Obtaining New Candidate Peptides for Biological Anticancer Drugs from Enzymatic Hydrolysis of Human and Bovine Hemoglobin
Abstract
:1. Introduction
2. Results
2.1. Effects of Bovine and Human Hemoglobin Hydrolysates and Their Peptide Fractions on the Inhibition of Protein Biosynthesis
2.1.1. Study of the Influence of Initial Bovine and Human Hemoglobin Concentration on Protein Biosynthesis
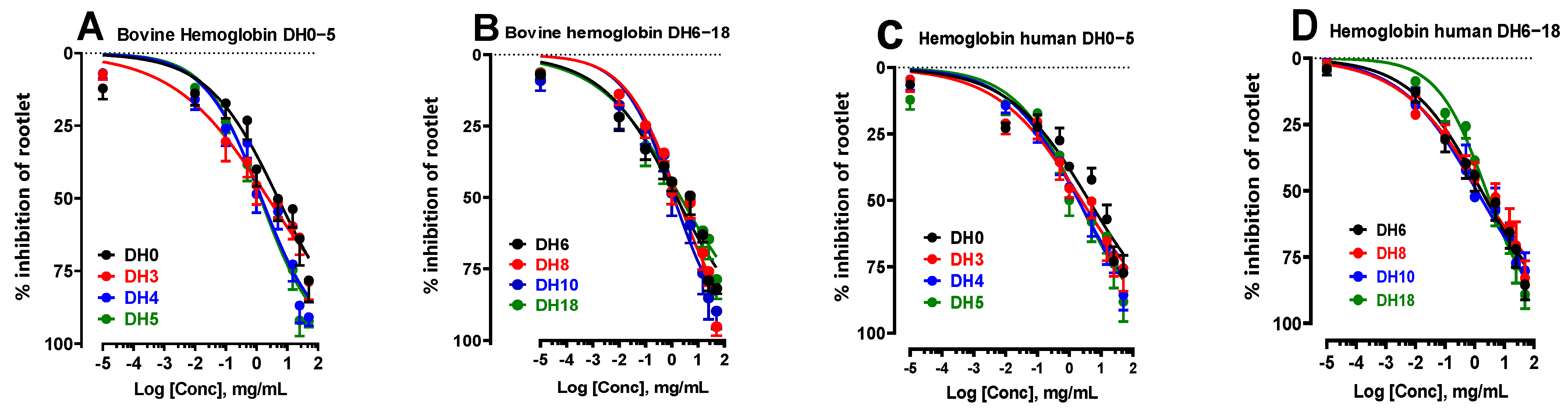
2.1.2. Study of the Inhibition of Rootlet Growth Activity by Bovine and Human Hemoglobin Hydrolysates According to Their Degree of Hydrolysis
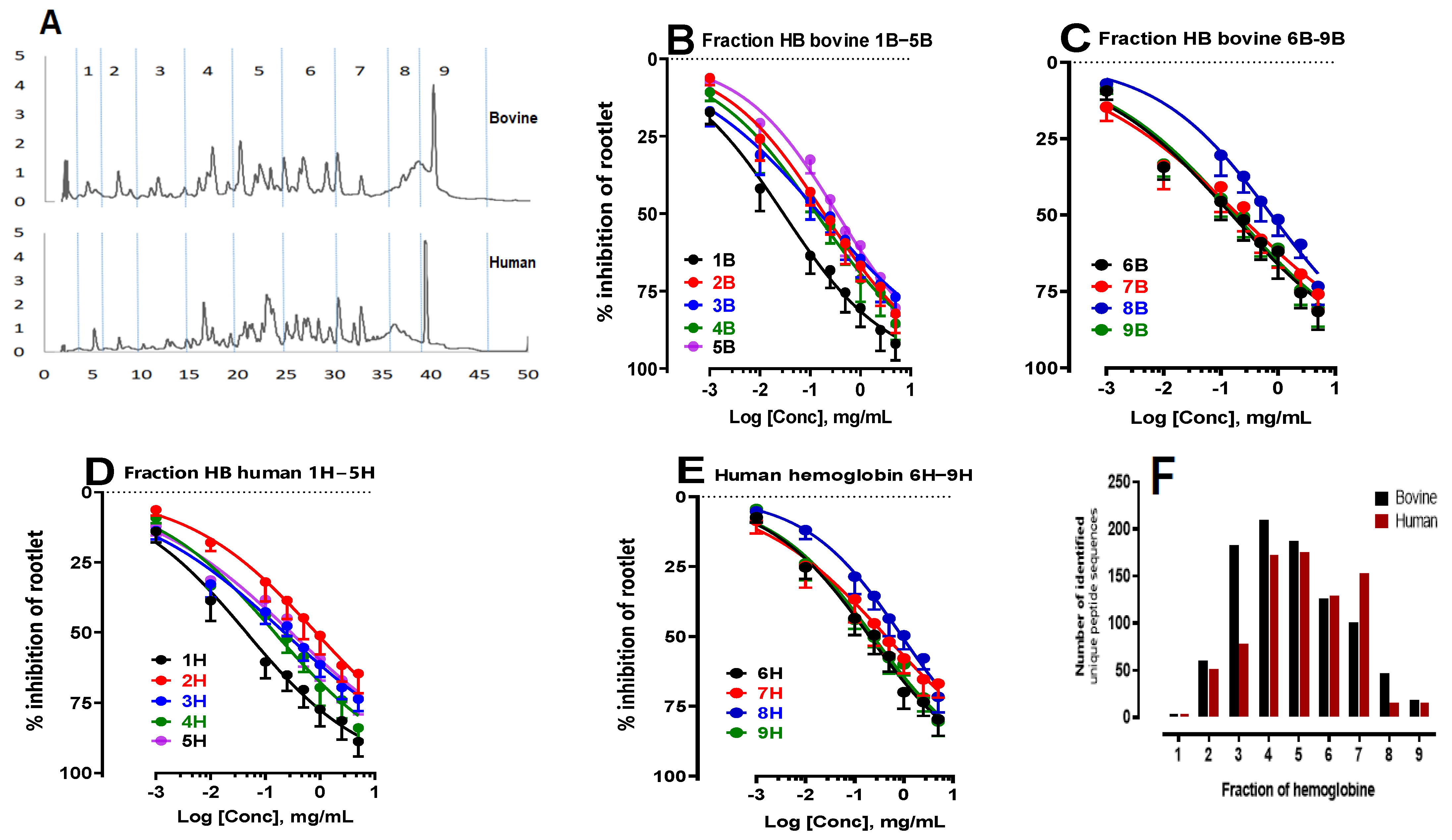
2.1.3. Study of the Inhibition of Rootlet Growth Activity of Bovine and Human Hemoglobin Hydrolysate Fractions and Peptidomics Approach
2.1.4. Study of the Inhibition of Rootlet Growth Activity of Pure Antimicrobial Peptides α137–141 and α1–32 with Different Structural Characteristics
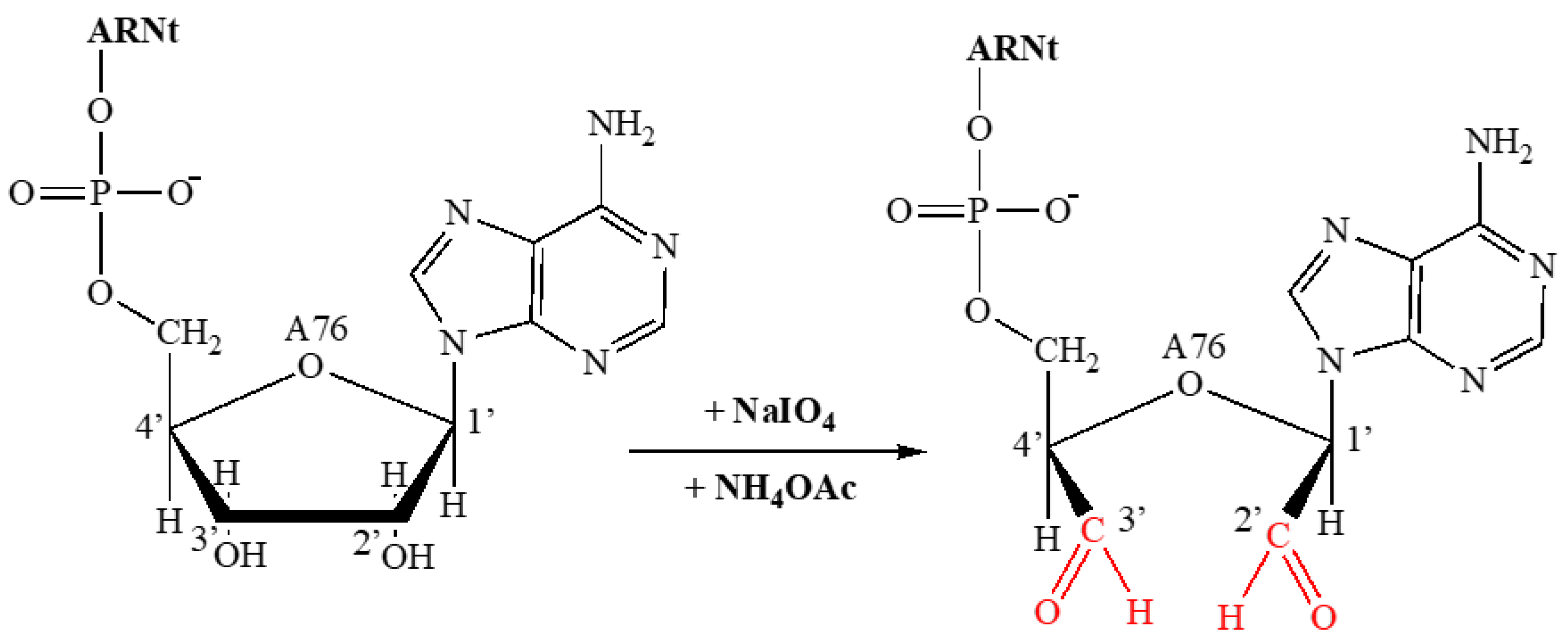
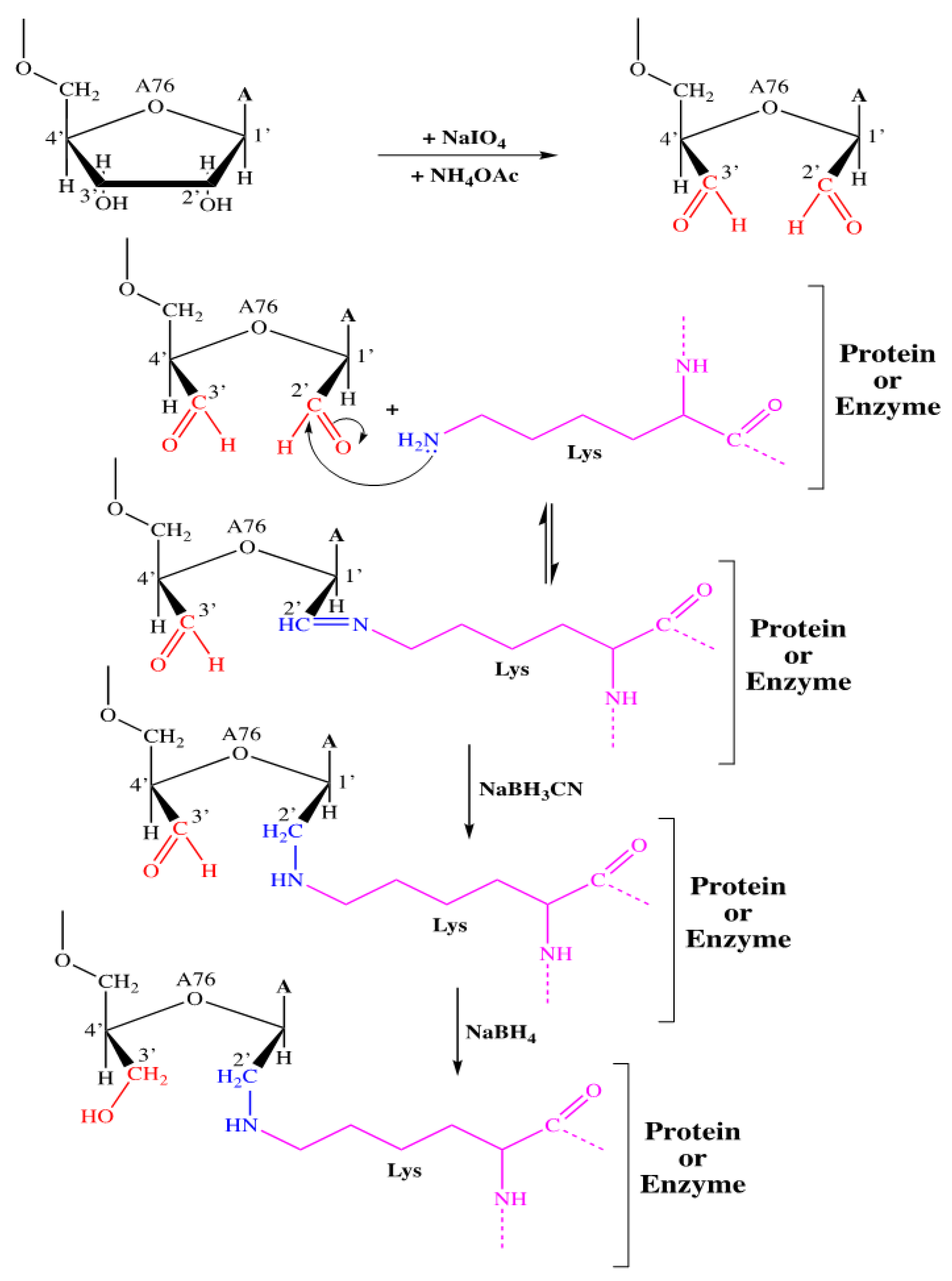
2.2. Study of the Inhibition of Rootlet Growth Activity of Bovine and Human Hemoglobinhydrolysates and Their Fractions: Covalent Labeling of Proteins Using tRNAox
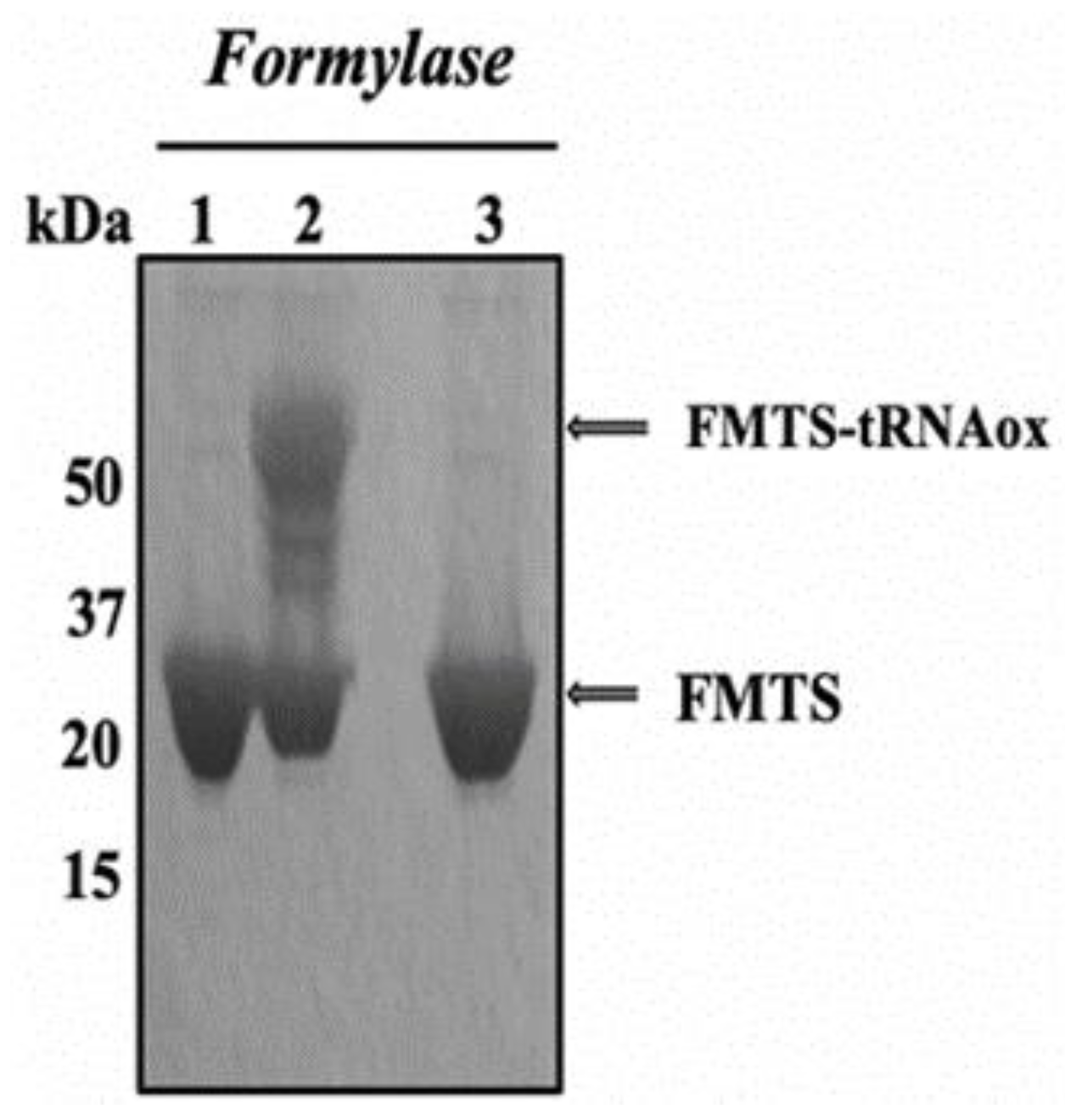
2.2.1. Covalent Labeling of Formylase by tRNAox
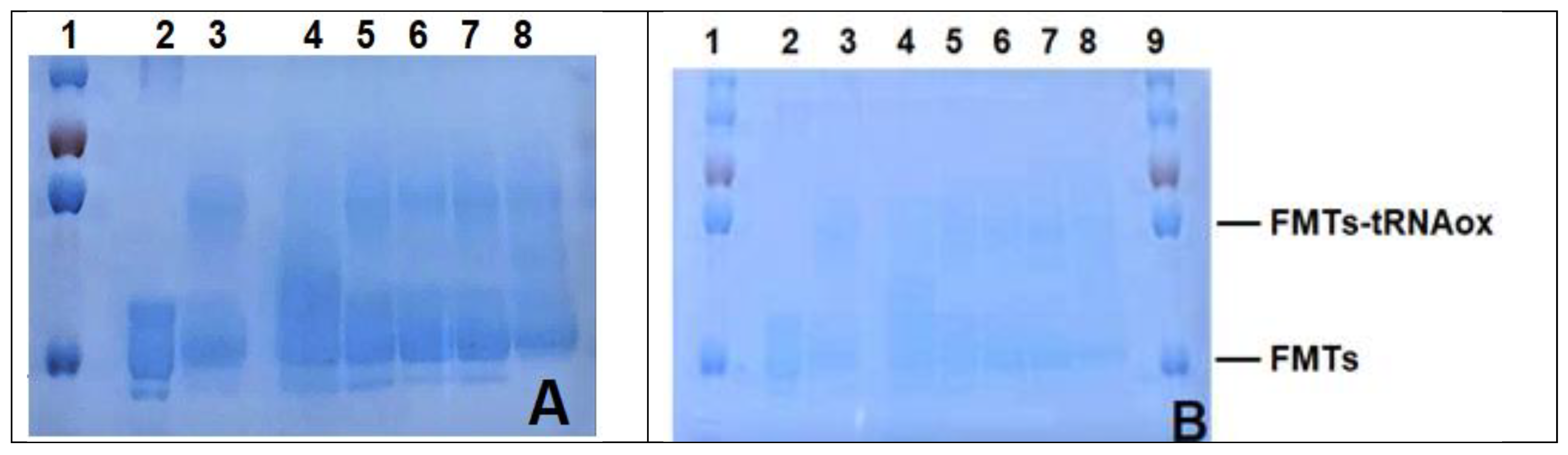
2.2.2. Testing of Bovine and Human Hemoglobin Peptide Hydrolysates and Their Peptide Fractions
3. Discussion
4. Materials and Methods
4.1. Materials: Reagents, Solvents, and Standards Used
4.2. Preparation of Bovine and Human Hemoglobin Hydrolysates
4.3. Preparation of the Stock Solution
4.4. Hydrolysis Process
4.5. Fractionation of Peptide Hydrolysates by Semipreparative HPLC
4.6. RP-UPLC Analysis and Mass Spectrometry
4.7. Determination of the Anticancer Activity of Hydrolysates and Peptide Fractions
4.7.1. Lepidium Sativum (LS) Rootlet Growth Test
Description of the Methods Used
Preparation of Vegetable Seeds
Preparation of Test Concentrations
4.7.2. Covalent Labeling of Proteins with tRNAox by Formylase
Preparation of tRNAox
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef] [PubMed]
- Bakare, O.O.; Gokul, A.; Wu, R.; Niekerk, L.-A.; Klein, A.; Keyster, M. Biomedical relevance of novel anticancer peptides in the sensitive treatment of cancer. Biomolecules 2021, 11, 1120. [Google Scholar] [CrossRef]
- Kolberg, H.C.; Villena-Heinsen, C.; Deml, M.M.; Kraemer, S.; Diedrich, K.; Friedrich, M. Relationship between chemotherapy with paclitaxel, cisplatin, vinorelbine and titanocene dichloride and expression of proliferation markers and tumour suppressor gene p53 in human ovarian cancer xenografts in nude mice. Eur. J. Gynaecol. Oncol. 2005, 26, 398–402. [Google Scholar] [PubMed]
- Pierard, G.E.; Focan, C.; Lapiere, C.M. Cell proliferation in a malignant angioendothelioma during sequential chemotherapy. J. Cutan. Pathol. 1979, 6, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A. Cancer chemotherapy and its side effect management. Nurs. J. India 2006, 97, 109–110. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Sato, J.; Kato, R.; Takata, R.; Obara, W. Side effect and supportive care to combination of gemcitabine and cisplatin chemotherapy for the advanced urothelial cancer. Nihon Rinsho. Jpn. J. Clin. Med. 2015, 73, 609–613. [Google Scholar]
- Chism, D.D.; De Silva, D.; Whang, Y.E. Mechanisms of acquired resistance to androgen receptor targeting drugs in castration-resistant prostate cancer. Expert Rev. Anticancer Ther. 2014, 14, 1369–1378. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017, 7, 339. [Google Scholar] [CrossRef]
- Deslouches, B.; Di, Y.P. Antimicrobial peptides with selective antitumor mechanisms: Prospect for anticancer applications. Oncotarget 2017, 8, 46635. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial peptides as anticancer agents: Functional properties and biological activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef]
- Kordi, M.; Borzouyi, Z.; Chitsaz, S.; Asmaei, M.H.; Salami, R.; Tabarzad, M. Antimicrobial peptides with anticancer activity: Today status, trends and their computational design. Arch. Biochem. Biophys. 2022, 733, 109484. [Google Scholar] [CrossRef] [PubMed]
- Bhat, Z.F.; Kumar, S.; Bhat, H.F. Bioactive peptides of animal origin: A review. J. Food Sci. Technol. 2015, 52, 5377–5392. [Google Scholar] [CrossRef] [PubMed]
- Chalamaiah, M.; Hemalatha, R.; Jyothirmayi, T. Fish protein hydrolysates: Proximate composition, amino acid composition, antioxidant activities and applications: A review. Food Chem. 2012, 135, 3020–3038. [Google Scholar] [CrossRef] [PubMed]
- García, M.C.; Puchalska, P.; Esteve, C.; Marina, M.L. Vegetable foods: A cheap source of proteins and peptides with antihypertensive, antioxidant, and other less occurrence bioactivities. Talanta 2013, 106, 328–349. [Google Scholar] [CrossRef]
- Bosso, M.; Ständker, L.; Kirchhoff, F.; Münch, J. Exploiting the human peptidome for novel antimicrobial and anticancer agents. Bioorg. Med. Chem. 2018, 26, 2719–2726. [Google Scholar] [CrossRef] [PubMed]
- Pimchan, T.; Tian, F.; Thumanu, K.; Rodtong, S.; Yongsawatdigul, J. Isolation, identification, and mode of action of antibacterial peptides derived from egg yolk hydrolysate. Poult. Sci. 2023, 102, 102695. [Google Scholar] [CrossRef] [PubMed]
- Zouari, O.; Przybylski, R.; Hannioui, M.; Sion, L.; Dhulster, P.; Nedjar-Arroume, N. High added-value co-product: The porcine cruor is an attractive source of active peptides. J. Nutr. Health Food Sci. 2020, 7, 1170. [Google Scholar]
- Lignot, B.; Froidevaux, R.; Nedjar-Arroume, N.; Guillochon, D. Solvent effect on kinetics of appearance of neokyotorphin, VV-haemorphin-4 and a bradykinin-potentiating peptide in the course of peptic hydrolysis of bovine haemoglobin. Biotechnol. Appl. Biochem. 1999, 30, 201–207. [Google Scholar]
- Zhao, Q.; Molina, P.; Marie Piot, J. Peptic peptide mapping by HPLC, on line with photodiode array detection, of a hemoglobin hydrolysate produced at pilot-plant scale from an ultrafiltration process. J. Liq. Chromatogr. Relat. Technol. 1997, 20, 1717–1739. [Google Scholar] [CrossRef]
- Vercaigne-Marko, D.; Kosciarz, E.; Nedjar-Arroume, N.; Guillochon, D. Improvement of Staphylococcus aureus-V8-protease hydrolysis of bovine haemoglobin by its adsorption on to a solid phase in the presence of SDS: Peptide mapping and obtention of two haemopoietic peptides. Biotechnol. Appl. Biochem. 2000, 31, 127–134. [Google Scholar] [CrossRef]
- Adje, E.Y.; Balti, R.; Kouach, M.; Guillochon, D.; Nedjar-Arroume, N. α 67-106 of bovine hemoglobin: A new family of antimicrobial and angiotensin I-converting enzyme inhibitory peptides. Eur. Food Res. Technol. 2011, 232, 637–646. [Google Scholar] [CrossRef]
- Daoud, R.; Dubois, V.; Bors-Dodita, L.; Nedjar-Arroume, N.; Krier, F.; Chihib, N.-E.; Mary, P.; Kouach, M.; Briand, G.; Guillochon, D. New antibacterial peptide derived from bovine hemoglobin. Peptides 2005, 26, 713–719. [Google Scholar] [CrossRef]
- Choisnard, L.; Froidevaux, R.; Nedjar-Arroume, N.; Lignot, B.; Vercaigne-Marko, D.; Krier, F.; Dhulster, P.; Guillochon, D. Kinetic study of the appearance of an anti-bacterial peptide in the course of bovine haemoglobin peptic hydrolysis. Biotechnol. Appl. Biochem. 2002, 36, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Przybylski, R.; Firdaous, L.; Châtaigné, G.; Dhulster, P.; Nedjar, N. Production of an antimicrobial peptide derived from slaughterhouse by-product and its potential application on meat as preservative. Food Chem. 2016, 211, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Parish, C.A.; Jiang, H.; Tokiwa, Y.; Berova, N.; Nakanishi, K.; McCabe, D.; Zuckerman, W.; Xia, M.M.; Gabay, J.E. Broad-spectrum antimicrobial activity of hemoglobin. Bioorg. Med. Chem. 2001, 9, 377–382. [Google Scholar] [CrossRef]
- Liepke, C.; Baxmann, S.; Heine, C.; Breithaupt, N.; Ständker, L.; Forssmann, W.-G. Human hemoglobin-derived peptides exhibit antimicrobial activity: A class of host defense peptides. J. Chromatogr. B 2003, 791, 345–356. [Google Scholar] [CrossRef]
- Outman, A.; Deracinois, B.; Flahaut, C.; Abou Diab, M.; Gressier, B.; Eto, B.; Nedjar, N. Potential of Human Hemoglobin as a Source of Bioactive Peptides: Comparative Study of Enzymatic Hydrolysis with Bovine Hemoglobin and Production of Active Peptide α137–141. Int. J. Mol. Sci. 2023, 24, 11921. [Google Scholar] [CrossRef]
- Outman, A.; Kouoh Elombo, F.; Abidi, C.; Bouhrim, M.; Hountondji, C.; Al-Zharani, M.; Nasr, F.A.; Aleissa, M.S.; Gressier, B.; Nedjar, N.; et al. Protein synthesis by the plant rootlet as a target for the rapid screening of anticancer drugs. J. Biol. Regul. Homeost. Agents 2023, 37, 3417–3420. [Google Scholar] [CrossRef]
- Outman, A.; Nedjar, N.; Gressier, B.; Eto, B. Method of Rapid and Simple Screening of Anticancer, Antibiotic, Antifungal and Antiparasitic Drugs and Results Obtained by this Process. Patent FR2213717, 18 December 2022. [Google Scholar]
- Kim, J.-H.; You, K.-R.; Kim, I.H.; Cho, B.-H.; Kim, C.-Y.; Kim, D.-G. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology 2004, 39, 129–138. [Google Scholar] [CrossRef]
- Hountondji, C.; Bulygin, K.; Woisard, A.; Tuffery, P.; Créchet, J.-B.; Pech, M.; Nierhaus, K.H.; Karpova, G.; Baouz, S. Lys53 of ribosomal protein L36AL and the CCA end of a tRNA at the P/E hybrid site are in close proximity on the human ribosome. Chembiochem 2012, 13, 1791–1797. [Google Scholar] [CrossRef]
- Blanquet, S.; Petrissant, G.; Waller, J.P. The Mechanism of Action of Methionyl-tRNA Synthetase: 2. Interaction of the Enzyme with Specific and Unspecific tRNAs. Eur. J. Biochem. 1973, 36, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Nedjar-Arroume, N.; Dubois-Delval, V.; Adje, E.Y.; Traisnel, J.; Krier, F.; Mary, P.; Kouach, M.; Briand, G.; Guillochon, D. Bovine hemoglobin: An attractive source of antibacterial peptides. Peptides 2008, 29, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Dubois, V. Préparation de Peptides Antimicrobiens à Partir de L’hydrolyse Enzymatique de deux Protéines: L’hémoglobine Bovine et l’α-lactalbumine Bovine. Ph.D. Thesis, Lille 1 University of Science and Technology, Villeneuve-d’Ascq, France, 2006. [Google Scholar]
- Hountondji, C.; Créchet, J.-B.; Tanaka, M.; Suzuki, M.; Nakayama, J.; Aguida, B.; Bulygin, K.; Cognet, J.; Karpova, G.; Baouz, S. Ribosomal protein eL42 contributes to the catalytic activity of the yeast ribosome at the elongation step of translation. Biochimie 2019, 158, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Catiau, L.; Traisnel, J.; Delval-Dubois, V.; Chihib, N.-E.; Guillochon, D.; Nedjar-Arroume, N. Minimal antimicrobial peptidic sequence from hemoglobin alpha-chain: KYR. Peptides 2011, 32, 633–638. [Google Scholar] [CrossRef]
- Nedjar-Arroume, N.; Dubois-Delval, V.; Miloudi, K.; Daoud, R.; Krier, F.; Kouach, M.; Briand, G.; Guillochon, D. Isolation and characterization of four antibacterial peptides from bovine hemoglobin. Peptides 2006, 27, 2082–2089. [Google Scholar] [CrossRef]
- Outman, A.; Deracinois, B.; Flahaut, C.; Diab, M.A.; Dhaouefi, J.; Gressier, B.; Eto, B.; Nedjar, N. Comparison of the Bioactive Properties of Human and Bovine Hemoglobin Hydrolysates Obtained by Enzymatic Hydrolysis: Antimicrobial and Antioxidant Potential of the Active Peptide α137–141. Int. J. Mol. Sci. 2023, 24, 13055. [Google Scholar] [CrossRef]
- Mak, P.; Wójcik, K.; Wicherek, Ł.; Suder, P.; Dubin, A. Antibacterial hemoglobin peptides in human menstrual blood. Peptides 2004, 25, 1839–1847. [Google Scholar] [CrossRef]
- Wang, R.E. Targeting heat shock proteins 70/90 and proteasome for cancer therapy. Curr. Med. Chem. 2011, 18, 4250–4264. [Google Scholar] [CrossRef]
- Kovalski, J.R.; Kuzuoglu-Ozturk, D.; Ruggero, D. Protein synthesis control in cancer: Selectivity and therapeutic targeting. EMBO J. 2022, 41, e109823. [Google Scholar] [CrossRef]
- Porse, B.T.; Kirillov, S.V.; Awayez, M.J.; Ottenheijm, H.C.; Garrett, R.A. Direct crosslinking of the antitumor antibiotic sparsomycin, and its derivatives, to A2602 in the peptidyl transferase center of 23S-like rRNA within ribosome-tRNA complexes. Proc. Natl. Acad. Sci. USA 1999, 96, 9003–9008. [Google Scholar] [CrossRef]
- Lockhead, S.; Moskaleva, A.; Kamenz, J.; Chen, Y.; Kang, M.; Reddy, A.R.; Santos, S.D.; Ferrell, J.E. The apparent requirement for protein synthesis during G2 phase is due to checkpoint activation. Cell Rep. 2020, 32, 107901. [Google Scholar] [CrossRef] [PubMed]
- Awad, D.; Prattes, M.; Kofler, L.; Rössler, I.; Loibl, M.; Pertl, M.; Zisser, G.; Wolinski, H.; Pertschy, B.; Bergler, H. Inhibiting eukaryotic ribosome biogenesis. BMC Biol. 2019, 17, 46. [Google Scholar] [CrossRef] [PubMed]
- Dinos, G.P. The macrolide antibiotic renaissance. Br. J. Pharmacol. 2017, 174, 2967–2983. [Google Scholar] [CrossRef]
- Zhong, W.; Chen, S.; Zhang, Q.; Xiao, T.; Qin, Y.; Gu, J.; Sun, B.; Liu, Y.; Jing, X.; Hu, X. Doxycycline directly targets PAR1 to suppress tumor progression. Oncotarget 2017, 8, 16829. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, X.; Zhang, Z.; Ma, C.; Wu, T.; Tang, S.; Zeng, Z.; Huang, S.; Gong, C.; Yuan, C. Monensin inhibits cell proliferation and tumor growth of chemo-resistant pancreatic cancer cells by targeting the EGFR signaling pathway. Sci. Rep. 2018, 8, 17914. [Google Scholar] [CrossRef]
- Kondo, J.; François, B.; Russell, R.J.; Murray, J.B.; Westhof, E. Crystal structure of the bacterial ribosomal decoding site complexed with amikacin containing the γ-amino-α-hydroxybutyryl (haba) group. Biochimie 2006, 88, 1027–1031. [Google Scholar] [CrossRef]
- Takagi, H.; Shiomi, H.; Fukui, K.; Hayashi, K.; Kiso, Y.; Kitagawa, K. Isolation of a novel analgesic pentapeptide, neo-kyotorphin, from bovine brain. Life Sci. 1982, 31, 1733–1736. [Google Scholar] [CrossRef]
- Zhao, Q.; Sannier, F.; Piot, J.M. Kinetics of appearance of four hemorphins from bovine hemoglobin peptic hydrolysates by HPLC coupled with photodiode array detection. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1996, 1295, 73–80. [Google Scholar] [CrossRef]


| IC50 (mg/mL) | ||
|---|---|---|
| (HB) | Bovine (B) | Human (H) |
| 1 | 1.47 ± 0.65 ns | 2.12 ± 0.86 ns |
| 2 | 1.69 ± 0.70 ns | 1.80 ± 0.74 ns |
| 8 | 1.53 ± 0.61 ns | 2.24 ± 0.80 ns |
| 10 | 1.23 ± 0.52 ns | 1.77 ± 0.74 ns |
| Hydrolysis | IC50 (mg/mL) | |
|---|---|---|
| DH | Bovine (B) | Human (H) |
| DH0 | 5.06 ± 2.00 ns | 3.97 ± 0.60 ns |
| DH3 | 2.42 ± 0.96 ns | 2.68 ± 0.67 ns |
| DH4 | 1.46 ± 0.59 ns | 1.94 ± 0.47 ns |
| DH5 | 1.35 ± 0.54 ns | 1.97 ± 0.84 ns |
| DH6 | 1.54 ± 0.60 ns | 1.68 ± 0.67 ns |
| DH8 | 1.68 ± 0.67 ns | 1.67 ± 0.65 ns |
| DH10 | 1.19 ± 0.47 ns | 1.17 ± 0.46 ns |
| DH18 | 2.25 ± 0.84 ns | 2.46 ± 1.06 ns |
| Comparison of IC50 (mg/mL) | |||
|---|---|---|---|
| Bovine (B) | Human (H) | ||
| 1B | 0.029 ± 0.001 * | 1H | 0.045 ± 0.002 * |
| 2B | 0.19 ± 0.08 * | 2H | 0.84 ± 0.35 * |
| 3B | 0.15 ± 0.06 ns | 3H | 0.22 ± 0.09 ns |
| 4B | 0.13 ± 0.05 ns | 4H | 0.15 ± 0.06 ns |
| 5B | 0.34 ± 0.14 ns | 5H | 0.29 ± 0.11 ns |
| 6B | 0.20 ± 0.08 ns | 6H | 0.20 ± 0.08 ns |
| 7B | 0.43 ± 0.17 ns | 7H | 0.48 ± 0.19 ns |
| 8B | 0.92 ± 0.40 ns | 8H | 0.92 ± 0.40 ns |
| 9B | 0.23 ± 0.10 ns | 9H | 0.22 ± 0.09 ns |
| Bioactive Peptides | IC50 (µg/mL) |
|---|---|
| α137−141 | 53 ± 9 * |
| α1−32 | 280 ± 31 ns |
| Strains | Gram − Escherichia. coli Salmonella. enteritidis | Gram + Staphylococcus. aureus Listeria. innocua Micrococcus. luteus | ||||
|---|---|---|---|---|---|---|
| Peptides | ||||||
| α1–32 | 154 | 54 | 38 | 38 | 90 | |
| α137–141 | 9 | 4.6 | 1 | 1 | 9 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Outman, A.; Bouhrim, M.; Hountondji, C.; Noman, O.M.; Alqahtani, A.S.; Gressier, B.; Nedjar, N.; Eto, B. Obtaining New Candidate Peptides for Biological Anticancer Drugs from Enzymatic Hydrolysis of Human and Bovine Hemoglobin. Int. J. Mol. Sci. 2023, 24, 15383. https://doi.org/10.3390/ijms242015383
Outman A, Bouhrim M, Hountondji C, Noman OM, Alqahtani AS, Gressier B, Nedjar N, Eto B. Obtaining New Candidate Peptides for Biological Anticancer Drugs from Enzymatic Hydrolysis of Human and Bovine Hemoglobin. International Journal of Molecular Sciences. 2023; 24(20):15383. https://doi.org/10.3390/ijms242015383
Chicago/Turabian StyleOutman, Ahlam, Mohamed Bouhrim, Codjo Hountondji, Omar M. Noman, Ali S. Alqahtani, Bernard Gressier, Naïma Nedjar, and Bruno Eto. 2023. "Obtaining New Candidate Peptides for Biological Anticancer Drugs from Enzymatic Hydrolysis of Human and Bovine Hemoglobin" International Journal of Molecular Sciences 24, no. 20: 15383. https://doi.org/10.3390/ijms242015383
APA StyleOutman, A., Bouhrim, M., Hountondji, C., Noman, O. M., Alqahtani, A. S., Gressier, B., Nedjar, N., & Eto, B. (2023). Obtaining New Candidate Peptides for Biological Anticancer Drugs from Enzymatic Hydrolysis of Human and Bovine Hemoglobin. International Journal of Molecular Sciences, 24(20), 15383. https://doi.org/10.3390/ijms242015383






