907 0.913 | 2018 | [65] | ||
| mTBI/HV | Saliva, Serum | miR-10b-5p↑ miR-30b-5p↑ miR-3678-3p↓ miR-455-5p↓ miR-5694↓ miR-6809-3p↓ miR-92a-3p↓ | Average 0.890 | 2019 | [66] | ||
| sTBI/HV | Serum | <24 h after trauma | miR-103a-3p↑ miR-219a-5p↑ miR-302d-3p↑ miR-422a↑ miR-518f-3p↑ miR-520d-3p↑ miR-627↑ | <0.050 | 2019 | [67] | |
| mTBI/HV | Serum | 24-48 h after trauma | miR-151-5p↑ miR-362-3p↑ miR-486↑ miR-505↑ miR-499↑ miR-625↑ miR-638↑ miR-381↑ | <0.050 | 2020 | [68] | |
| TBI/HV | Serum | <24 h after trauma | miR-124-3p↑ miR-219a-5p↑ miR-9-5p↑ miR-9-3p↑ miR-137↑ miR-128-3p↑ | 0.710 0.890 0.890 0.780 0.810 0.740 | 2020 | [69] | |
| Isolated TBI/HV | Plasma/Serum | 6 h after trauma | miR-423-3p↑ | 0.790 | 2020 | [70] | |
| TBI/HV | Serum | <24 h after trauma | miR-93↑ miR-191↑ | 0.712 0.660 | 2020 | [71] | |
| TBI/HV | Plasma | <24 h after trauma | miR-203b-5p↓ miR-203a-3p↓ miR-206↑ miR-185-5p↑ | 0.840 | 2020 | [72] | |
| modTBI/HV | Plasma | less than 1 day, 7, 28 days after trauma | miR-32-5p↑ | 0.700–0.900 | 2022 | [60] |
3.3. Circulating miRNAs in Ischemic Stroke
| Experimental Design | Sample | Sampling Time | MiRNAs | AUC | p-Value | Year | Link |
|---|---|---|---|---|---|---|---|
| IS/HV | Serum | 3, 4, 7 days after attack | miR-210↓ | 0.001 | 2011 | [81] | |
| IS/A/HV | Serum | miR-21↑ miR-221↓ | <0.0001 0.0002 | 2013 | [82] | ||
| IS/HV | Serum | 24 h, 1 week, 4 weeks, 24 weeks and 48 weeks after attack. | 24 h–24 weeks: miR-30a↓ miR-126↓ let-7b↑ (but let-7b ↓ for atherosclerosis of large vessels) | 0.910–0.930 0.920–0.940 0.920–0.930 | 2013 | [78] | |
| IS/HV | Serum | miR-145↑ | 2014 | [83] | |||
| IS/HV | Serum | 72 h after attack | miR-223↑ | <0.050 | 2014 | [84] | |
| IS/HV with vascular risk factors | Serum | >2 weeks after attack | miR-122↓ miR-148a↓ let-7i↓ miR-19a↓ miR-320d↓ miR-4429↓ miR-363↑ miR-487b↑ | 0.047 0.009 0.023 0.030 0.020 0.034 0.037 0.044 | 2014 | [85] | |
| ACI/HV | Plasma | miR-21↑ miR-24↓ | <0.050 | 2014 | [86] | ||
| AS/HV | Plasma | 0–24 h after attack | hsa-miR106b-5p↑ hsa-miR-4306↑ hsa-miR-320e↓ hsa-miR320d↓ | 0.962 0.877 0.981 0.987 | 2014 | [87] | |
| IS/HS | Plasma | miR-124-3p↓ miR-16↑ | 0.010 0.039 | 2014 | [88] | ||
| AcuteIS/HV | Serum | 24 h after attack | miR-124↓ miR-9↓ | 0.011 <0.001 | 2015 | [89] | |
| IS/HV | Serum | 24 h after attack | miR-32-3p↑ miR-106-5p↑ miR-532-5p↓ | <0.050 <0.010 <0.010 | 2015 | [90] | |
| AS/IS/HV | Serum | 24 h after attack | miR-145↑ miR-23a↓ miR-221↓ | 0.794 0.816 0.819 | <0.001 <0.001 <0.001 | 2015 | [91] |
| IS/HV | Serum | miR-15a↑ miR-16↑ miR-17-5p↑ | 0.698 0.820 0.784 | 2015 | [92] | ||
| IS/HV | Serum, CSF | let-7e↑ | 0.033 | 2015 | [93] | ||
| IS/HV | Serum | miR-9,-22,-23,-27,-125,-524↑ miR-9,-30,-33,-124,-135, -181,-197,-218,-330,-452↓ | <0.100 <0.150 | 2015 | [94] | ||
| IS/HV | Serum | miR-107↑ miR-128b↑ miR-153↑ | 0.970 0.903 0.893 | 2016 | [95] | ||
| PSCI/PSCN/HV | Serum | - | miR-132↑ | 0.961 | 2016 | [79] | |
| Fatal post-stroke/survivors | Plasma | <24 h after attack | miR-124-3p↑ miR-16↓ | 0.750 0.690 | 2016 | [80] | |
| IS/HV | Serum | <72 h after attack | let-7i↓ | <0.00001 | 2016 | [96] | |
| IS/HV TIA/HV | Plasma | <24 h after attack | miR-125a-5p↑ miR-125b-5p↑ miR-143-3p↑ | IS/HV 1.1 × 10−7 8.2 × 10−9, 2.4 × 10−9 TIS/HV 0.003 0.003 0.005 | 2017 | [97] | |
| IS/HV | Plasma | 1–3 days after attack 4–14 days after attack | miR-422a↑ miR-125b-2-3p↓ miR-422a↓ miR-125b-2-3p↓ | 0.769 0.971 0.889 | 2017 | [98] | |
| IS/HV | Serum | <24 h after attack | miR-221-3p↓ miR-382-5p↓ | 0.8106 0.7483 | 2017 | [99] | |
| IS/TIA/HV | Serum | <24 h after attack | IS: miR-23b-3p↑ miR-29b-3p↑ miR-181a-5p↑ miR-21-5p↑ TIS: miR-23b-3p↑ miR-29b-3p↑ miR-181a-5p↑ | <0.001 <0.001 0.027 <0.001 <0.001 0.001 0.007 | 2017 | [100] | |
| IS/HV | Serum | 1–3 days after attack | miR-146a↓ | 0.910 | 2017 | [101] | |
| IS/HV | CSF | <48 h after attack | miR-9-5p↑ miR-128-3p↑ | <0.050 | 2017 | [102] | |
| IS/HV | Serum | <48 h after attack | miR-4656,-432, -503↑ miR-874↓ | <0.050 | 2018 | [103] | |
| IS/HV | Serum | <24 h after attack | miR-146b↑ | 0.863 | 2018 | [104] | |
| IS/HV | Serum | <72 h after attack | miR-15a↑ | <0.050 | 2018 | [105] | |
| IS/HV | Serum | <24 h after attack | miR-148b-3p↓ miR-151b↑ miR-27b-3p↑ | 0.665 0.685 0.666 | 2018 | [106] | |
| IS/HV | Serum | <24 h after attack | miR-186-5p↓ miR-19a-3p↓ miR-32-5p↓ miR-340-5p↓ miR-579-3p↓ let-7e↑ miR-362-3p↑ miR-1238-5p↓ | <0.050 | 2019 | [107] | |
| Acute IS/HV | Serum | <24 h after attack | miR-124↓ | <0.001 | 2019 | [108] | |
| IS/HV | Serum | <24 h after attack | miR-146a↓ | <0.001 | 2019 | [109] | |
| IS/HV | Serum | miR-451↑ | 0.912 | 2019 | [110] | ||
| Acute IS/HV | Serum | <24 h after attack | miR-124↑ | 0.007 | 2020 | [111] | |
| IS/HV | Serum | miR-24↓ miR-29b↓ | 0.802 0.835 | 2020 | [112] | ||
| IS/HV | Serum | <24 h 14 days after attack | miR-135b↑ (24 h after attack) miR-135b↑ (14 days after attack) | 0.780 | 2020 | [113] | |
| IS/HV | Serum | <24 h after attack | miR-602↓ | 0.817 | [114] | ||
| IS/HV | Plasma | miR-155↑ | 0.851 | 2020 | [115] | ||
| IS/HV | Serum | miR-128↑ | <0.001 | 2020 | [116] | ||
| IS/HV | Serum | <72 h after attack | miR-124↓ | 0.953 | 2021 | [117] | |
| IS/HV | Serum | <72 h after attack | miR-148a↑ miR-342-3p↑ miR-19a↑ miR-320d↑ | 0.872 0.844 0.721 0.673 | 2021 | [118] | |
| Acute IS/HV | Serum | miR-9-5p↑ miR-128-3p↑ | 0.946 0.929 | 2021 | [119] | ||
| IS/HV | Serum | miR-659-5p↑ miR-151a-3p↑ miR-29c-5p↑ | 0.003 0.0007 0.0007 | 2022 | [120] | ||
| IS/A/HV | Plasma | IS <6 h after attack | miR-129-1-3p↑ miR-4312↑ miR-5196-3p(IS↑, A↓) | <0.001 | 2022 | [121] | |
| IS/HV | Serum | <24 h after attack | miR-451a↑ miR-574-5p↓ miR-142-3p↓ | 0.970 | 2022 | [122] | |
| IS/HV | Serum | miR-15b-5p↑ miR-184↑ miR-16-5p↑ | 0.581 0.607 0.719 | 2022 | [123] | ||
| IS/HV | Serum | miR-6089↓ | <0.050 | 2023 | [124] |
3.4. Discussions
3.4.1. miR-21
3.4.2. miR-181a
3.4.3. miR-155
3.4.4. Other Potential miRNA-Based Markers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fisher, R.S.; Acevedo, C.; Arzimanoglou, A.; Bogacz, A.; Cross, J.H.; Elger, C.E.; Engel, J., Jr.; Forsgren, L.; French, J.A.; Glynn, M.; et al. ILAE Official Report: A practical clinical definition of epilepsy. Epilepsia 2014, 55, 475–482. [Google Scholar] [CrossRef]
- Epilepsy. Available online: https://www.who.int/news-room/fact-sheets/detail/epilepsy (accessed on 28 July 2023).
- Pitkänen, A.; Immonen, R. Epilepsy Related to Traumatic Brain Injury. Neurotherapeutics 2014, 11, 286–296. [Google Scholar] [CrossRef]
- Golub, V.M.; Reddy, D.S. Post-Traumatic Epilepsy and Comorbidities: Advanced Models, Molecular Mechanisms, Biomarkers, and Novel Therapeutic Interventions. Pharmacol. Rev. 2022, 74, 387–438. [Google Scholar] [CrossRef] [PubMed]
- Dudek, F.E.; Staley, K.J. The Time Course and Circuit Mechanisms of Acquired Epileptogenesis. In Jasper’s Basic Mechanisms of the Epilepsies [Internet]; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2012; pp. 405–415. [Google Scholar] [CrossRef]
- Kadam, S.D.; White, A.M.; Staley, K.J.; Dudek, F.E. Continuous Electroencephalographic Monitoring with Radio-Telemetry in a Rat Model of Perinatal Hypoxia–Ischemia Reveals Progressive Post-Stroke Epilepsy. J. Neurosci. 2010, 30, 404. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.A.; White, A.M.; Clark, S.; Ferraro, D.J.; Swiercz, W.; Staley, K.J.; Dudek, F.E. Development of spontaneous recurrent seizures after kainate-induced status epilepticus. J. Neurosci. 2009, 29, 2103–2112. [Google Scholar] [CrossRef]
- Pitkänen, A.; Lukasiuk, K.; Dudek, F.E.; Staley, K.J. Epileptogenesis. Cold Spring Harb. Perspect. Med. 2015, 5, a022822. [Google Scholar] [CrossRef] [PubMed]
- Avakyan, G.N.; Blinov, D.V.; Alikhanov, A.A.; Perepelova, E.M.; Perepelov, V.A.; Burd, S.G.; Lebedeva, A.V. Recommendations of the Russian League Against Epilepsy (RLAE) on the use of magnetic resonance imaging in the diagnosis of epilepsy. Epilepsy Paroxysmal Cond. 2019, 11, 208–232. [Google Scholar] [CrossRef]
- BEST (Biomarkers, EndpointS, and Other Tools) Resource; Food and Drug Administration (US): Silver Spring, MD, USA; National Institutes of Health (US): Bethesda, MD, USA, 2016. Available online: https://www.ncbi.nlm.nih.gov/books/NBK326791/ (accessed on 28 July 2023).
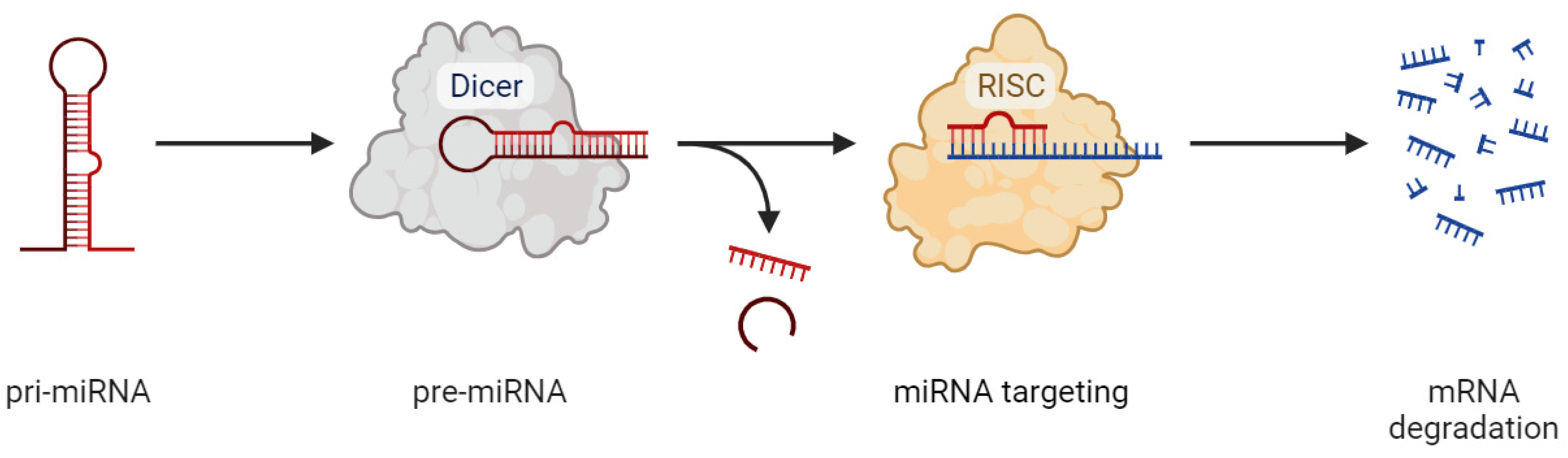
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 388354. [Google Scholar] [CrossRef]
- Simonato, M.; Agoston, D.V.; Brooks-Kayal, A.; Dulla, C.; Fureman, B.; Henshall, D.C.; Pitkänen, A.; Theodore, W.H.; Twyman, R.E.; Kobeissy, F.H.; et al. Identification of clinically relevant biomarkers of epileptogenesis—A strategic roadmap. Nat. Rev. Neurol. 2021, 17, 231–242. [Google Scholar] [CrossRef]
- Gallego, J.A.; Gordon, M.L.; Claycomb, K.; Bhatt, M.; Lencz, T.; Malhotra, A.K. In Vivo microRNA detection and quantitation in cerebrospinal fluid. J. Mol. Neurosci. 2012, 47, 243–248. [Google Scholar] [CrossRef]
- Raoof, R.; Bauer, S.; El Naggar, H.; Connolly, N.M.; Brennan, G.P.; Brindley, E.; Hill, T.; McArdle, H.; Spain, E.; Forster, R.J.; et al. Dual-center, dual-platform microRNA profiling identifies potential plasma biomarkers of adult temporal lobe epilepsy. EBioMedicine 2018, 38, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Surges, R.; Kretschmann, A.; Abnaof, K.; van Rikxoort, M.; Ridder, K.; Fröhlich, H.; Danis, B.; Kaminski, R.M.; Foerch, P.; Elger, C.E.; et al. Changes in serum miRNAs following generalized convulsive seizures in human mesial temporal lobe epilepsy. Biochem. Biophys. Res. Commun. 2016, 481, 13–18. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, J. MicroRNA Dysregulation in Epilepsy: From Pathogenetic Involvement to Diagnostic Biomarker and Therapeutic Agent Development. Front. Mol. Neurosci. 2021, 14, 650372. [Google Scholar] [CrossRef]
- Brennan, G.P.; Henshall, D.C. microRNAs in the pathophysiology of epilepsy. Neurosci. Lett. 2018, 667, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.-H.; Zhang, Y.-X.; Zheng, Y.; Yang, F.; Hu, Y.; Xu, S.; Yan, S.-Q.; Ding, Y.; Guo, Y.; Ding, M.-P. Expression of plasma microRNA-145-5p and its correlation with clinical features in patients with refractory epilepsy. Epilepsy Res. 2019, 154, 21–25. [Google Scholar] [CrossRef]
- Antônio, L.G.L.; Freitas-Lima, P.; Pereira-Da-Silva, G.; Assirati, J.A.; Matias, C.M.; Cirino, M.L.A.; Tirapelli, L.F.; Velasco, T.R.; Sakamoto, A.C.; Carlotti, C.G.; et al. Expression of MicroRNAs miR-145, miR-181c, miR-199a and miR-1183 in the Blood and Hippocampus of Patients with Mesial Temporal Lobe Epilepsy. J. Mol. Neurosci. 2019, 69, 580–587. [Google Scholar] [CrossRef]
- Zheng, H.; Tang, R.; Yao, Y.; Ji, Z.; Cao, Y.; Liu, Z.; Peng, F.; Wang, W.; Can, D.; Xing, H.; et al. MiR-219 Protects Against Seizure in the Kainic Acid Model of Epilepsy. Mol. Neurobiol. 2016, 53, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lu, C.; Wang, H.; Lin, Q.; Cai, L.; Meng, F.; Tesfaye, E.B.; Lai, H.-C.; Tzeng, C.-M. Identification of hsa-miR-1275 as a Novel Biomarker Targeting MECP2 for Human Epilepsy of Unknown Etiology. Mol. Ther. Methods Clin. Dev. 2020, 19, 398–410. [Google Scholar] [CrossRef]
- Raoof, R.; Jimenez-Mateos, E.M.; Bauer, S.; Tackenberg, B.; Rosenow, F.; Lang, J.; Onugoren, M.D.; Hamer, H.; Huchtemann, T.; Körtvélyessy, P.; et al. Cerebrospinal fluid microRNAs are potential biomarkers of temporal lobe epilepsy and status epilepticus. Sci. Rep. 2017, 7, 3328. [Google Scholar] [CrossRef]
- Coppola, A.; Moshé, S.L. Why is the developing brain more susceptible to status epilepticus? Epilepsia 2009, 50 (Suppl. S12), 25–26. [Google Scholar] [CrossRef]
- An, N.; Zhao, W.; Liu, Y.; Yang, X.; Chen, P. Elevated serum miR-106b and miR-146a in patients with focal and generalized epilepsy. Epilepsy Res. 2016, 127, 311–316. [Google Scholar] [CrossRef]
- Sun, J.; Cheng, W.; Liu, L.; Tao, S.; Xia, Z.; Qi, L.; Huang, M. Identification of serum miRNAs differentially expressed in human epilepsy at seizure onset and post-seizure. Mol. Med. Rep. 2016, 14, 5318–5324. [Google Scholar] [CrossRef] [PubMed]
- Elnady, H.G.; Abdelmoneam, N.; Eissa, E.; Hamid, E.R.A.; Abu Zeid, D.; Shanab, A.M.A.; Atta, H.; Kholoussi, N.M. MicroRNAs as Potential Biomarkers for Childhood Epilepsy. Open Access Maced. J. Med. Sci. 2019, 7, 3965–3969. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Chen, Y.; Wang, X. MicroRNA-155 contributes to the occurrence of epilepsy through the PI3K/Akt/mTOR signaling pathway. Int. J. Mol. Med. 2018, 42, 1577–1584. [Google Scholar] [CrossRef]
- Martins-Ferreira, R.; Chaves, J.; Carvalho, C.; Bettencourt, A.; Chorão, R.; Freitas, J.; Samões, R.; Boleixa, D.; Lopes, J.; Ramalheira, J.; et al. Circulating microRNAs as potential biomarkers for genetic generalized epilepsies: A three microRNA panel. Eur. J. Neurol. 2020, 27, 660–666. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, G.; Ding, Y.-Y.; Zhang, Y.-X. Expression of miR-155 in Serum Exosomes in Children with Epilepsy and Its Diagnostic Value. Dis. Markers 2022, 2022, 7979500. [Google Scholar] [CrossRef]
- Wang, J.; Tan, L.; Tan, L.; Tian, Y.; Ma, J.; Tan, C.-C.; Wang, H.-F.; Liu, Y.; Tan, M.-S.; Jiang, T.; et al. Circulating microRNAs are promising novel biomarkers for drug-resistant epilepsy. Sci. Rep. 2015, 5, 10201. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, Y.; Tan, Z.; Che, N.; Ji, A.; Luo, X.; Sun, X.; Li, X.; Yang, K.; Wang, G.; et al. Serum MicroRNA-4521 is a Potential Biomarker for Focal Cortical Dysplasia with Refractory Epilepsy. Neurochem. Res. 2016, 41, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, C.; Feng, P.; Jiang, Y.; Wang, W.; Zhou, D.; Chen, L. Aberrant expression of miR-153 is associated with overexpression of hypoxia-inducible factor-1α in refractory epilepsy. Sci. Rep. 2016, 6, 32091. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, X.; Wang, Z.; Zhang, Y.; Che, N.; Luo, X.; Tan, Z.; Sun, X.; Li, X.; Yang, K.; et al. Expression of microRNA-129-2-3p and microRNA-935 in plasma and brain tissue of human refractory epilepsy. Epilepsy Res. 2016, 127, 276–283. [Google Scholar] [CrossRef]
- Che, N.; Zu, G.; Zhou, T.; Wang, X.; Sun, Y.; Tan, Z.; Liu, Y.; Wang, D.; Luo, X.; Zhao, Z.; et al. Aberrant Expression of miR-323a-5p in Patients with Refractory Epilepsy Caused by Focal Cortical Dysplasia. Genet. Test. Mol. Biomarkers 2017, 21, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Zhang, H.; Xie, W.; Meng, F.; Zhang, K.; Jiang, Y.; Zhang, X.; Zhang, J. Altered microRNA profiles in plasma exosomes from mesial temporal lobe epilepsy with hippocampal sclerosis. Oncotarget 2017, 8, 4136–4146. [Google Scholar] [CrossRef] [PubMed]
- Avansini, S.H.; de Sousa Lima, B.P.; Secolin, R.; Santos, M.L.; Coan, A.C.; Vieira, A.S.; Torres, F.R.; Carvalho, B.S.; Alvim, M.K.M.; Morita, M.E.; et al. MicroRNA hsa-miR-134 is a circulating biomarker for mesial temporal lobe epilepsy. PLoS ONE 2017, 12, e0173060. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.-H.; An, F.-M.; Wang, Y.; Bian, M.; Wang, D.; Wei, C.-X. MiR-153 regulates expression of hypoxia-inducible factor-1α in refractory epilepsy. Oncotarget 2018, 9, 8542–8547. [Google Scholar] [CrossRef][Green Version]
- Leontariti, M.; Avgeris, M.; Katsarou, M.; Drakoulis, N.; Siatouni, A.; Verentzioti, A.; Alexoudi, A.; Fytraki, A.; Patrikelis, P.; Vassilacopoulou, D.; et al. Circulating miR-146a and miR-134 in predicting drug-resistant epilepsy in patients with focal impaired awareness seizures. Epilepsia 2020, 61, 959–970. [Google Scholar] [CrossRef]
- Raouf, H.A.; Kholoussi, N.M.; Eissa, E.; El Nady, H.G.; Fayed, D.B.; Abdelkawy, R.F.M. MicroRNAs as Immune Regulators of Inflammation in Children with Epilepsy. Int. J. Mol. Cell Med. 2020, 9, 188–197. [Google Scholar] [CrossRef]
- Ioriatti, E.S.; Cirino, M.L.A.; Neto, F.S.L.; Velasco, T.R.; Sakamoto, A.C.; Freitas-Lima, P.; Tirapelli, D.P.C.; Carlotti, C.G. Expression of circulating microRNAs as predictors of diagnosis and surgical outcome in patients with mesial temporal lobe epilepsy with hippocampal sclerosis. Epilepsy Res. 2020, 166, 106373. [Google Scholar] [CrossRef]
- De Benedittis, S.; Fortunato, F.; Cava, C.; Gallivanone, F.; Iaccino, E.; Caligiuri, M.E.; Castiglioni, I.; Bertoli, G.; Manna, I.; Labate, A.; et al. Circulating microRNA: The Potential Novel Diagnostic Biomarkers to Predict Drug Resistance in Temporal Lobe Epilepsy, a Pilot Study. Int. J. Mol. Sci. 2021, 22, 702. [Google Scholar] [CrossRef]
- Niu, X.; Zhu, H.-L.; Liu, Q.; Yan, J.-F.; Li, M.-L. MiR-194-5p serves as a potential biomarker and regulates the proliferation and apoptosis of hippocampus neuron in children with temporal lobe epilepsy. J. Chin. Med. Assoc. 2021, 84, 510–516. [Google Scholar] [CrossRef]
- Huang, H.; Cui, G.; Tang, H.; Kong, L.; Wang, X.; Cui, C.; Xiao, Q.; Ji, H. Relationships between plasma expression levels of microRNA-146a and microRNA-132 in epileptic patients and their cognitive, mental and psychological disorders. Bioengineered 2022, 13, 941–9492. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Chen, Y.; Hua, Y.; Xu, L.; Zhu, M.; Zhao, C.; Zhang, W.; Sheng, G.; Liu, L.; et al. Circulating MicroRNAs From Plasma Small Extracellular Vesicles as Potential Diagnostic Biomarkers in Pediatric Epilepsy and Drug-Resistant Epilepsy. Front. Mol. Neurosci. 2022, 15, 823802. [Google Scholar] [CrossRef] [PubMed]
- Ünalp, A.; Coskunpinar, E.; Gunduz, K.; Pekuz, S.; Baysal, B.T.; Edizer, S.; Hayretdag, C.; Gudeloglu, E. Detection of Deregulated miRNAs in Childhood Epileptic Encephalopathies. J. Mol. Neurosci. 2022, 72, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gao, Z.; Ma, M.L.; Li, L.; Guo, S. Identification of serum miR-378 and miR-575 as diagnostic indicators and predicting surgical prognosis in human epilepsy. J. Med. Biochem. 2022, 41, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Gattás, D.; Neto, F.; Freitas-Lima, P.; Bonfim-Silva, R.; de Almeida, S.M.; Cirino, M.d.A.; Tiezzi, D.G.; Tirapelli, L.; Velasco, T.; Sakamoto, A.; et al. MicroRNAs miR-629-3p, miR-1202 and miR-1225-5p as potential diagnostic and surgery outcome biomarkers for mesial temporal lobe epilepsy with hippocampal sclerosis. Neurochirurgie 2022, 68, 583–588. [Google Scholar] [CrossRef]
- Tubi, M.A.; Lutkenhoff, E.; Blanco, M.B.; McArthur, D.; Villablanca, P.; Ellingson, B.; Diaz-Arrastia, R.; Van Ness, P.; Real, C.; Shrestha, V.; et al. Early seizures and temporal lobe trauma predict post-traumatic epilepsy: A longitudinal study. Neurobiol. Dis. 2019, 123, 115. [Google Scholar] [CrossRef]
- Silva, L.F.A.; Hoffmann, M.S.; Gerbatin, R.d.R.; Fiorin, F.d.S.; Dobrachinski, F.; Mota, B.C.; Wouters, A.T.B.; Pavarini, S.P.; Soares, F.A.A.; Fighera, M.R.; et al. Treadmill exercise protects against pentylenetetrazol-induced seizures and oxidative stress after traumatic brain injury. J. Neurotrauma 2013, 30, 1278–1287. [Google Scholar] [CrossRef]
- Kori, P.; Garg, R.K.; Malhotra, H.S.; Gupta, R.K.; Verma, R.; Singh, M.K.; Rathore, R.K.S.; Gupta, P.K. Evaluation of cerebral white-matter micro-structural alterations in patients with medically refractory epilepsy using diffusion tensor tractography. Epilepsy Res. 2013, 107, 82–90. [Google Scholar] [CrossRef]
- Babb, T.; Kupfer, W.; Pretorius, J.; Crandall, P.; Levesque, M. Synaptic reorganization by mossy fibers in human epileptic fascia dentata. Neuroscience 1991, 42, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Cole-Edwards, K.K.; Musto, A.E.; Bazan, N.G. c-Jun N-terminal kinase activation responses induced by hippocampal kindling are mediated by reactive astrocytes. J. Neurosci. 2006, 26, 8295–8304. [Google Scholar] [CrossRef]
- Mishra, M.; Singh, R.; Mukherjee, S.; Sharma, D. Dehydroepiandrosterone’s antiepileptic action in FeCl3-induced epileptogenesis involves upregulation of glutamate transporters. Epilepsy Res. 2013, 106, 83–91. [Google Scholar] [CrossRef]
- Alyu, F.; Dikmen, M. Inflammatory aspects of epileptogenesis: Contribution of molecular inflammatory mechanisms. Acta Neuropsychiatr. 2017, 29, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Jeter, C.B.; Hergenroeder, G.W.; Hylin, M.J.; Redell, J.B.; Moore, A.N.; Dash, P.K.; Baugh, C.M.; Kroshus, E.; Kiernan, P.T.; Mendel, D.; et al. Biomarkers for the diagnosis and prognosis of mild traumatic brain injury/concussion. J. Neurotrauma 2013, 30, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Bhomia, M.; Balakathiresan, N.S.; Wang, K.K.; Papa, L.; Maheshwari, R.K. A Panel of Serum MiRNA Biomarkers for the Diagnosis of Severe to Mild Traumatic Brain Injury in Humans. Sci. Rep. 2016, 6, 28148. [Google Scholar] [CrossRef]
- Yang, T.; Song, J.; Bu, X.; Wang, C.; Wu, J.; Cai, J.; Wan, S.; Fan, C.; Zhang, C.; Wang, J. Elevated serum miR-93, miR-191, and miR-499 are noninvasive biomarkers for the presence and progression of traumatic brain injury. J. Neurochem. 2016, 137, 122–129. [Google Scholar] [CrossRef]
- Taheri, S.; Tanriverdi, F.; Zararsiz, G.; Elbuken, G.; Ulutabanca, H.; Karaca, Z.; Selcuklu, A.; Unluhizarci, K.; Tanriverdi, K.; Kelestimur, F.; et al. Circulating MicroRNAs as Potential Biomarkers for Traumatic Brain Injury-Induced Hypopituitarism. J. Neurotrauma 2016, 33, 1818–1825. [Google Scholar] [CrossRef]
- Di Pietro, V.; Ragusa, M.; Davies, D.; Su, Z.; Hazeldine, J.; Lazzarino, G.; Hill, L.J.; Crombie, N.; Foster, M.; Purrello, M.; et al. MicroRNAs as Novel Biomarkers for the Diagnosis and Prognosis of Mild and Severe Traumatic Brain Injury. J. Neurotrauma 2017, 34, 1948–1956. [Google Scholar] [CrossRef] [PubMed]
- Mitra, B.; Reyes, J.; O’Brien, W.T.; Surendran, N.; Carter, A.; Bain, J.; McEntaggart, L.; Sorich, E.; Shultz, S.R.; O’Brien, T.J.; et al. Micro-RNA levels and symptom profile after mild traumatic brain injury: A longitudinal cohort study. J. Clin. Neurosci. 2022, 95, 81–87. [Google Scholar] [CrossRef]
- Redell, J.B.; Moore, A.N.; Ward, N.H.; Hergenroeder, G.W.; Dash, P.K. Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma 2010, 27, 2147–2156. [Google Scholar] [CrossRef]
- You, W.-D.; Tang, Q.-L.; Wang, L.; Lei, J.; Feng, J.-F.; Mao, Q.; Gao, G.-Y.; Jiang, J.-Y. Alteration of microRNA expression in cerebrospinal fluid of unconscious patients after traumatic brain injury and a bioinformatic analysis of related single nucleotide polymorphisms. Chin. J. Traumatol. 2016, 19, 11–15. [Google Scholar] [CrossRef]
- Pan, P.; Bai, L.; Hua, X.; Wang, Y.; Jiang, X.; Cheng, X.; Song, Y.; Yu, X. miR-155 Regulates claudin1 Expression in Humans With Intestinal Mucosa Dysfunction After Brain Injury. Transplant. Proc. 2019, 51, 3474–3480. [Google Scholar] [CrossRef]
- Hicks, S.D.; Johnson, J.; Carney, M.C.; Bramley, H.; Olympia, R.P.; Loeffert, A.C.; Thomas, N.J. Overlapping MicroRNA Expression in Saliva and Cerebrospinal Fluid Accurately Identifies Pediatric Traumatic Brain Injury. J. Neurotrauma 2018, 35, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Li, L.; Lv, Q.; Shu, Q.; Zhang, Y.; Wang, Y. Expression profile of plasma microRNAs and their roles in diagnosis of mild to severe traumatic brain injury. PLoS ONE 2018, 13, e0204051. [Google Scholar] [CrossRef]
- LaRocca, D.; Barns, S.; Hicks, S.D.; Brindle, A.; Williams, J.; Uhlig, R.; Johnson, P.; Neville, C.; Middleton, F.A. Comparison of serum and saliva miRNAs for identification and characterization of mTBI in adult mixed martial arts fighters. PLoS ONE 2019, 14, e0207785. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Bu, X.; Li, Z.; Wu, J.; Wang, C.; Li, D.; Song, J.; Wang, J. Screening the expression of several miRNAs from TaqMan Low Density Array in traumatic brain injury: miR-219a-5p regulates neuronal apoptosis by modulating CCNA2 and CACUL1. J. Neurochem. 2019, 150, 202–217. [Google Scholar] [CrossRef] [PubMed]
- Polito, F.; Famà, F.; Oteri, R.; Raffa, G.; Vita, G.; Conti, A.; Daniele, S.; Macaione, V.; Passalacqua, M.; Cardali, S.; et al. Circulating miRNAs expression as potential biomarkers of mild traumatic brain injury. Mol. Biol. Rep. 2020, 47, 2941–2949. [Google Scholar] [CrossRef]
- O’connell, G.C.; Smothers, C.G.; Winkelman, C. Bioinformatic analysis of brain-specific miRNAs for identification of candidate traumatic brain injury blood biomarkers. Brain Inj. 2020, 34, 965–974. [Google Scholar] [CrossRef]
- Schindler, C.R.; Woschek, M.; Vollrath, J.T.; Kontradowitz, K.; Lustenberger, T.; Störmann, P.; Marzi, I.; Henrich, D. miR-142-3p Expression Is Predictive for Severe Traumatic Brain Injury (TBI) in Trauma Patients. Int. J. Mol. Sci. 2020, 21, 5381. [Google Scholar] [CrossRef] [PubMed]
- Tas, D.; Kaplan, O.; Sogut, O. Validity of Serum miRNA 93 and miRNA 191 to Reduce Unnecessary Computed Tomography in Patients with Mild Head Trauma. J. Clin. Med. Res. 2020, 12, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Hemphill, M.; Yang, Z.; Beard, K.; Sewell, E.; Shallcross, J.; Schweizer, M.; Sandsmark, D.K.; Diaz-Arrastia, R.; Kim, J.; et al. Multi-Dimensional Mapping of Brain-Derived Extracellular Vesicle MicroRNA Biomarker for Traumatic Brain Injury Diagnostics. J. Neurotrauma 2020, 37, 2424–2434. [Google Scholar] [CrossRef]
- Pitkänen, A.; Roivainen, R.; Lukasiuk, K. Development of epilepsy after ischaemic stroke. Lancet Neurol. 2016, 15, 185–197. [Google Scholar] [CrossRef]
- Tröscher, A.R.; Gruber, J.; Wagner, J.N.; Böhm, V.; Wahl, A.-S.; von Oertzen, T.J. Inflammation Mediated Epileptogenesis as Possible Mechanism Underlying Ischemic Post-stroke Epilepsy. Front. Aging Neurosci. 2021, 13, 781174. [Google Scholar] [CrossRef] [PubMed]
- Myint, P.K.; Staufenberg, E.F.A.; Sabanathan, K. Post-stroke seizure and post-stroke epilepsy. Postgrad. Med. J. 2006, 82, 568. [Google Scholar] [CrossRef] [PubMed]
- Phan, J.; Ramos, M.; Soares, T.; Parmar, M.S. Poststroke Seizure and Epilepsy: A Review of Incidence, Risk Factors, Diagnosis, Pathophysiology, and Pharmacological Therapies. Oxidative Med. Cell. Longev. 2022, 2022, 7692215. [Google Scholar] [CrossRef]
- Khoshnam, S.E.; Winlow, W.; Farbood, Y.; Moghaddam, H.F.; Farzaneh, M. Emerging Roles of microRNAs in Ischemic Stroke: As Possible Therapeutic Agents. J. Stroke 2017, 19, 166–187. [Google Scholar] [CrossRef] [PubMed]
- Long, G.; Wang, F.; Li, H.; Yin, Z.; Sandip, C.; Lou, Y.; Wang, Y.; Chen, C.; Wang, D.W. Circulating miR-30a, miR-126 and let-7b as biomarker for ischemic stroke in humans. BMC Neurol. 2013, 13, 178. [Google Scholar] [CrossRef]
- Huang, S.; Zhao, J.; Huang, D.; Zhuo, L.; Liao, S.; Jiang, Z. Serum miR-132 is a risk marker of post-stroke cognitive impairment. Neurosci. Lett. 2016, 615, 102–106. [Google Scholar] [CrossRef]
- Rainer, T.H.; Leung, L.Y.; Chan, C.P.Y.; Leung, Y.K.; Abrigo, J.M.; Wang, D.; Graham, C.A. Plasma miR-124-3p and miR-16 concentrations as prognostic markers in acute stroke. Clin. Biochem. 2016, 49, 663–668. [Google Scholar] [CrossRef]
- Zeng, L.; Liu, J.; Wang, Y.; Wang, L.; Weng, S.; Tang, Y.; Zheng, C.; Cheng, Q.; Chen, S.; Yang, G.-Y. MicroRNA-210 as a novel blood biomarker in acute cerebral ischemia. Front. Biosci. 2011, 3, 1265–1272. [Google Scholar] [CrossRef]
- Tsai, P.-C.; Liao, Y.-C.; Wang, Y.-S.; Lin, H.-F.; Lin, R.-T.; Juo, S.-H.H. Serum microRNA-21 and microRNA-221 as potential biomarkers for cerebrovascular disease. J. Vasc. Res. 2013, 50, 346–354. [Google Scholar] [CrossRef]
- Gan, C.; Wang, C.; Tan, K. Circulatory microRNA-145 expression is increased in cerebral ischemia. Genet. Mol. Res. 2012, 11, 147–152. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Huang, J.; Chen, X.; Gu, X.; Wang, Y.; Zeng, L.; Yang, G.-Y. Increase of circulating miR-223 and insulin-like growth factor-1 is associated with the pathogenesis of acute ischemic stroke in patients. BMC Neurol. 2014, 14, 77. [Google Scholar] [CrossRef] [PubMed]
- Jickling, G.C.; Ander, B.P.; Zhan, X.; Noblett, D.; Stamova, B.; Liu, D. microRNA expression in peripheral blood cells following acute ischemic stroke and their predicted gene targets. PLoS ONE 2014, 9, e99283. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, J. Identification of miRNA-21 and miRNA-24 in plasma as potential early stage markers of acute cerebral infarction. Mol. Med. Rep. 2014, 10, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Sun, G.; Zhang, L.; Shi, L.; Zeng, Y. Circulating microRNAs as novel potential biomarkers for early diagnosis of acute stroke in humans. J. Stroke Cerebrovasc. Dis. 2014, 23, 2607–2613. [Google Scholar] [CrossRef]
- Leung, L.Y.; Chan, C.P.Y.; Leung, Y.K.; Jiang, H.L.; Abrigo, J.M.; Wang, D.F.; Chung, J.S.H.; Rainer, T.H.; Graham, C.A. Comparison of miR-124-3p and miR-16 for early diagnosis of hemorrhagic and ischemic stroke. Clin. Chim. Acta 2014, 433, 139–144. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Han, R.; Liu, H.; Sun, D.; Liu, X. Downregulation of serum brain specific microRNA is associated with inflammation and infarct volume in acute ischemic stroke. J. Clin. Neurosci. 2015, 22, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Teng, F.; Gao, F.; Zhang, M.; Wu, J.; Zhang, C. Identification of circulating microRNAs as potential biomarkers for detecting acute ischemic stroke. Cell. Mol. Neurobiol. 2015, 35, 433–447. [Google Scholar] [CrossRef]
- Jia, L.; Hao, F.; Wang, W.; Qu, Y. Circulating miR-145 is associated with plasma high-sensitivity C-reactive protein in acute ischemic stroke patients. Cell Biochem. Funct. 2015, 33, 314–319. [Google Scholar] [CrossRef]
- Wu, J.; Du, K.; Lu, X. Elevated expressions of serum miR-15a, miR-16, and miR-17-5p are associated with acute ischemic stroke. Int. J. Clin. Exp. Med. 2015, 8, 21071. [Google Scholar]
- Peng, G.; Yuan, Y.; Wu, S.; He, F.; Hu, Y.; Luo, B. MicroRNA let-7e Is a Potential Circulating Biomarker of Acute Stage Ischemic Stroke. Transl. Stroke Res. 2015, 6, 437–445. [Google Scholar] [CrossRef]
- Zeng, Y.; Liu, J.-X.; Yan, Z.-P.; Yao, X.-H.; Liu, X.-H. Potential microRNA biomarkers for acute ischemic stroke. Int. J. Mol. Med. 2015, 36, 1639–1647. [Google Scholar] [CrossRef]
- Yang, Z.-B.; Li, T.-B.; Zhang, Z.; Ren, K.-D.; Zheng, Z.-F.; Peng, J.; Luo, X.-J. The Diagnostic Value of Circulating Brain-specific MicroRNAs for Ischemic Stroke. Intern. Med. 2016, 55, 1279–1286. [Google Scholar] [CrossRef]
- Jickling, G.C.; Ander, B.P.; Shroff, N.; Orantia, M.; Stamova, B.; Dykstra-Aiello, C.; Hull, H.; Zhan, X.; Liu, D.; Sharp, F.R. Leukocyte response is regulated by microRNA let7i in patients with acute ischemic stroke. Neurology 2016, 87, 2198–2205. [Google Scholar] [CrossRef]
- Tiedt, S.; Prestel, M.; Malik, R.; Schieferdecker, N.; Duering, M.; Kautzky, V.; Stoycheva, I.; Böck, J.; Northoff, B.H.; Klein, M.; et al. RNA-Seq Identifies Circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as Potential Biomarkers for Acute Ischemic Stroke. Circ. Res. 2017, 121, 970–980. [Google Scholar] [CrossRef]
- Li, D.-B.; Liu, J.-L.; Wang, W.; Li, R.-Y.; Yu, D.-J.; Lan, X.-Y.; Li, J.-P. Plasma Exosomal miR-422a and miR-125b-2-3p Serve as Biomarkers for Ischemic Stroke. Curr. Neurovascular Res. 2017, 14, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, Z.; Kan, P.; Zhang, B. The Diagnostic Value of Serum miRNA-221-3p, miRNA-382-5p, and miRNA-4271 in Ischemic Stroke. J. Stroke Cerebrovasc. Dis. 2017, 26, 1055–1060. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Fan, C.L.; Ma, L.J.; Liu, T.; Wang, C.; Song, J.-X.; Lv, Q.-S.; Pan, H.; Zhang, C.; Wu, J.; et al. Distinctive expression signatures of serum microRNAs in ischaemic stroke and transient ischaemic attack patients. Thromb. Haemost. 2017, 117, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-H.; Chen, L.; Pang, X.-M.; Su, S.-Y.; Zhou, X.; Chen, C.-Y.; Huang, L.-G.; Li, J.-P.; Liu, J.-L. Decreased miR-146a expression in acute ischemic stroke directly targets the Fbxl10 mRNA and is involved in modulating apoptosis. Neurochem. Int. 2017, 107, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, S.S.; Nygaard, A.-B.; Carlsen, A.L.; Heegaard, N.H.H.; Bak, M.; Christensen, T. Elevation of brain-enriched miRNAs in cerebrospinal fluid of patients with acute ischemic stroke. Biomark. Res. 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Bam, M.; Yang, X.; Sen, S.; Zumbrun, E.E.; Dennis, L.; Zhang, J.; Nagarkatti, P.S.; Nagarkatti, M. Characterization of Dysregulated miRNA in Peripheral Blood Mononuclear Cells from Ischemic Stroke Patients. Mol. Neurobiol. 2018, 55, 1419–1429. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, K.; Huang, J.; Zheng, G.; Lv, Y.; Luo, N.; Liang, M.; Huang, L. Upregulated Serum MiR-146b Serves as a Biomarker for Acute Ischemic Stroke. Cell. Physiol. Biochem. 2018, 45, 397–405. [Google Scholar] [CrossRef]
- Lu, W.-J.; Zeng, L.-L.; Wang, Y.; Zhang, Y.; Liang, H.-B.; Tu, X.-Q.; He, J.-R.; Yang, G.-Y. Blood microRNA-15a Correlates with IL-6, IGF-1 and Acute Cerebral Ischemia. Curr. Neurovascular Res. 2018, 15, 63–71. [Google Scholar] [CrossRef]
- Cheng, X.; Kan, P.; Ma, Z.; Wang, Y.; Song, W.; Huang, C.; Zhang, B. Exploring the potential value of miR-148b-3p, miR-151b and miR-27b-3p as biomarkers in acute ischemic stroke. Biosci. Rep. 2018, 38, 20181033. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.; Kim, J.; Lee, A.-R.; Lee, J.-M.; Kim, O.-J.; Kim, J.-K.; Oh, S.-H. Alteration of microRNA 340-5p and Arginase-1 Expression in Peripheral Blood Cells during Acute Ischemic Stroke. Mol. Neurobiol. 2019, 56, 3211–3221. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Hou, X.; Ren, G.; Zhang, Y.; Cheng, H. Dynamic changes in miR-124 levels in patients with acute cerebral infarction. Int. J. Neurosci. 2019, 129, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Kotb, H.G.; Ibrahim, A.H.; Mohamed, E.F.; Ali, O.M.; Hassanein, N.; Badawy, D.; Aly, E.A. The expression of microRNA 146a in patients with ischemic stroke: An observational study. Int. J. Gen. Med. 2019, 12, 273–278. [Google Scholar] [CrossRef]
- Liu, G.; Cao, C.; Zhu, M. Peripheral Blood miR-451 May Serve as a Biomarker of Ischemic Stroke. Clin. Lab. 2019, 65, 1713–1718. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Q.; Ding, J.; Wang, X. Elevated serum levels of brain-derived neurotrophic factor and miR-124 in acute ischemic stroke patients and the molecular mechanism. 3 Biotech 2019, 9, 386. [Google Scholar] [CrossRef]
- Guo, C.; Zhong, C.; Li, Q.; Gao, Y.; Li, W.; Ou, Y. Expressions and neural function prognostic evaluation of serum microRNA-24 and microRNA-29b in elderly patients with acute ischemic stroke. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 2020, 32, 78–82. [Google Scholar] [CrossRef]
- Yang, S.; Zhan, X.; He, M.; Wang, J.; Qiu, X. miR-135b levels in the peripheral blood serve as a marker associated with acute ischemic stroke. Exp. Ther. Med. 2020, 19, 3551–3558. [Google Scholar] [CrossRef]
- Rahmati, M.; Azarpazhooh, M.R.; Ehteram, H.; Ferns, G.A.; Ghayour-Mobarhan, M.; Ghannadan, H.; Mobarra, N. The elevation of S100B and downregulation of circulating miR-602 in the sera of ischemic stroke (IS) patients: The emergence of novel diagnostic and prognostic markers. Neurol. Sci. 2020, 41, 2185–2192. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, G.; Qiu, W.; Pan, Q.; Chen, Y.; Chen, Y.; Ma, X. Plasma endothelial microvesicles and their carrying miRNA-155 serve as biomarkers for ischemic stroke. J. Neurosci. Res. 2020, 98, 2290–2301. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, Y.; Zhao, J.; Mou, L. Expression and clinical value of miR-128 and IGF-1 in patients with acute ischemic stroke. Minerva Medica 2020, 111, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Qi, L. miR-124 Is Downregulated in Serum of Acute Cerebral Infarct Patients and Shows Diagnostic and Prognostic Value. Clin. Appl. Thromb./Hemost. 2021, 27, 10760296211035446. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, X.; Wu, X.; Zhu, L.; Long, J.; Su, L.; Gu, L. Machine Learning Analysis of MicroRNA Expression Data Reveals Novel Diagnostic Biomarker for Ischemic Stroke. J. Stroke Cerebrovasc. Dis. 2021, 30, 105825. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, F.; Fu, F.; Liu, J.; Sun, W.; Chen, Y. Diagnostic and prognostic value of serum miR-9-5p and miR-128-3p levels in early-stage acute ischemic stroke. Clinics 2021, 76, e2958. [Google Scholar] [CrossRef]
- Li, K.; Shen, L.; Zheng, P.; Wang, Y.; Wang, L.; Meng, X.; Lv, Y.; Xue, Z.; Guo, X.; Zhang, A.; et al. IIdentification of MicroRNAs as potential biomarkers for detecting ischemic stroke. Genes Genom. 2022, 44, 9–17. [Google Scholar] [CrossRef]
- Zhou, B.; Li, B.; Feng, P.; Wang, X.; Gao, H.; Xu, L.; Wang, T.; Guo, X. Identification of a miRNA biomarker for the large artery atherosclerosis subtype of acute ischemic stroke. Folia Neuropathol. 2022, 60, 210–220. [Google Scholar] [CrossRef]
- Aldous, E.K.; Toor, S.M.; Parray, A.; Al-Sarraj, Y.; Diboun, I.; Abdelalim, E.M.; Arredouani, A.; El-Agnaf, O.; Thornalley, P.J.; Akhtar, N.; et al. Identification of Novel Circulating miRNAs in Patients with Acute Ischemic Stroke. Int. J. Mol. Sci. 2022, 23, 3387. [Google Scholar] [CrossRef]
- SJiang, S.; Wu, J.; Geng, Y.; Zhang, Y.; Wang, Y.; Wu, J.; Lu, C.; Luo, G.; Zan, J.; Zhang, Y. Identification of Differentially Expressed microRNAs Associated with Ischemic Stroke by Integrated Bioinformatics Approaches. Int. J. Genom. 2022, 2022, 9264555. [Google Scholar] [CrossRef]
- Zhang, J.; Shen, Y.; Kang, K.; Lin, J.; Wang, A.; Li, S.; Wu, S.; Zhao, X.; Zhang, Q. Circulating MicroRNAs as Potential Biomarkers for Ischemic Stroke in Patients with Asymptomatic Intracranial Artery Stenosis. Cell. Mol. Neurobiol. 2023, 43, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef]
- Sheedy, F.J. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front. Immunol. 2015, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The Role of miR-21 in Cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Kura, B.; Kalocayova, B.; Devaux, Y.; Bartekova, M. Potential Clinical Implications of miR-1 and miR-21 in Heart Disease and Cardioprotection. Int. J. Mol. Sci. 2020, 21, 700. [Google Scholar] [CrossRef]
- Chen, C.-Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.-B.; Lu, Y.; Yue, S.; Xu, L.-J.; Xiong, X.-X.; White, R.E.; Sun, X.; Giffard, R.G. miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol. Dis. 2012, 45, 555–563. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, Z.; Yang, F.; Gao, Y.; Song, J.; Wan, Y. microRNA-181a is involved in insulin-like growth factor-1-mediated regulation of the transcription factor CREB1. J. Neurochem. 2013, 126, 771–780. [Google Scholar] [CrossRef]
- Ouyang, Y.-B.; Lu, Y.; Yue, S.; Giffard, R.G. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion 2012, 12, 213–219. [Google Scholar] [CrossRef]
- Mahesh, G.; Biswas, R. MicroRNA-155: A Master Regulator of Inflammation. J. Interf. Cytokine Res. 2019, 39, 321–330. [Google Scholar] [CrossRef]
- Hu, J.; Huang, S.; Liu, X.; Zhang, Y.; Wei, S.; Hu, X. miR-155: An Important Role in Inflammation Response. J. Immunol. Res. 2022, 2022, 7437281. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Wu, L.; Yuan, H. miR-30b-5p targeting GRIN2A inhibits hippocampal damage in epilepsy. Open Med. 2023, 18, 20230675. [Google Scholar] [CrossRef] [PubMed]
- Fu, N.; Yu, J.; Zhu, L.; Yang, L.; Ma, L.; He, J.; Yu, H.; Liu, J.; Tian, Y.; Xu, J. Role of miR-219a-5p in regulating NMDAR in nonylphenol-induced synaptic plasticity damage. Ecotoxicol. Environ. Saf. 2023, 252, 114576. [Google Scholar] [CrossRef] [PubMed]
- Heiland, M.; Connolly, N.M.C.; Mamad, O.; Nguyen, N.T.; Kesavan, J.C.; Langa, E.; Fanning, K.; Sanfeliu, A.; Yan, Y.; Su, J.; et al. MicroRNA-335-5p suppresses voltage-gated sodium channel expression and may be a target for seizure control. Proc. Natl. Acad. Sci. USA 2023, 120, e2216658120. [Google Scholar] [CrossRef]
- Pala, M.; Meral, I.; Acikgoz, N.P.; Yilmaz, S.G.; Okur, S.K.; Acar, S.; Polat, Y.; Akbas, F. Downregulatory effect of miR-342-3p on epileptogenesis in the PTZ-kindling model. Mol. Biol. Rep. 2022, 49, 11997–12006. [Google Scholar] [CrossRef]
- Wang, Z.; Xie, W.; Guan, H. The diagnostic, prognostic role and molecular mechanism of miR-328 in human cancer. Biomed. Pharmacother. 2023, 157, 114031. [Google Scholar] [CrossRef]
- Jiang, L.-H.; Zhang, H.-D.; Tang, J.-H. MiR-30a: A Novel Biomarker and Potential Therapeutic Target for Cancer. J. Oncol. 2018, 2018, 5167829. [Google Scholar] [CrossRef]
- Wang, T.; Li, F.; Tang, S. MiR-30a upregulates BCL2A1, IER3 and cyclin D2 expression by targeting FOXL2. Oncol. Lett. 2015, 9, 967–971. [Google Scholar] [CrossRef]
- Liu, T.; Xu, Z.; Ou, D.; Liu, J.; Zhang, J. The miR-15a/16 gene cluster in human cancer: A systematic review. J. Cell. Physiol. 2019, 234, 5496–5506. [Google Scholar] [CrossRef]
- Yin, H.; Sun, Y.; Wang, X.; Park, J.; Zhang, Y.; Li, M.; Yin, J.; Liu, Q.; Wei, M. Progress on the relationship between miR-125 family and tumorigenesis. Exp. Cell Res. 2015, 339, 252–260. [Google Scholar] [CrossRef]
- Li, S.-H.; Su, S.-Y.; Liu, J.-L. Differential Regulation of microRNAs in Patients with Ischemic Stroke. Curr. Neurovascular Res. 2015, 12, 214–221. [Google Scholar] [CrossRef]
- Climent, M.; Quintavalle, M.; Miragoli, M.; Chen, J.; Condorelli, G.; Elia, L. TGFβ triggers miR-143/145 transfer from smooth muscle cells to endothelial cells, thereby modulating vessel stabilization. Circ. Res. 2015, 116, 1753–1764. [Google Scholar] [CrossRef] [PubMed]
- Harraz, M.M.; Eacker, S.M.; Wang, X.; Dawson, T.M.; Dawson, V.L. MicroRNA-223 is neuroprotective by targeting glutamate receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 18962–18967. [Google Scholar] [CrossRef] [PubMed]
- Henshall, D.C.; Araki, T.; Schindler, C.K.; Lan, J.-Q.; Tiekoter, K.L.; Taki, W.; Simon, R.P. Activation of Bcl-2-Associated Death Protein and Counter-Response of Akt within Cell Populations during Seizure-Induced Neuronal Death. J. Neurosci. 2002, 22, 8458–8465. [Google Scholar] [CrossRef] [PubMed]
- Aronica, E.; Gorter, J.; Jansen, G.; van Veelen, C.; van Rijen, P.; Leenstra, S.; Ramkema, M.; Scheffer, G.; Scheper, R.; Troost, D. Expression and cellular distribution of multidrug transporter proteins in two major causes of medically intractable epilepsy: Focal cortical dysplasia and glioneuronal tumors. Neuroscience 2003, 118, 417–429. [Google Scholar] [CrossRef]
- Suzuki, A.; Fukushima, H.; Mukawa, T.; Toyoda, H.; Wu, L.-J.; Zhao, M.-G.; Xu, H.; Shang, Y.; Endoh, K.; Iwamoto, T.; et al. Upregulation of CREB-Mediated Transcription Enhances Both Short- and Long-Term Memory. J. Neurosci. 2011, 31, 8786–8802. [Google Scholar] [CrossRef]

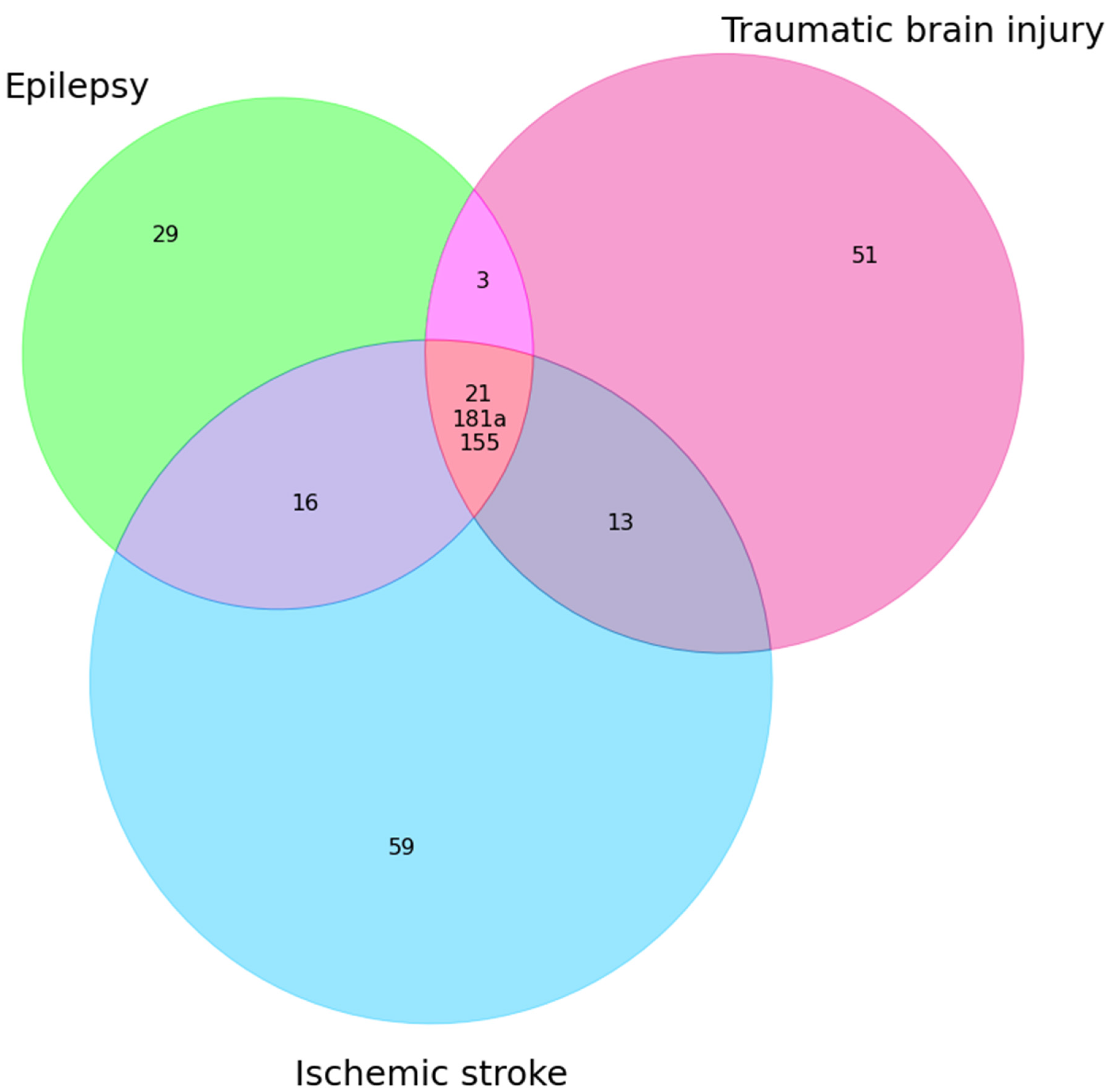
| Epilepsy + TBI | Epilepsy + IS | Epilepsy + TBI + IS |
|---|---|---|
| miR-30b↑ miR-21(E↓TBI↑) miR-328-3p↑ miR-335(E↓ TBI ↑) miR-155 (E↑ TBI↓) miR-181a(E↓ TBI↑) | miR-342(E—3p,IS—5p)↑ miR-153 (E↓ IS↑) miR-106b↑ miR-146a(E↑ AIS↓) miR-30a (E↑ IS↓) miR-15a(E↑ IS↓) miR-143↑ miR-145(E↑↓ IS↓) miR-21(E↓ IS ↑↓) miR-223↑ miR-451a ↑ miR-155↑ miR-132 miR-125a(E↓ IS↑) miR-181a(E↓ IS↑) miR-142 (E↑ IS↓) | miR-21 miR-181a miR-155 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasilieva, A.A.; Timechko, E.E.; Lysova, K.D.; Paramonova, A.I.; Yakimov, A.M.; Kantimirova, E.A.; Dmitrenko, D.V. MicroRNAs as Potential Biomarkers of Post-Traumatic Epileptogenesis: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 15366. https://doi.org/10.3390/ijms242015366
Vasilieva AA, Timechko EE, Lysova KD, Paramonova AI, Yakimov AM, Kantimirova EA, Dmitrenko DV. MicroRNAs as Potential Biomarkers of Post-Traumatic Epileptogenesis: A Systematic Review. International Journal of Molecular Sciences. 2023; 24(20):15366. https://doi.org/10.3390/ijms242015366
Chicago/Turabian StyleVasilieva, Anastasia A., Elena E. Timechko, Kristina D. Lysova, Anastasia I. Paramonova, Alexey M. Yakimov, Elena A. Kantimirova, and Diana V. Dmitrenko. 2023. "MicroRNAs as Potential Biomarkers of Post-Traumatic Epileptogenesis: A Systematic Review" International Journal of Molecular Sciences 24, no. 20: 15366. https://doi.org/10.3390/ijms242015366
APA StyleVasilieva, A. A., Timechko, E. E., Lysova, K. D., Paramonova, A. I., Yakimov, A. M., Kantimirova, E. A., & Dmitrenko, D. V. (2023). MicroRNAs as Potential Biomarkers of Post-Traumatic Epileptogenesis: A Systematic Review. International Journal of Molecular Sciences, 24(20), 15366. https://doi.org/10.3390/ijms242015366