Non-Steroidal Estrogens Inhibit Influenza Virus by Interacting with Hemagglutinin and Preventing Viral Fusion
Abstract
:1. Introduction
2. Results
2.1. DES Is the Top Hit of a Screening of the Prestwick Library against Influenza A Virus
2.2. Dienestol, Hexestrol, and Bakuchiol Show Comparable Antiviral Activity
2.3. The Activity of DES Is Maintained against Multiple Laboratory and Clinical Strains
2.4. DES Is Active in Different Cell Lines and Human-Derived Respiratory Tissues
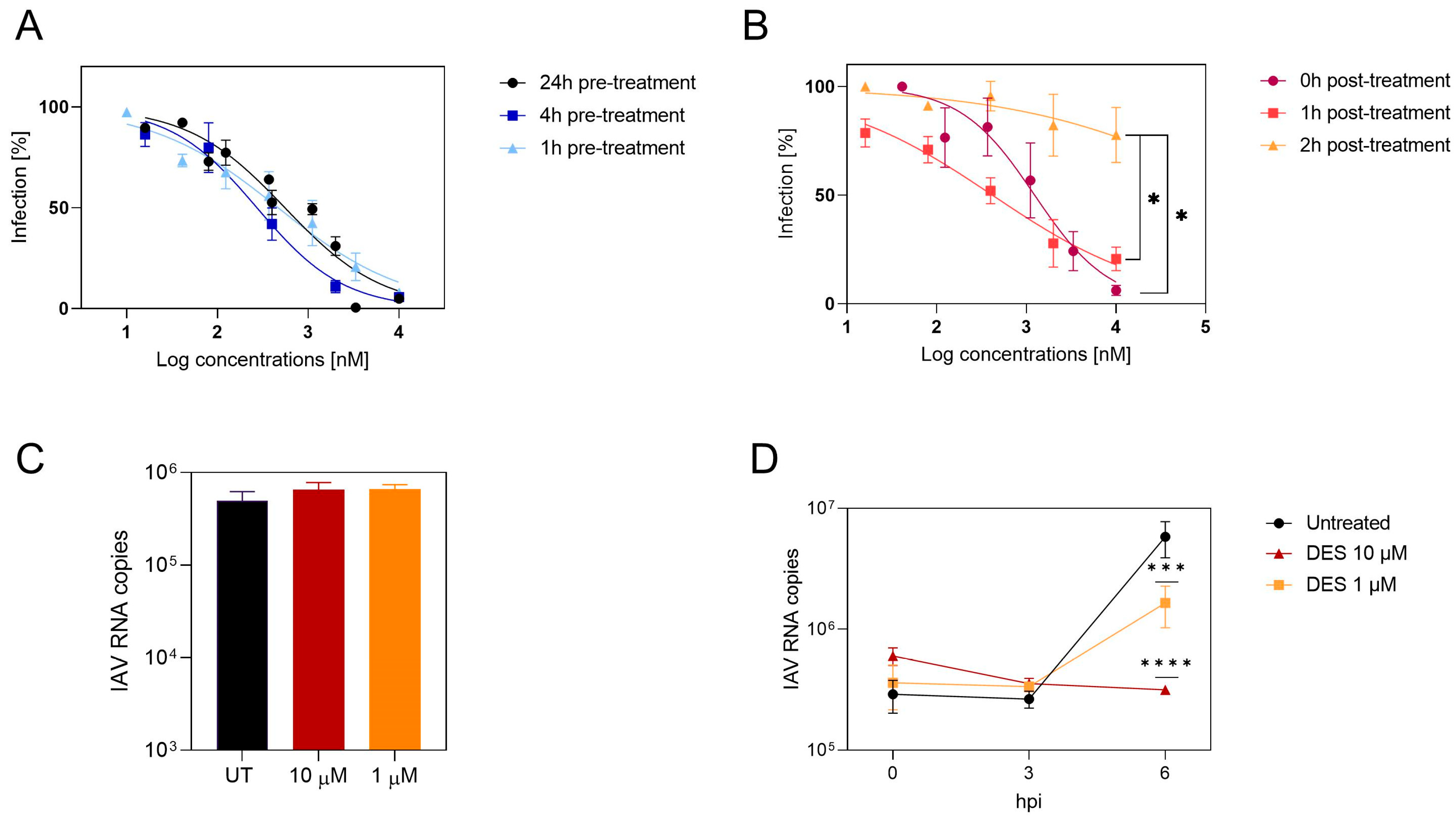
2.5. DES Acts on an Initial Phase of Viral Infection
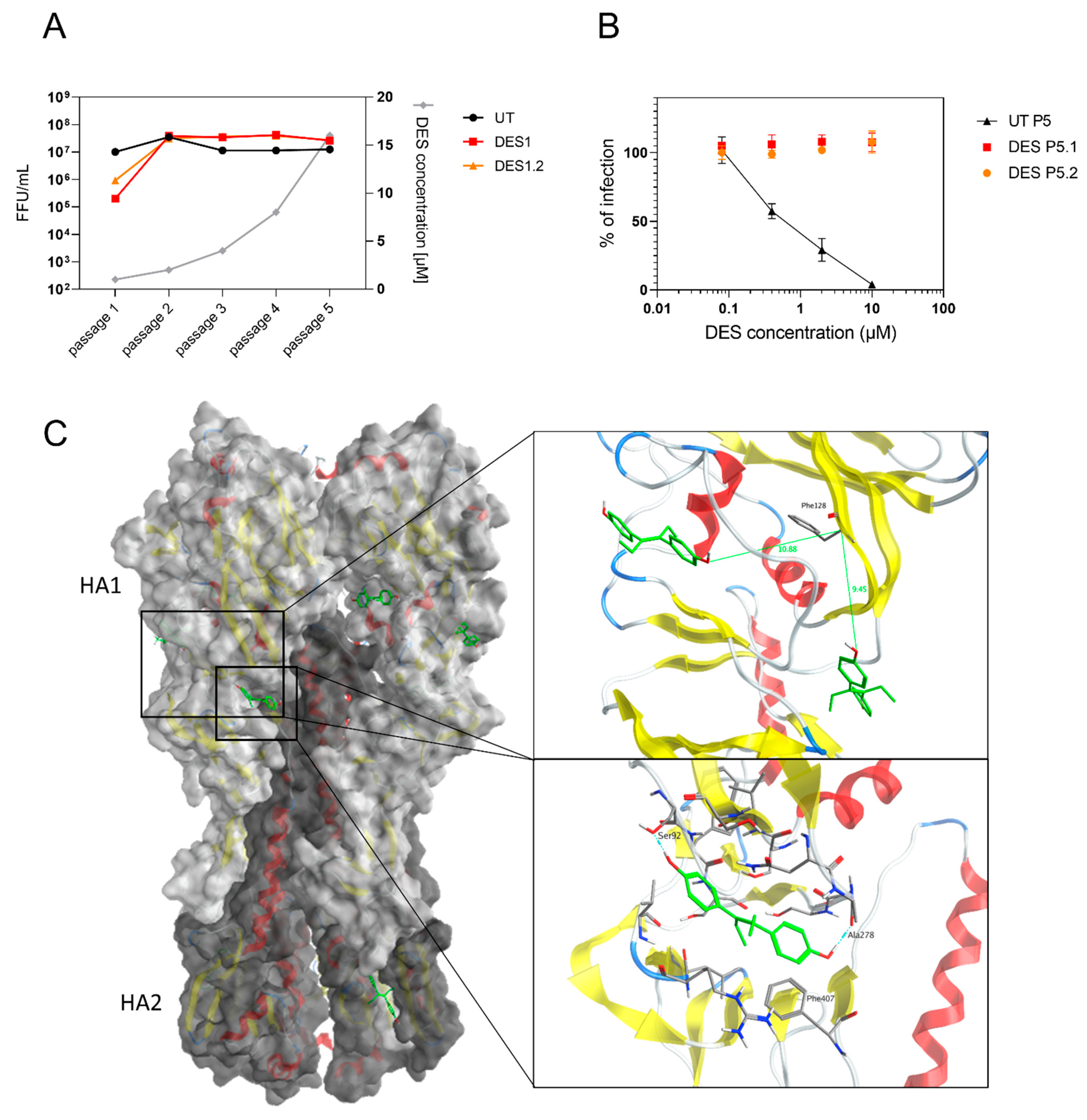
2.6. The Target of DES Is the Viral Hemagglutinin
2.7. In Vivo Testing
3. Discussion
4. Materials and Methods
4.1. Cells and Viruses
4.2. Molecules
4.3. HTS Screening
4.4. Hit Selection
4.5. Toxicity Assays
4.6. Antiviral Assays
4.7. Mechanism of Action
4.8. Kinetics
4.9. Binding
4.10. Resistance
4.11. Tissues: Toxicity Assay
4.12. Tissues: Antiviral Assay
4.13. Analysis of Resistant Mutation Frequency
4.14. In Silico Methods
4.15. In Vivo Experiments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control; European Union Reference Laboratory for Avian Influenza; Adlhoch, C.; Fusaro, A.; Gonzales, J.L.; Kuiken, T.; Marangon, S.; Mirinaviciute, G.; Niqueux, É.; et al. Avian influenza overview December 2022–March 2023. EFSA J. 2023, 21, e07917. [Google Scholar] [CrossRef]
- Price, A.M.; Flannery, B.; Talbot, H.K.; Grijalva, C.G.; Wernli, K.J.; Phillips, C.H.; Monto, A.S.; Martin, E.T.; Belongia, E.A.; McLean, H.Q.; et al. Influenza Vaccine Effectiveness Against Influenza A(H3N2)-Related Illness in the United States During the 2021-2022 Influenza Season. Clin. Infect. Dis. 2023, 76, 1358–1363. [Google Scholar] [CrossRef]
- Jalily, P.H.; Duncan, M.C.; Fedida, D.; Wang, J.; Tietjen, I. Put a cork in it: Plugging the M2 viral ion channel to sink influenza. Antivir. Res. 2020, 178, 104780. [Google Scholar] [CrossRef] [PubMed]
- Memoli, M.J.; Hrabal, R.J.; Hassantoufighi, A.; Eichelberger, M.C.; Taubenberger, J.K. Rapid selection of oseltamivir- and peramivir-resistant pandemic H1N1 virus during therapy in 2 immunocompromised hosts. Clin. Infect. Dis. 2010, 50, 1252–1255. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G.; Sugaya, N.; Hirotsu, N.; Lee, N.; de Jong, M.D.; Hurt, A.C.; Ishida, T.; Sekino, H.; Yamada, K.; Portsmouth, S.; et al. Baloxavir Marboxil for Uncomplicated Influenza in Adults and Adolescents. N. Engl. J. Med. 2018, 379, 913–923. [Google Scholar] [CrossRef]
- Uehara, T.; Hayden, F.G.; Kawaguchi, K.; Omoto, S.; Hurt, A.C.; De Jong, M.D.; Hirotsu, N.; Sugaya, N.; Lee, N.; Baba, K.; et al. Treatment-Emergent Influenza Variant Viruses with Reduced Baloxavir Susceptibility: Impact on Clinical and Virologic Outcomes in Uncomplicated Influenza. J. Infect. Dis. 2020, 221, 346–355. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Bakowski, M.A.; Beutler, N.; Wolff, K.C.; Kirkpatrick, M.G.; Chen, E.; Nguyen, T.H.; Riva, L.; Shaabani, N.; Parren, M.; Ricketts, J.; et al. Drug repurposing screens identify chemical entities for the development of COVID-19 interventions. Nat. Commun. 2021, 12, 3309. [Google Scholar] [CrossRef]
- Riva, L.; Yuan, S.; Yin, X.; Martin-Sancho, L.; Matsunaga, N.; Pache, L.; Burgstaller-Muehlbacher, S.; De Jesus, P.D.; Teriete, P.; Hull, M.V.; et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 2020, 586, 113–119. [Google Scholar] [CrossRef]
- Bragina, M.E.; Daina, A.; Perez, M.A.S.; Michielin, O.; Zoete, V. The SwissSimilarity 2021 Web Tool: Novel Chemical Libraries and Additional Methods for an Enhanced Ligand-Based Virtual Screening Experience. Int. J. Mol. Sci. 2022, 23, 811. [Google Scholar] [CrossRef]
- Cagno, V.; Gasbarri, M.; Medaglia, C.; Gomes, D.; Clement, S.; Stellacci, F.; Tapparel, C. Sulfonated Nanomaterials with Broad-Spectrum Antiviral Activity Extending beyond Heparan Sulfate-Dependent Viruses. Antimicrob. Agents Chemother. 2020, 64, e02001-20. [Google Scholar] [CrossRef]
- Limburg, H.; Harbig, A.; Bestle, D.; Stein, D.A.; Moulton, H.M.; Jaeger, J.; Janga, H.; Hardes, K.; Koepke, J.; Schulte, L.; et al. TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes. J. Virol. 2019, 93, e00649-19. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; McBride, R.; Nycholat, C.M.; Paulson, J.C.; Wilson, I.A. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J. Virol. 2012, 86, 982–990. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.; Kondrashkina, E.; Wang, Q. Structural basis for the divergent evolution of influenza B virus hemagglutinin. Virology 2013, 446, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Kocabiyik, O.; Cagno, V.; Silva, P.J.; Zhu, Y.; Sedano, L.; Bhide, Y.; Mettier, J.; Medaglia, C.; Da Costa, B.; Constant, S.; et al. Non-Toxic Virucidal Macromolecules Show High Efficacy Against Influenza Virus Ex Vivo and In Vivo. Adv. Sci. 2021, 8, 2001012. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.P.; Lorenzo, M.E.; Jian, W.; Klein, S.L. Elevated 17beta-estradiol protects females from influenza A virus pathogenesis by suppressing inflammatory responses. PLoS Pathog. 2011, 7, e1002149. [Google Scholar] [CrossRef]
- Vermillion, M.S.; Ursin, R.L.; Attreed, S.E.; Klein, S.L. Estriol Reduces Pulmonary Immune Cell Recruitment and Inflammation to Protect Female Mice from Severe Influenza. Endocrinology 2018, 159, 3306–3320. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Leon, P. Are the Effects of DES Over? A Tragic Lesson from the Past. Int. J. Environ. Res. Public Health 2021, 18, 10309. [Google Scholar] [CrossRef]
- Kuhl, H. Pharmacology of estrogens and progestogens: Influence of different routes of administration. Climacteric 2005, 8 (Suppl. S1), 3–63. [Google Scholar] [CrossRef] [PubMed]
- Medaglia, C.; Zwygart, A.C.; Silva, P.J.; Constant, S.; Huang, S.; Stellacci, F.; Tapparel, C. Interferon Lambda Delays the Emergence of Influenza Virus Resistance to Oseltamivir. Microorganisms 2021, 9, 1196. [Google Scholar] [CrossRef] [PubMed]
- Shoji, M.; Arakaki, Y.; Esumi, T.; Kohnomi, S.; Yamamoto, C.; Suzuki, Y.; Takahashi, E.; Konishi, S.; Kido, H.; Kuzuhara, T. Bakuchiol Is a Phenolic Isoprenoid with Novel Enantiomer-selective Anti-influenza A Virus Activity Involving Nrf2 Activation. J. Biol. Chem. 2015, 290, 28001–28017. [Google Scholar] [CrossRef] [PubMed]
- Palamara, A.T.; Nencioni, L.; Aquilano, K.; De Chiara, G.; Hernandez, L.; Cozzolino, F.; Ciriolo, M.R.; Garaci, E. Inhibition of influenza A virus replication by resveratrol. J. Infect. Dis. 2005, 191, 1719–1729. [Google Scholar] [CrossRef]
- Zhu, X.; Guo, Y.H.; Jiang, T.; Wang, Y.D.; Chan, K.H.; Li, X.F.; Yu, W.; McBride, R.; Paulson, J.C.; Yuen, K.Y.; et al. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J. Virol. 2013, 87, 12619–12635. [Google Scholar] [CrossRef]
- Van Dongen, M.J.P.; Kadam, R.U.; Juraszek, J.; Lawson, E.; Brandenburg, B.; Schmitz, F.; Schepens, W.B.G.; Stoops, B.; van Diepen, H.A.; Jongeneelen, M.; et al. A small-molecule fusion inhibitor of influenza virus is orally active in mice. Science 2019, 363, eaar6221. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cui, Q.; Caffrey, M.; Rong, L.; Du, R. Small Molecule Inhibitors of Influenza Virus Entry. Pharmaceuticals 2021, 14, 587. [Google Scholar] [CrossRef]
- Mathez, G.; Cagno, V. Clinical severe acute respiratory syndrome coronavirus 2 isolation and antiviral testing. Antivir. Chem. Chemother 2021, 29, 20402066211061063. [Google Scholar] [CrossRef]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Varricchio, C.; Mathez, G.; Pillonel, T.; Bertelli, C.; Kaiser, L.; Tapparel, C.; Brancale, A.; Cagno, V. Geneticin shows selective antiviral activity against SARS-CoV-2 by interfering with programmed -1 ribosomal frameshifting. Antivir. Res. 2022, 208, 105452. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [PubMed]
- Vila, I.K.; Fretaud, M.; Vlachakis, D.; Laguette, N.; Langevin, C. Animal Models for the Study of Nucleic Acid Immunity: Novel Tools and New Perspectives. J. Mol. Biol. 2020, 432, 5529–5543. [Google Scholar] [CrossRef] [PubMed]


| Compound | Inhibition (%) | Viability (%) |
|---|---|---|
| Alverine citrate salt | 99 | 145 |
| Dextromethorphan hydrobromide monohydrate | 100 | 103 |
| Camylofine chlorhydrate | 97 | 97 |
| Procyclidine hydrochloride | 97 | 91 |
| Diethylstilbestrol | 114 | 72 |
| Structure | EC50 (95% CI) (µM) | CC50 (µM) | SI | |
|---|---|---|---|---|
| Diethylstilbestrol | 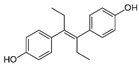 | 0.12 (0.06–0.17) | 9.81 | 89.2 |
| Dienestrol | 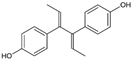 | 3.13 (2.09–4.78) | 32.6 | 10.4 |
| Resveratrol | 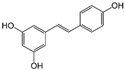 | >10 | 52.0 | n.a. |
| Hexestrol | 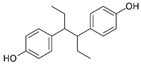 | 0.14 (0.05–0.25) | 46.9 | 335 |
| Bakuchiol | 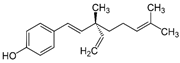 | 0.94 (0.65–1.35) | 46.2 | 49.1 |
| Masoprocol | 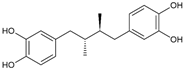 | >10 | 84.9 | n.a. |
| CD-MUS | 13.4 (10.6–17.3) | >100 | >7.46 |
| Strains | EC50 (95% CI) (µM) |
|---|---|
| H1N1 A/Puerto Rico/8/34 | 1.64 (1.18–2.25) |
| H1N1 A/WSN/1933 | 1.11 (0.79–1.59) |
| H1N1 A/Netherlands/602/2009 | 0.12 (0.06–0.17) |
| H1N1 A/Lausanne/2022 Clinical 1 | 0.56 (0.48–0.65) |
| H1N1 A/Lausanne/2022 Clinical 2 | 0.37 (0.28–0.48) |
| H3N2 A/Wyoming/2003/3 | 3.81 (2.73–5.16) |
| H3N2 A/Lausanne/2022 Clinical 3 | 4.71 (3.27–6.83) |
| H5N1 A/Viet Nam/1203/2004 | 3.84 (2.64–5.89) |
| B/Washington/02/2019 | 4.15 (1.84–17.1) |
| SARS-CoV-2 | n.a. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franzi, E.; Mathez, G.; Dinant, S.; Deloizy, C.; Kaiser, L.; Tapparel, C.; Le Goffic, R.; Cagno, V. Non-Steroidal Estrogens Inhibit Influenza Virus by Interacting with Hemagglutinin and Preventing Viral Fusion. Int. J. Mol. Sci. 2023, 24, 15382. https://doi.org/10.3390/ijms242015382
Franzi E, Mathez G, Dinant S, Deloizy C, Kaiser L, Tapparel C, Le Goffic R, Cagno V. Non-Steroidal Estrogens Inhibit Influenza Virus by Interacting with Hemagglutinin and Preventing Viral Fusion. International Journal of Molecular Sciences. 2023; 24(20):15382. https://doi.org/10.3390/ijms242015382
Chicago/Turabian StyleFranzi, Elisa, Gregory Mathez, Soraya Dinant, Charlotte Deloizy, Laurent Kaiser, Caroline Tapparel, Ronan Le Goffic, and Valeria Cagno. 2023. "Non-Steroidal Estrogens Inhibit Influenza Virus by Interacting with Hemagglutinin and Preventing Viral Fusion" International Journal of Molecular Sciences 24, no. 20: 15382. https://doi.org/10.3390/ijms242015382
APA StyleFranzi, E., Mathez, G., Dinant, S., Deloizy, C., Kaiser, L., Tapparel, C., Le Goffic, R., & Cagno, V. (2023). Non-Steroidal Estrogens Inhibit Influenza Virus by Interacting with Hemagglutinin and Preventing Viral Fusion. International Journal of Molecular Sciences, 24(20), 15382. https://doi.org/10.3390/ijms242015382







