Comparative Genomics Identifies the Evolutionarily Conserved Gene TPM3 as a Target of eca-miR-1 Involved in the Skeletal Muscle Development of Donkeys
Abstract
:1. Introduction
2. Results
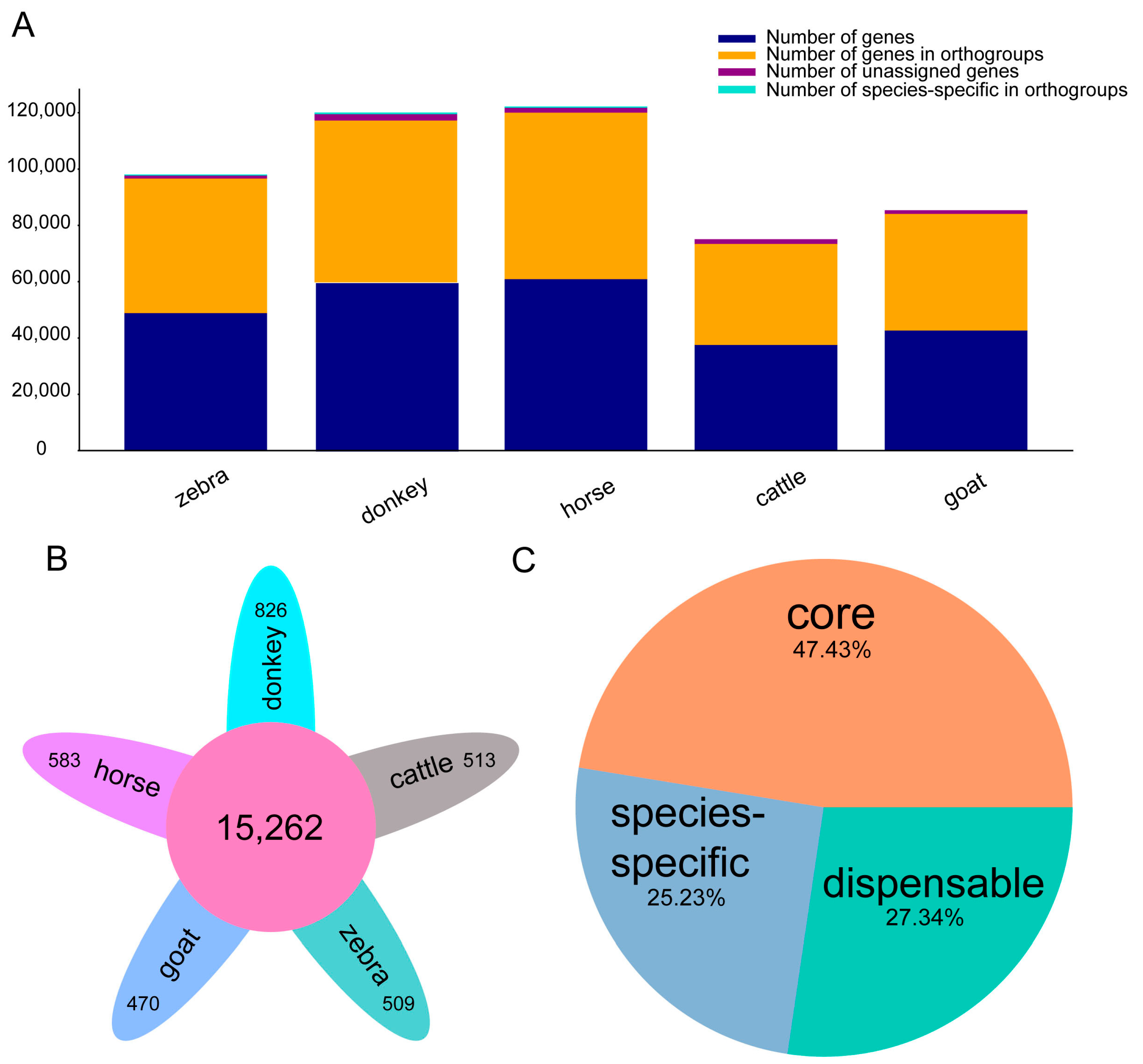
2.1. Comparative Genomics of Equine Genomes and Outgroup (Goat and Cattle)
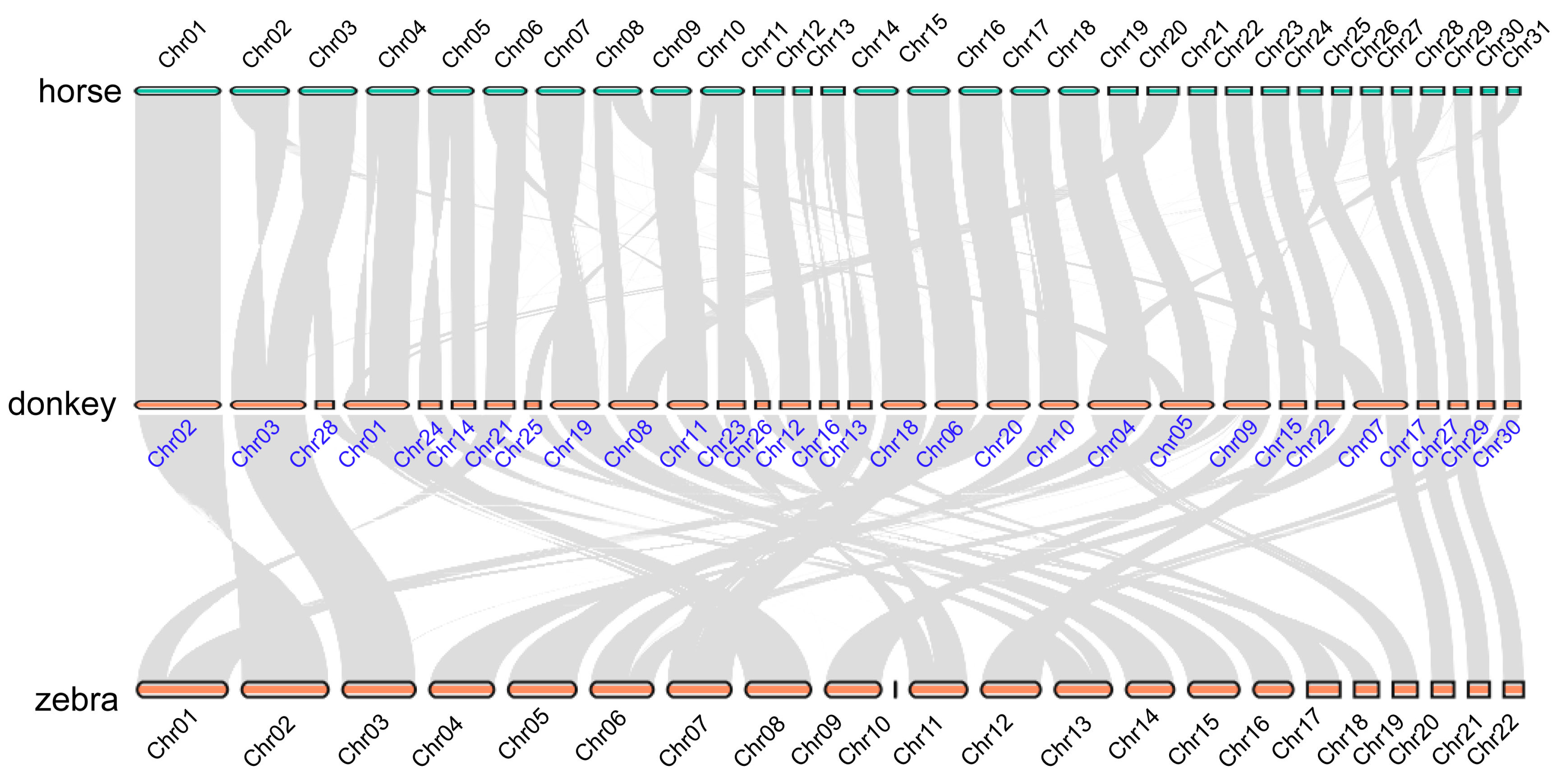
2.2. The Karyotype Evolution of Equine Genomes
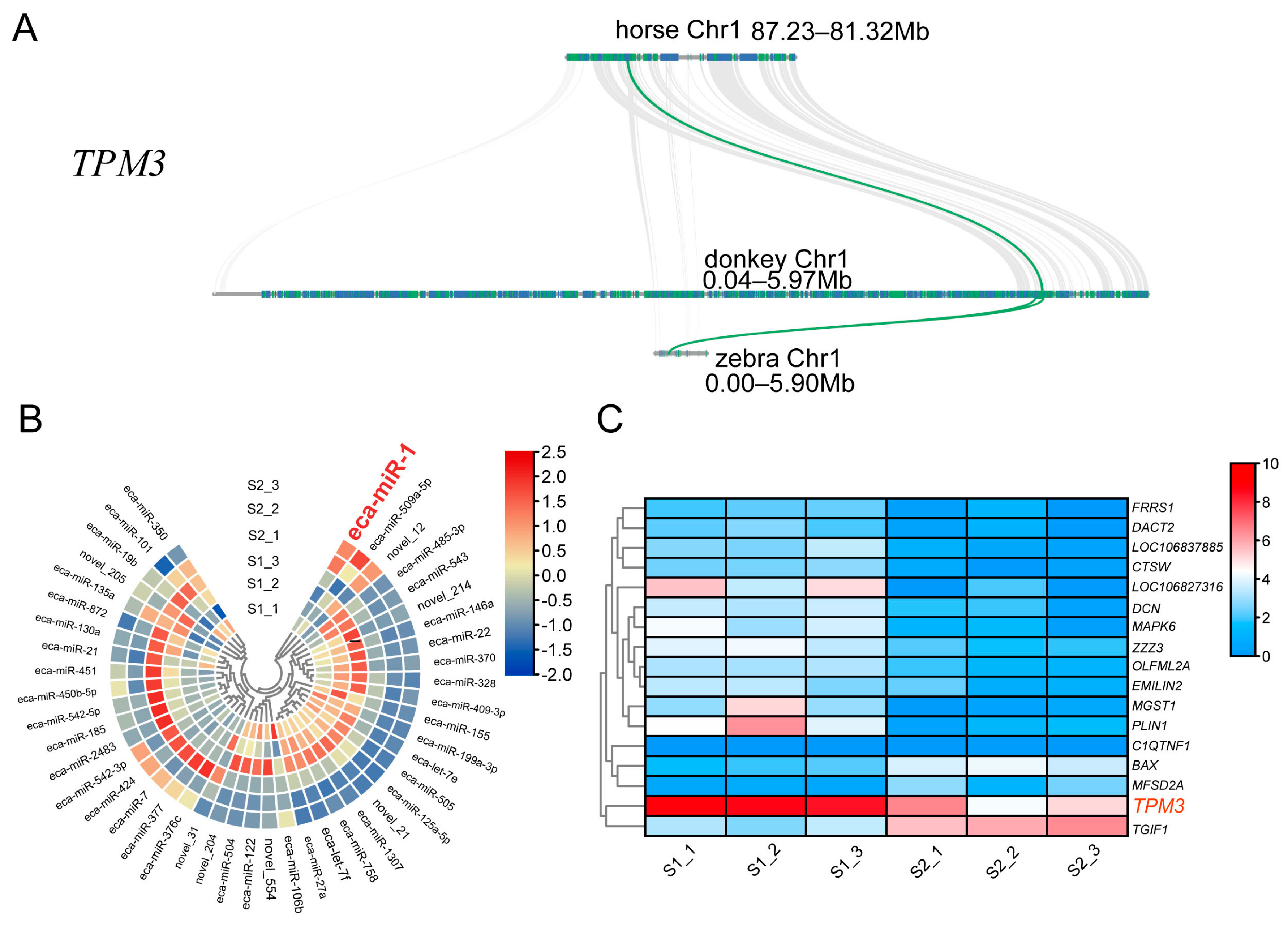
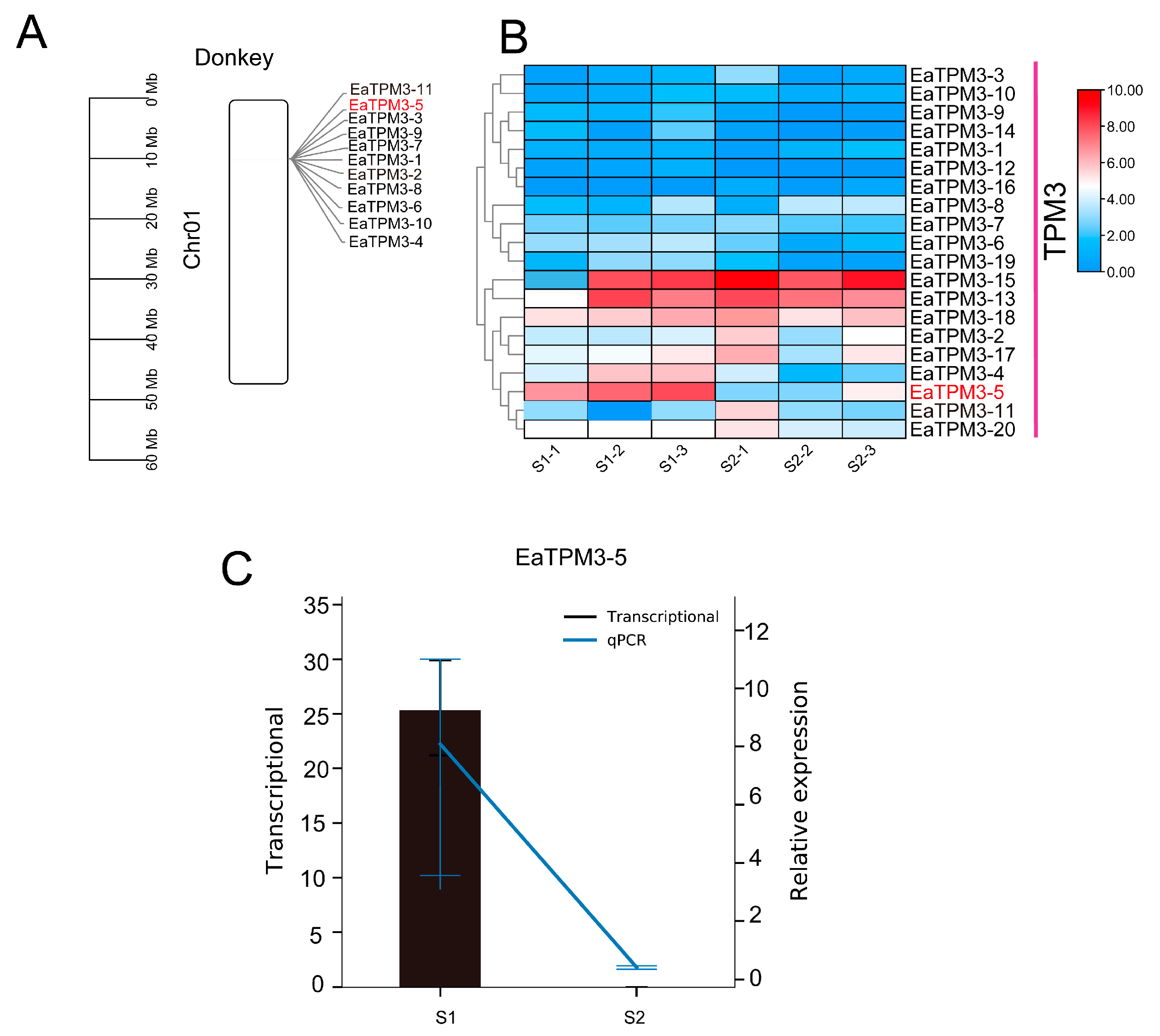
2.3. TPM3 Evolutionarily Conserved in Equus and Differentially Expressed in S1 and S2 Muscle
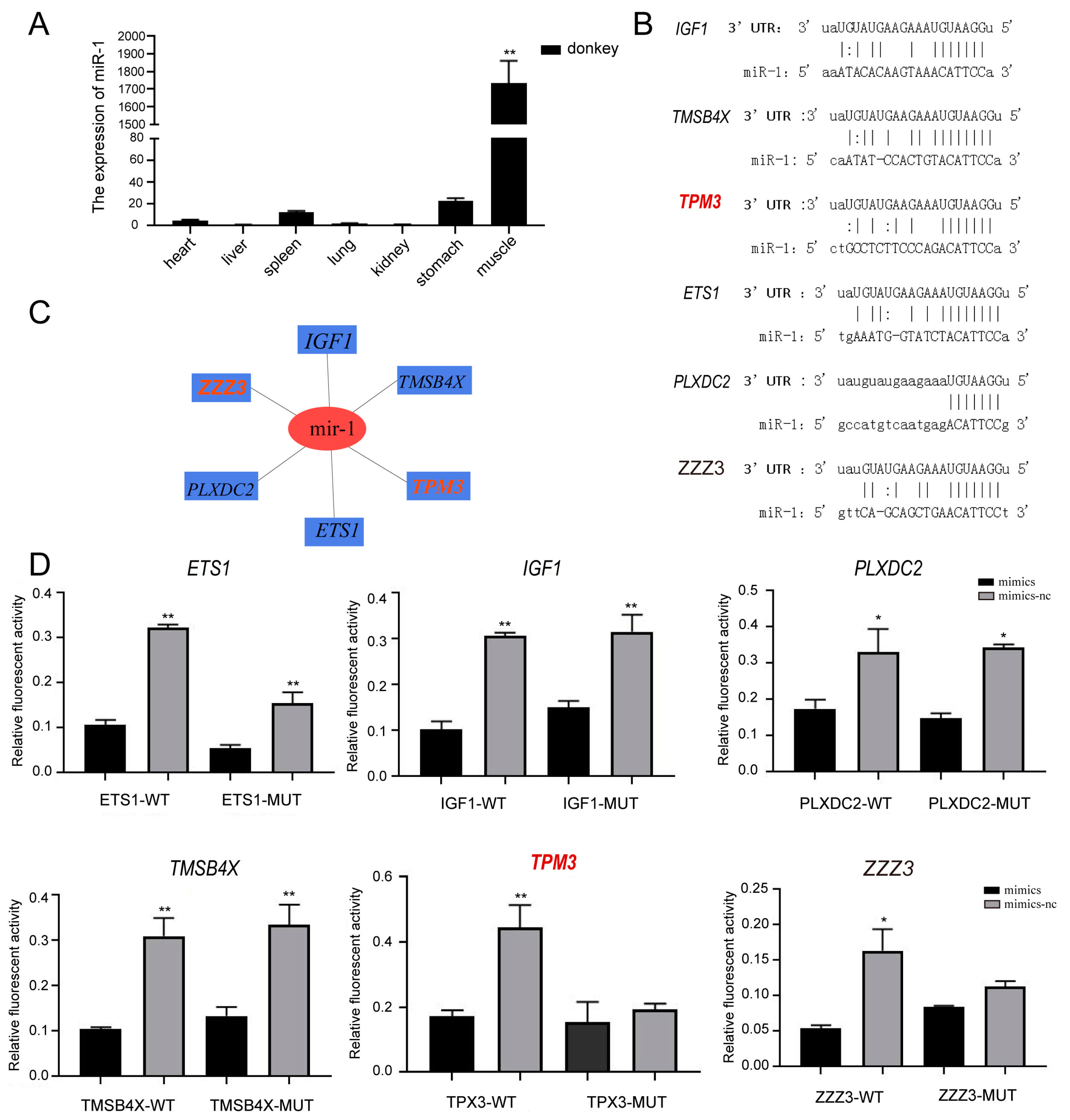
2.4. Eca-miR-1 Targeting the TPM3 Gene
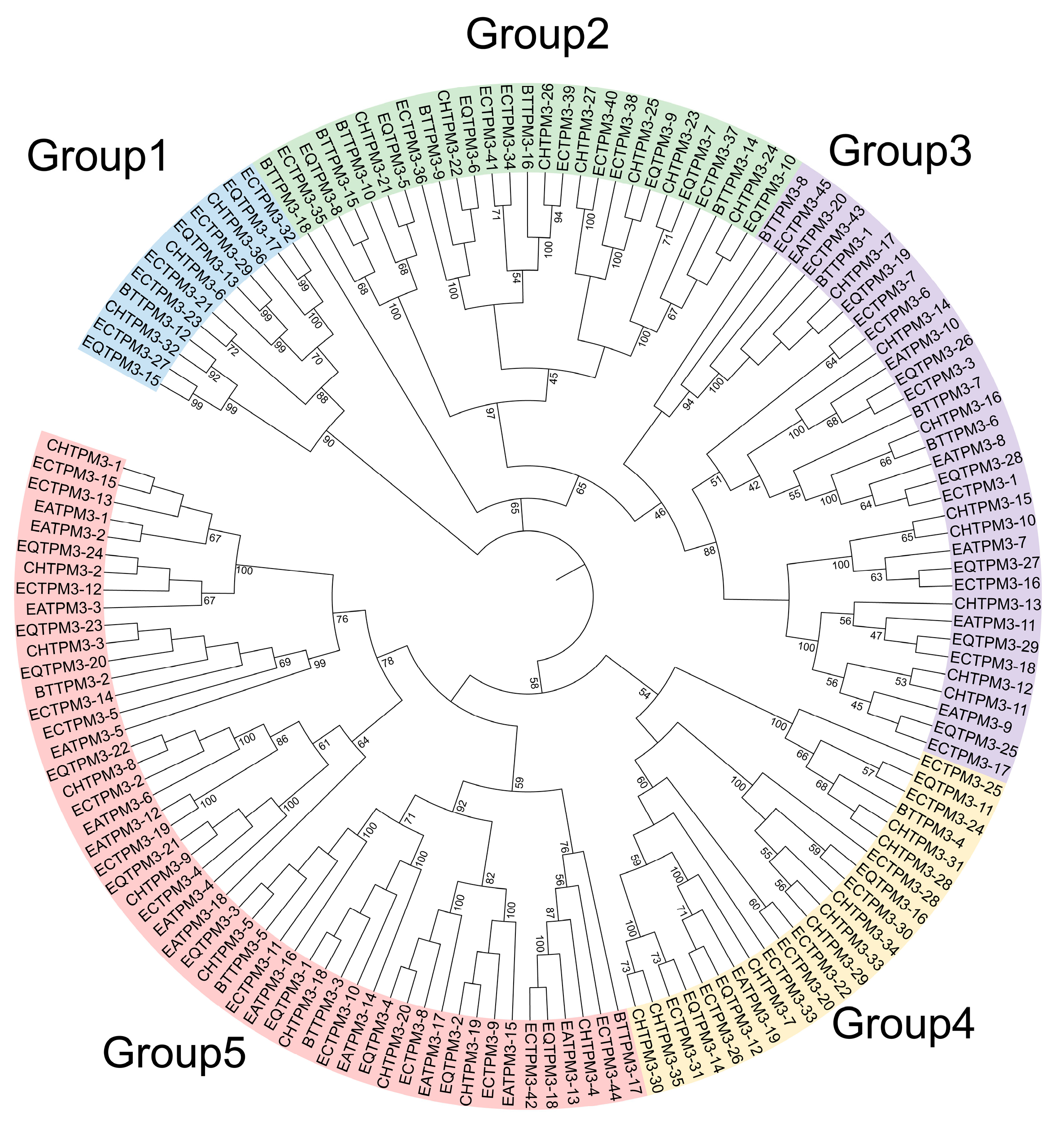
2.5. The TPM3 Gene Family Evolution and Expression Profile
3. Discussion
3.1. Comparative Genomics and Collinearity Analysis Identify Muscle Development Gene TPM3 in Equus
3.2. Comparative Transcriptomics Reveals the TPM3 Gene Potentially Involved in Muscle Development in Donkeys
3.3. TPM3 Regulates Muscle Development Targeted by eca-miR-1
4. Materials and Methods
4.1. Ethics Statement
4.2. Comparative Genomics Analysis
4.3. Chromosome Collinearity Analysis Rearrangement Analysis
4.4. Sample Collection
4.5. RNA Extraction and Sequencing Data Processing
4.6. Identification of the miRNAs
4.7. Prediction and Validation of the miRNA Target Genes
4.8. Vector Construction, Cell Transfection, and Dual-Luciferase Reporter Assay
4.9. Gene Family Identification Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brosnahan, M.M. Genetics, Evolution, and Physiology of Donkeys and Mules. The Veterinary clinics of North America. Equine Pract. 2019, 35, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, H.; Guo, Y.; Huang, J.; Sun, Y.; Min, J.; Wang, J.; Fang, X.; Zhao, Z.; Wang, S.; et al. Donkey genomes provide new insights into domestication and selection for coat color. Nat. Commun. 2020, 11, 6014. [Google Scholar] [CrossRef] [PubMed]
- Kalbfleisch, T.S.; Rice, E.S.; DePriest, M.S., Jr.; Walenz, B.P.; Hestand, M.S.; Vermeesch, J.R.; O′connell, B.L.; Fiddes, I.T.; Vershinina, A.O.; Saremi, N.F.; et al. Improved reference genome for the domestic horse increases assembly contiguity and composition. Commun. Biol. 2018, 1, 197. [Google Scholar] [CrossRef]
- Conte, M.A.; Kocher, T.D. An improved genome reference for the African cichlid, Metriaclima zebra. BMC Genom. 2015, 16, 724. [Google Scholar] [CrossRef]
- Alföldi, J.; Lindblad-Toh, K. Comparative genomics as a tool to understand evolution and disease. Genome Res. 2013, 23, 1063–1068. [Google Scholar] [CrossRef]
- Fulton, T.M.; Hoeven, R.V.; Eannetta, N.T.; Tanksley, S.D. Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 2002, 14, 1457–1467. [Google Scholar] [CrossRef]
- Liu, Z.; Qanmber, G.; Lu, L.; Qin, W.; Liu, J.; Li, J.; Ma, S.; Yang, Z.; Yang, Z. Genome-wide analysis of BES1 genes in Gossypium revealed their evolutionary conserved roles in brassinosteroid signaling. Sci. China Life Sci. 2018, 61, 1566–1582. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cui, Y.; Chen, Y.; Zhang, D.; Liang, Y.; Zhang, D.; Wu, Q.; Xie, J.; Ouyang, S.; Li, D.; et al. Comparative genetic mapping and genomic region collinearity analysis of the powdery mildew resistance gene Pm41. Theor. Appl. Genet. 2014, 127, 1741–1751. [Google Scholar] [CrossRef]
- Cappai, M.G.; Picciau, M.; Pinna, W. An integrated approach towards the nutritional assessment of the Sardinian donkey: A tool for clinical nutritionists. Ital. J. Anim. Sci. 2013, 12, 182–185. [Google Scholar] [CrossRef]
- Yang, G.; Wu, M.; Liu, X.; Wang, F.; Li, M.; An, X.; Bai, F.; Lei, C.; Dang, R. MiR-24-3p conservatively regulates muscle cell proliferation and apoptosis by targeting common gene camk2b in rat and cattle. Animals 2022, 12, 505. [Google Scholar] [CrossRef]
- Jing, L.; Hou, Y.; Wu, H.; Miao, Y.; Li, X.; Cao, J.; Brameld, J.M.; Parr, T.; Zhao, S. Transcriptome analysis of mRNA and miRNA in skeletal muscle indicates an important network for differential Residual Feed Intake in pigs. Sci. Rep. 2015, 5, 11953. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Q.; Chamba, Y.; Zhang, B.; Shang, P.; Zhang, H.; Wu, C. Identification of genes related to growth and lipid deposition from transcriptome profiles of pig muscle tissue. PLoS ONE 2015, 10, e0141138. [Google Scholar] [CrossRef]
- Liu, J.; Li, F.; Hu, X.; Cao, D.; Liu, W.; Han, H.; Zhou, Y.; Lei, Q. Deciphering the miRNA transcriptome of breast muscle from the embryonic to post-hatching periods in chickens. BMC Genom. 2021, 22, 64. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, M.; Shan, Y.; Ji, G.; Ju, X.; Tu, Y.; Sheng, Z.; Xie, J.; Zou, J.; Shu, J. miRNA-mRNA network regulation in the skeletal muscle fiber phenotype of chickens revealed by integrated analysis of miRNAome and transcriptome. Sci. Rep. 2020, 10, 10619. [Google Scholar] [CrossRef] [PubMed]
- Dube, D.K.; Dube, S.; Abbott, L.; Elsekaily, O.; Sanger, J.W.; Sanger, J.M.; Poiesz, B.J. Sarcomeric TPM3 expression in human heart and skeletal muscle. Cytoskeleton 2020, 77, 313–328. [Google Scholar] [CrossRef]
- Matyushenko, A.M.; Nefedova, V.V.; Shchepkin, D.V.; Kopylova, G.V.; Berg, V.Y.; Pivovarova, A.V.; Kleymenov, S.Y.; Bershitsky, S.Y.; Levitsky, D.I. Mechanisms of disturbance of the contractile function of slow skeletal muscles induced by myopathic mutations in the tropomyosin TPM3 gene. FASEB J. 2020, 34, 13507–13520. [Google Scholar] [CrossRef] [PubMed]
- Gonchar, A.D.; Kopylova, G.V.; Kochurova, A.M.; Berg, V.Y.; Shchepkin, D.V.; Koubasova, N.A.; Tsaturyan, A.K.; Kleymenov, S.Y.; Matyushenko, A.M.; Levitsky, D.I. Effects of myopathy-causing mutations R91P and R245G in the TPM3 gene on structural and functional properties of slow skeletal muscle tropomyosin. Biochem. Biophys. Res. Commun. 2021, 534, 8–13. [Google Scholar] [CrossRef]
- Chai, W.; Qu, H.; Ma, Q.; Zhu, M.; Li, M.; Zhan, Y.; Liu, Z.; Xu, J.; Yao, H.; Li, Z.; et al. RNA-seq analysis identifies differentially expressed gene in different types of donkey skeletal muscles. Anim. Biotechnol. 2023, 34, 1786–1795. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Hu, W.; Hou, H.; Zhao, Z.; Shang, M.; Zhang, L. Identification and comparative analysis of long non-coding rna in the skeletal muscle of two dezhou donkey strains. Genes 2020, 11, 508. [Google Scholar] [CrossRef]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5’-UTR and 3’-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef]
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008, 22, 3172–3183. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Donadeu, F.X.; Sanchez, J.M.; Mohammed, B.T.; Ioannidis, J.; Stenhouse, C.; Maioli, M.A.; Esteves, C.L.; Lonergan, P. Relationships between size, steroidogenesis and miRNA expression of the bovine corpus luteum. Theriogenology 2020, 145, 226–230. [Google Scholar] [CrossRef]
- Chen, K.; Yu, Y.; Sun, K.; Xiong, H.; Yu, C.; Chen, P.; Chen, J.; Gao, G.; Zhu, A. The miRNAome of ramie (Boehmeria nivea L.): Identification, expression, and potential roles of novel microRNAs in regulation of cadmium stress response. BMC Plant Biol. 2018, 18, 369. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.R.; Williams, C.M.; Yang, Z. MicroRNA-27b targets gremlin 1 to modulate fibrotic responses in pulmonary cells. J. Cell. Biochem. 2014, 115, 1539–1548. [Google Scholar] [CrossRef]
- O’Rourke, J.R.; Georges, S.A.; Seay, H.R.; Tapscott, S.J.; McManus, M.T.; Goldhamer, D.J.; Swanson, M.S.; Harfe, B.D. Essential role for Dicer during skeletal muscle development. Dev. Biol. 2007, 311, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef]
- Sabater-Arcis, M.; Bargiela, A.; Furling, D.; Artero, R. miR-7 Restores Phenotypes in Myotonic Dystrophy Muscle Cells by Repressing Hyperactivated Autophagy. Molecular therapy. Nucleic Acids 2020, 19, 278–292. [Google Scholar] [CrossRef] [PubMed]
- Pardo, P.S.; Hajira, A.; Boriek, A.M.; Mohamed, J.S. MicroRNA-434-3p regulates age-related apoptosis through eIF5A1 in the skeletal muscle. Aging 2017, 9, 1012–1029. [Google Scholar] [CrossRef]
- Ge, Y.; Sun, Y.; Chen, J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol. 2011, 192, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, M.; Zhou, P.; Yan, H.; Zhang, Z.; Zhang, H.; Qi, R.; Liu, J. β-Hydroxy-β-methylbutyrate-Induced Upregulation of miR-199a-3p Contributes to Slow-To-Fast Muscle Fiber Type Conversion in Mice and C2C12 Cells. J. Agric. Food Chem. 2020, 68, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; He, H.B.; Zhang, W.Y.; Zhang, H.X.; Bai, J.B.; Liu, H.Z.; Cao, J.H.; Chang, K.C.; Li, X.Y.; Zhao, S.H. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013, 4, e668. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, H.; Yu, C.; Lu, R.; Song, T.; Wang, X.; Tang, W.; Gao, Y. MiR-638 Repressed Vascular Smooth Muscle Cell Glycolysis by Targeting LDHA. Open Med. 2019, 14, 663–672. [Google Scholar] [CrossRef]
- Kang, X.; Cao, S.; Ji, Z.; Zhang, Y.; Sun, S.; Shang, X. miR-3646 promotes vascular inflammation and augments vascular smooth muscle cell proliferation and migration in progression of coronary artery disease by directly targeting RHOH. Int. J. Clin. Exp. Pathol. 2018, 11, 5830–5839. [Google Scholar]
- Bao, Q.; Jia, H.; Rong, A.; Cao, Z.; Zhang, Y. MiR-210 inhibits hypoxia-induced apoptosis of smooth muscle cells via targeting MEF2C. Int. J. Clin. Exp. Pathol. 2019, 12, 1846–1858. [Google Scholar]
- Insausti, K.; Beldarrain, L.R.; Lavín, M.P.; Aldai, N.; Mantecón, Á.R.; Sáez, J.L.; Canals, R.M. Horse meat production in northern Spain: Ecosystem services and sustainability in High Nature Value farmland. Anim. Front. Rev. Mag. Anim. Agric. 2021, 11, 47–54. [Google Scholar] [CrossRef]
- Polidori, P.; Cammertoni, N.; Santini, G.; Klimanova, Y.; Zhang, J.J.; Vincenzetti, S. Effects of Donkeys Rearing System on Performance Indices, Carcass, and Meat Quality. Foods 2021, 10, 3119. [Google Scholar] [CrossRef]
- Valberg, S.J.; Carlson, G.P.; Cardinet, G.H., 3rd; Birks, E.K.; Jones, J.H.; Chomyn, A.; DiMauro, S. Skeletal muscle mitochondrial myopathy as a cause of exercise intolerance in a horse. Muscle Nerve 1994, 17, 305–312. [Google Scholar] [CrossRef]
- Wang, M.; Li, W.; Chang, G.Q.; Ye, C.S.; Ou, J.S.; Li, X.X.; Liu, Y.; Cheang, T.Y.; Huang, X.L.; Wang, S.M. MicroRNA-21 regulates vascular smooth muscle cell function via targeting tropomyosin 1 in arteriosclerosis obliterans of lower extremities. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2044–2053. [Google Scholar] [CrossRef]
- Duan, Z.; Choy, E.; Nielsen, G.P.; Rosenberg, A.; Iafrate, J.; Yang, C.; Schwab, J.; Mankin, H.; Xavier, R.; Hornicek, F.J. Differential expression of microRNA (miRNA) in chordoma reveals a role for miRNA-1 in Met expression. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2010, 28, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Xiong, X.; Liu, Y.; Wang, J. miRNA-1: Functional roles and dysregulation in heart disease. Mol. Biosyst. 2014, 10, 2775–2782. [Google Scholar] [CrossRef]
- Sun, Y.; Ge, Y.; Drnevich, J.; Zhao, Y.; Band, M.; Chen, J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J. Cell Biol. 2010, 189, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Kiełbasa, S.M.; Wan, R.; Sato, K.; Horton, P.; Frith, M.C. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011, 21, 487–493. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Simms, D.; Chomczynski, P. TRIzolTM: A new reagent for optimal single-step isolation of RNA. Focus 1992, 15, 532–535. [Google Scholar]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Gouda, M.A. Common Pitfalls in Reporting the Use of SPSS Software. Med. Princ. Pract. 2015, 24, 300. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, G.; Sun, M.; Wang, Z.; Hu, Q.; Guo, J.; Yu, J.; Lei, C.; Dang, R. Comparative Genomics Identifies the Evolutionarily Conserved Gene TPM3 as a Target of eca-miR-1 Involved in the Skeletal Muscle Development of Donkeys. Int. J. Mol. Sci. 2023, 24, 15440. https://doi.org/10.3390/ijms242015440
Yang G, Sun M, Wang Z, Hu Q, Guo J, Yu J, Lei C, Dang R. Comparative Genomics Identifies the Evolutionarily Conserved Gene TPM3 as a Target of eca-miR-1 Involved in the Skeletal Muscle Development of Donkeys. International Journal of Molecular Sciences. 2023; 24(20):15440. https://doi.org/10.3390/ijms242015440
Chicago/Turabian StyleYang, Ge, Minhao Sun, Zhaofei Wang, Qiaoyan Hu, Jiajun Guo, Jie Yu, Chuzhao Lei, and Ruihua Dang. 2023. "Comparative Genomics Identifies the Evolutionarily Conserved Gene TPM3 as a Target of eca-miR-1 Involved in the Skeletal Muscle Development of Donkeys" International Journal of Molecular Sciences 24, no. 20: 15440. https://doi.org/10.3390/ijms242015440





