The Functionality of IbpA from Acholeplasma laidlawii Is Governed by Dynamic Rearrangement of Its Globular–Fibrillar Quaternary Structure
Abstract
:1. Introduction
2. Results
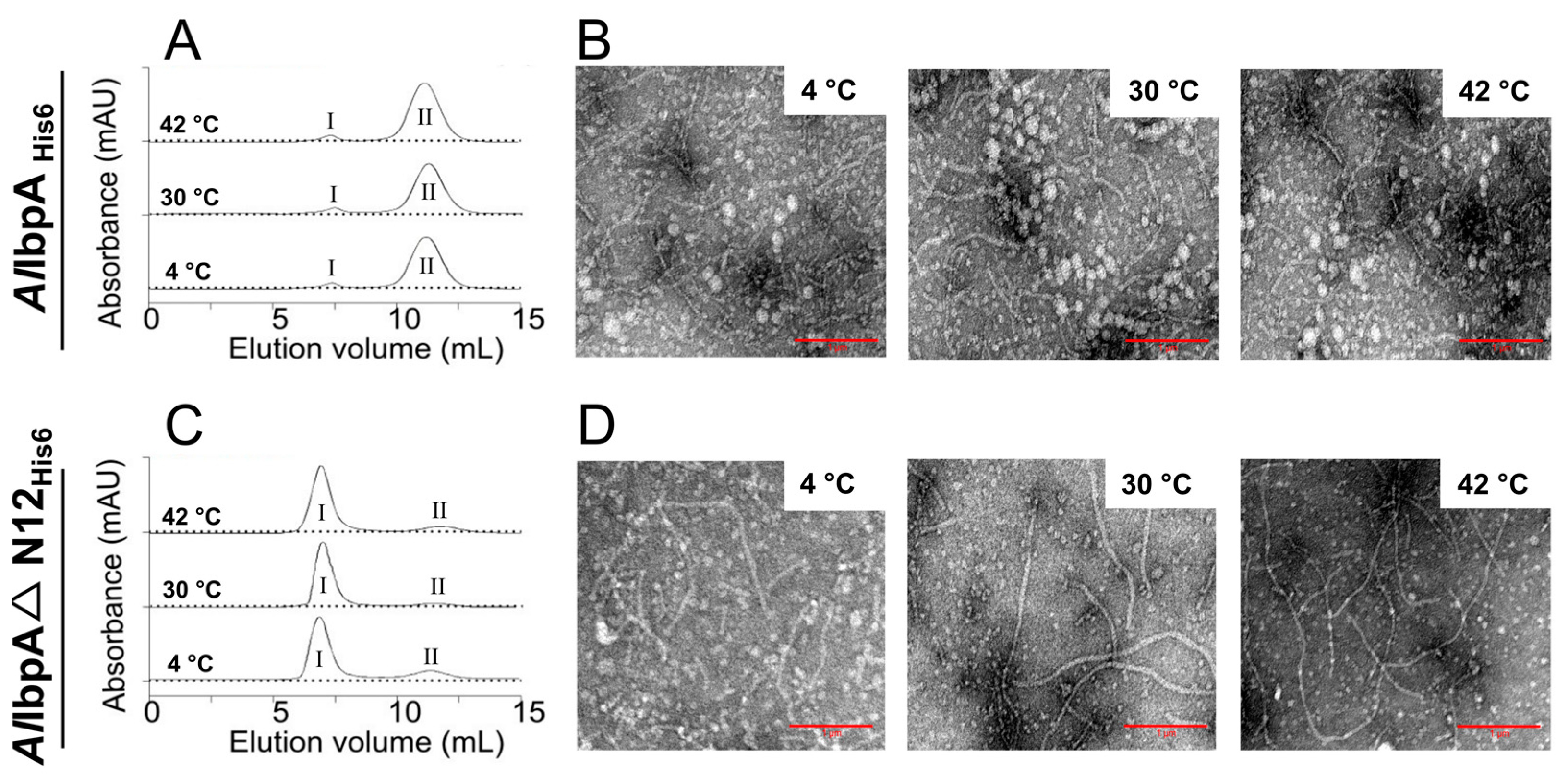
2.1. AlIbpA in Active (Substrate-Binding) Form Is Present as Fibrils In Vitro
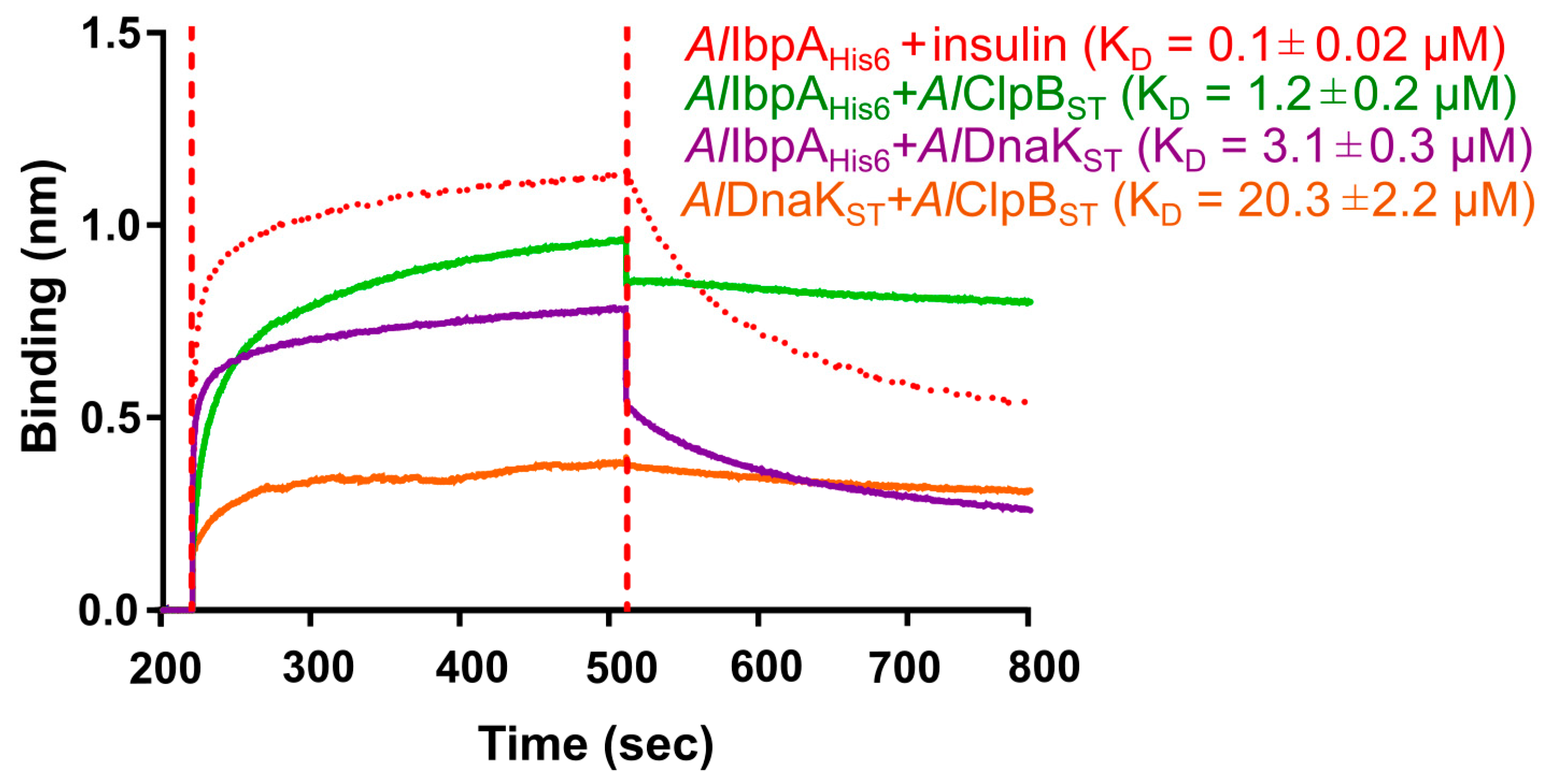
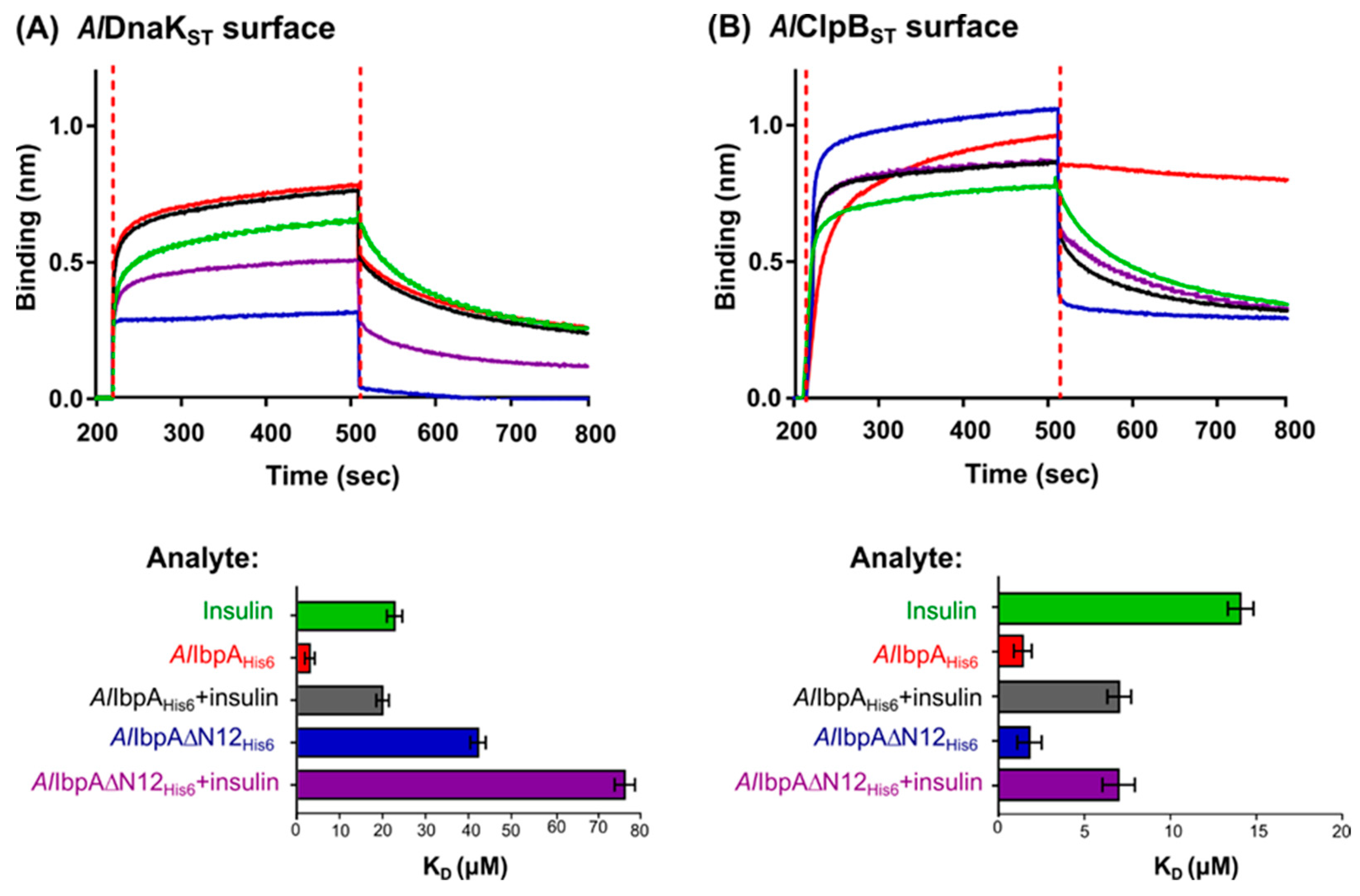
2.2. AlIbpA Interacts with AlDnaK and AlClpB In Vitro
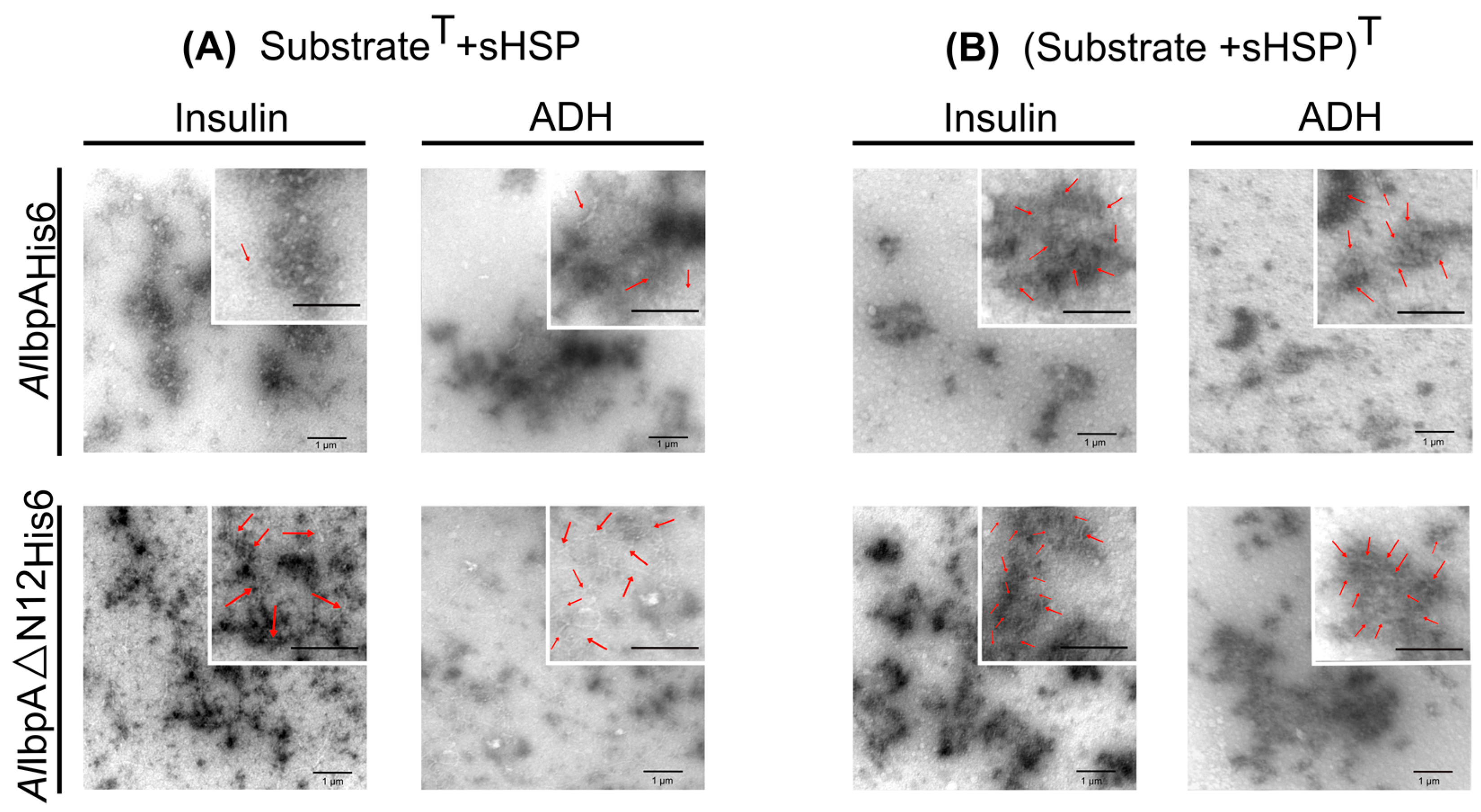
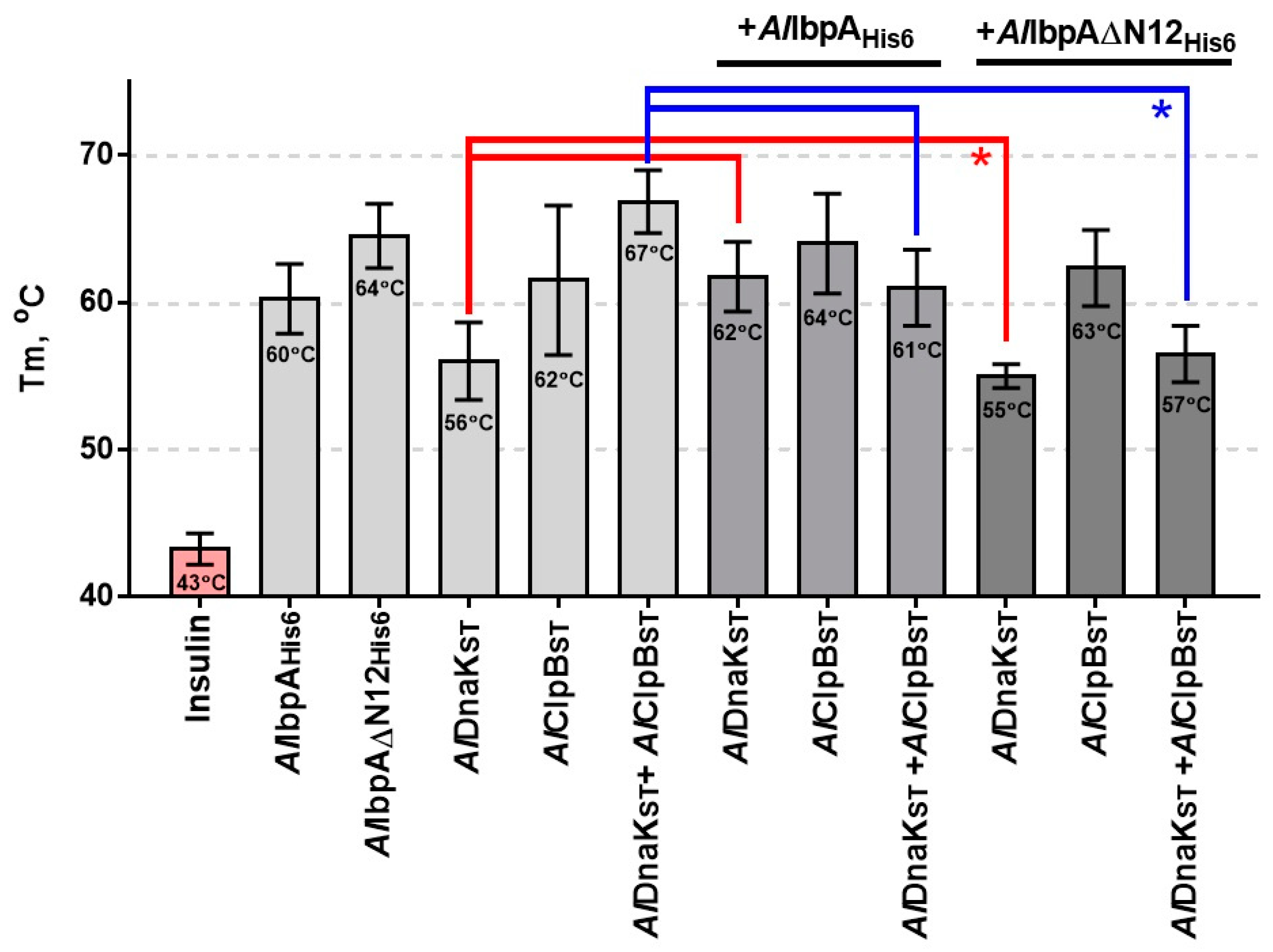
2.3. The Impact of AlIbpA in Fibrillar Form on Its Interaction with AlDnaK and AlClpB and Substrate Transfer to HSPs
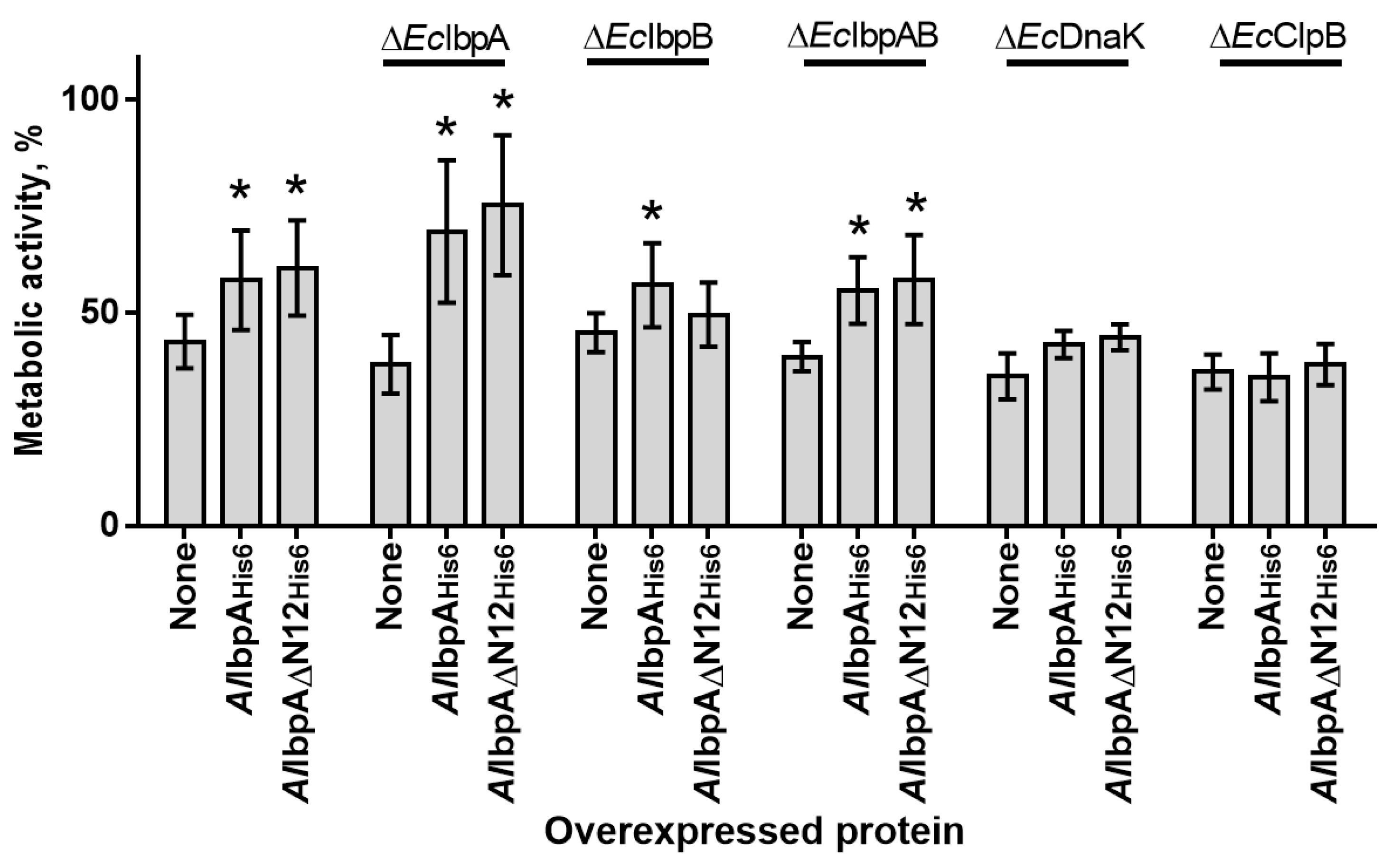
2.4. The In Vivo Assessment of the Role of HSP20, HSP70 and HSP100 in Heat Resistance of Cells
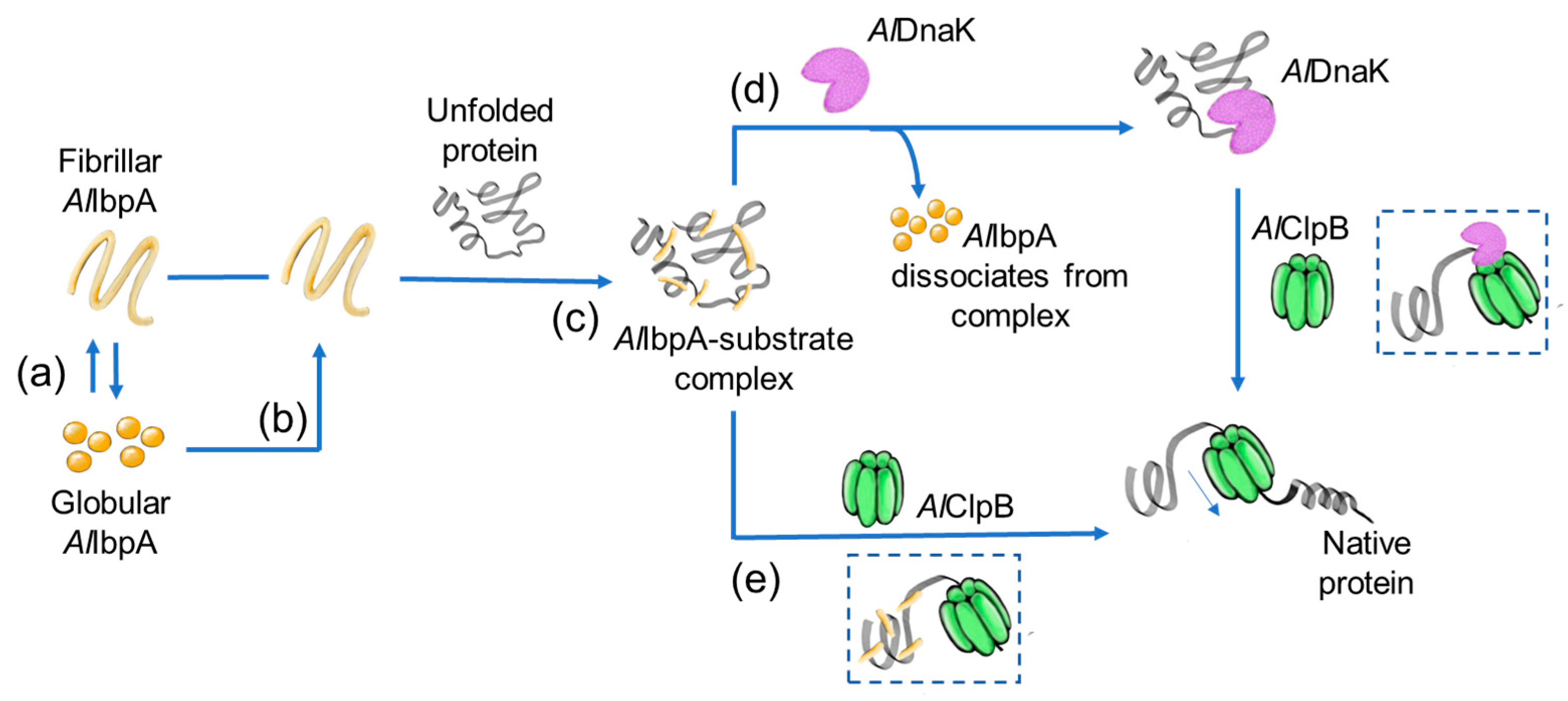
3. Discussion
4. Materials and Methods
4.1. Protein Alignment and Structure Analyses
4.2. Strains and Plasmids
4.3. Protein Purification
4.4. In Vitro Cross-Linking of Proteins
4.5. Size-Exclusion Chromatography
4.6. Pull Down Assay
4.7. Transmission Electron Microscopy
4.8. Bio-Layer Interferometry
4.9. In Vitro Chaperone-like Activity Assay
4.10. Cell Viability Test
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubrez, L.; Causse, S.; Borges Bonan, N.; Dumétier, B.; Garrido, C. Heat-shock proteins: Chaperoning DNA repair. Oncogene 2020, 39, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Jee, H. Size dependent classification of heat shock proteins: A mini-review. J. Exerc. Rehabil. 2016, 12, 255–259. [Google Scholar] [CrossRef]
- Liberek, K.; Lewandowska, A.; Zietkiewicz, S. Chaperones in control of protein disaggregation. EMBO J. 2008, 27, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Deuerling, E.; Vorderwülbecke, S.; Vierling, E.; Bukau, B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 2003, 50, 585–595. [Google Scholar] [CrossRef]
- Ul Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-X.; Zhang, H.-X.; Wei, A.-M.; Gong, Z.-H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef]
- Ehrnsperger, M.; Hergersberg, C.; Wienhues, U.; Nichtl, A.; Buchner, J. Stabilization of proteins and peptides in diagnostic immunological assays by the molecular chaperone Hsp25. Anal. Biochem. 1998, 259, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Bukau, B. Role of sHsps in organizing cytosolic protein aggregation and disaggregation. Cell Stress Chaperones 2017, 22, 493–502. [Google Scholar] [CrossRef]
- De Graff, A.M.; Mosedale, D.E.; Sharp, T.; Dill, K.A.; Grainger, D.J. Proteostasis is adaptive: Balancing chaperone holdases against foldases. PLoS Comput. Biol. 2020, 16, e1008460. [Google Scholar] [CrossRef]
- Bandyopadhyay, B.; Das Gupta, T.; Roy, D.; Das Gupta, S.K. DnaK dependence of the mycobacterial stress-responsive regulator HspR is mediated through its hydrophobic C-terminal tail. J. Bacteriol. 2012, 194, 4688–4697. [Google Scholar] [CrossRef]
- Raman, S.; Song, T.; Puyang, X.; Bardarov, S.; Jacobs, W.R.; Husson, R.N. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 2001, 183, 6119–6125. [Google Scholar] [CrossRef]
- Ullers, R.S.; Luirink, J.; Harms, N.; Schwager, F.; Georgopoulos, C.; Genevaux, P. SecB is a bona fide generalized chaperone in Escherichia coli. Proc. Natl. Acad. Sci. USA 2004, 101, 7583–7588. [Google Scholar] [CrossRef] [PubMed]
- Vorderwülbecke, S.; Kramer, G.; Merz, F.; Kurz, T.A.; Rauch, T.; Zachmann-Brand, B.; Bukau, B.; Deuerling, E. Low temperature of GroEL/ES overproduction permits growth of Escherichia coli cells lacking trigger factor DnaK. FEBS Lett. 2005, 579, 181–187. [Google Scholar] [CrossRef]
- Målen, H.; Berven, F.S.; Fladmark, K.E.; Wiker, H.G. Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 2007, 7, 1702–1718. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.R.; Snewin, V.A.; Walzl, G.; Hussell, T.; Tormay, P.; O’Gaora, P.; Goyal, M.; Betts, J.; Brown, I.N.; Young, D.B. Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat. Med. 2001, 7, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Franck, E.; Madsen, O.; van Rheede, T.; Ricard, G.; Huynen, M.A.; de Jong, W.W. Evolutionary diversity of vertebrate small heat shock proteins. J. Mol. Evol. 2004, 59, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Basha, E.; O’Neill, H.; Vierling, E. Small heat shock proteins and α-crystallins: Dynamic proteins with flexible functions. Trends Biochem. Sci. 2012, 37, 106–117. [Google Scholar] [CrossRef]
- Lindner, R.A.; Carver, J.A.; Ehrnsperger, M.; Buchner, J.; Esposito, G.; Behlke, J.; Lutsch, G.; Kotlyarov, A.; Gaestel, M. Mouse Hsp25, a small shock protein. The role of its C-terminal extension in oligomerization and chaperone action. Eur. J. Biochem. 2000, 267, 1923–1932. [Google Scholar] [CrossRef]
- Strózecka, J.; Chrusciel, E.; Górna, E.; Szymanska, A.; Ziętkiewicz, S.; Liberek, K. Importance of N- and C-terminal regions of IbpA, Escherichia coli small heat shock protein, for chaperone function and oligomerization. J. Biol. Chem. 2012, 287, 2843–2853. [Google Scholar] [CrossRef]
- Sun, Y.; MacRae, T.H. Small heat shock proteins: Molecular structure and chaperone function. Cell Mol. Life Sci. 2005, 62, 2460–2476. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, V.; Houry, W.A. Substrate interaction networks of the Escherichia coli chaperones: Trigger factor, DnaK and GroEL. Adv. Exp. Med. Biol. 2015, 883, 271–294. [Google Scholar] [CrossRef]
- Clerico, E.M.; Tilitsky, J.M.; Meng, W.; Gierasch, L.M. How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 2015, 427, 1575–1588. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Gierasch, L.M. Correction: Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 2020, 295, 288. [Google Scholar] [CrossRef]
- Kravats, A.N.; Doyle, S.M.; Hoskins, J.R.; Genest, O.; Doody, E.; Wickner, S. Interaction of E. coli Hsp90 with DnaK Involves the DnaJ Binding Region of DnaK. J. Mol. Biol. 2017, 429, 858–872. [Google Scholar] [CrossRef]
- Mayer, M.P. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013, 38, 507–514. [Google Scholar] [CrossRef]
- Rüdiger, S.; Buchberger, A.; Bukau, B. Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 1997, 4, 342–349. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Farber, P.; Velyvis, A.; Rennella, E.; Latham, M.P.; Kay, L.E. ClpB N-terminal domain plays a regulatory role in protein disaggregation. Proc. Natl. Acad. Sci. USA 2015, 112, E6872–E6881. [Google Scholar] [CrossRef]
- Squires, C.L.; Pedersen, S.; Ross, B.M.; Squires, C. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 1991, 173, 4254–4262. [Google Scholar] [CrossRef]
- Goloubinoff, P.; Mogk, A.; Zvi, A.P.; Tomoyasu, T.; Bukau, B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. USA 1999, 96, 13732–13737. [Google Scholar] [CrossRef]
- Hanson, P.I.; Whiteheart, S.W. AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 2005, 6, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Hodson, S.; Marshall, J.J.T.; Burston, S.G. Mapping the road to recovery: The ClpB/Hsp104 molecular chaperone. J. Struct. Biol. 2012, 179, 161–171. [Google Scholar] [CrossRef]
- Neuwald, A.F.; Aravind, L.; Spouge, J.L.; Koonin, E.V. AAA+: A Class of Chaperone-Like ATPases Associated with the Assembly, Operation, and Disassembly of Protein Complexes. Genome Res. 1999, 9, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Zolkiewski, M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J. Biol. Chem. 1999, 274, 28083–28086. [Google Scholar] [CrossRef] [PubMed]
- Zolkiewski, M.; Zhang, T.; Nagy, M. Aggregate reactivation mediated by the Hsp100 chaperones. Arch. Biochem. Biophys. 2012, 520, 1–6. [Google Scholar] [CrossRef]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef]
- Doyle, S.M.; Shorter, J.; Zolkiewski, M.; Hoskins, J.R.; Lindquist, S.; Wickner, S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat. Struct. Mol. Biol. 2007, 14, 114–122. [Google Scholar] [CrossRef]
- Zhang, T.; Kedzierska-Mieszkowska, S.; Liu, H.; Cheng, C.; Ganta, R.R.; Zolkiewski, M. Aggregate-reactivation activity of the molecular chaperone ClpB from Ehrlichia chaffeensis. PLoS ONE 2013, 8, e62454. [Google Scholar] [CrossRef]
- Miot, M.; Reidy, M.; Doyle, S.M.; Hoskins, J.R.; Johnston, D.M.; Genest, O.; Vitery, M.-C.; Masison, D.C.; Wickner, S. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc. Natl. Acad. Sci. USA 2011, 108, 6915–6920. [Google Scholar] [CrossRef] [PubMed]
- Schlee, S.; Beinker, P.; Akhrymuk, A.; Reinstein, J. A chaperone network for the resolubilization of protein aggregates: Direct interaction of ClpB and DnaK. J. Mol. Biol. 2004, 336, 275–285. [Google Scholar] [CrossRef]
- Obuchowski, I.; Piróg, A.; Stolarska, M.; Tomiczek, B.; Liberek, K. Duplicate divergence of two bacterial small heat shock proteins reduces the demand for Hsp70 in refolding of substrates. PLoS Genet. 2019, 15, e1008479. [Google Scholar] [CrossRef]
- Ratajczak, E.; Zietkiewicz, S.; Liberek, K. Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J. Mol. Biol. 2009, 386, 178–189. [Google Scholar] [CrossRef]
- Bissonnette, S.A.; Rivera-Rivera, I.; Sauer, R.T.; Baker, T.A. The IbpA and IbpB small heat-shock proteins are substrates of the AAA+ Lon protease. Mol. Microbiol. 2010, 75, 1539–1549. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E. A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Ruger-Herreros, C.; Bukau, B. Cellular functions and mechanisms of action of small heat shock proteins. Annu. Rev. Microbiol. 2019, 73, 89–110. [Google Scholar] [CrossRef]
- Żwirowski, S.; Kłosowska, A.; Obuchowski, I.; Nillegoda, N.B.; Piróg, A.; Ziętkiewicz, S.; Bukau, B.; Mogk, A.; Liberek, K. Hsp70 displaces small heat shock proteins from aggregates to initiate protein refolding. EMBO J. 2017, 36, 783–796. [Google Scholar] [CrossRef]
- Vishnyakov, I.E.; Borchsenius, S.N. Mycoplasma heat shock proteins and their genes. Microbiology 2013, 82, 643–659. [Google Scholar] [CrossRef]
- Vishnyakov, I.E.; Levitskii, S.A.; Manuvera, V.A.; Lazarev, V.N.; Ayala, J.A.; Ivanov, V.A.; Snigirevskaya, E.S.; Komissarchik, Y.Y.; Borchsenius, S.N. The identification and characterization of IbpA, a novel α-crystallin-type heat shock protein from mycoplasma. Cell Stress Chaperones 2012, 17, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Chernova, L.S.; Bogachev, M.I.; Chasov, V.V.; Vishnyakov, I.E.; Kayumov, A.R. N- and C-terminal regions of the small heat shock protein IbpA from Acholeplasma laidlawii competitively govern its oligomerization pattern and chaperone-like activity. RSC Adv. 2020, 10, 8364–8376. [Google Scholar] [CrossRef] [PubMed]
- Bukach, O.V.; Seit-Nebi, A.S.; Marston, S.B.; Gusev, N.B. Some properties of human small heat shock protein Hsp20 (HspB6). Eur. J. Biochem. 2004, 271, 291–302. [Google Scholar] [CrossRef]
- Horwitz, J.; Huang, Q.L.; Ding, L.; Bova, M.P. Lens α-crystallin: Chaperone-like properties. Methods Enzymol. 1998, 290, 365–383. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.K.; Kumar, R.S.; Kumar, G.S.; Quinn, P.T. Synthesis and characterization of a peptide identified as a functional element in αA-crystallin. J. Biol. Chem. 2000, 275, 3767–3771. [Google Scholar] [CrossRef] [PubMed]
- Haslbeck, M.; Weinkauf, S.; Buchner, J. Small heat shock proteins: Simplicity meets complexity. J. Biol. Chem. 2019, 294, 2121–2132. [Google Scholar] [CrossRef]
- Ratajczak, E.; Strózecka, J.; Matuszewska, M.; Zietkiewicz, S.; Kuczyńska-Wiśnik, D.; Laskowska, E.; Liberek, K. IbpA the small heat shock protein from Escherichia coli forms fibrils in the absence of its cochaperone IbpB. FEBS Lett. 2010, 584, 2253–2257. [Google Scholar] [CrossRef]
- Kitagawa, M.; Matsumura, Y.; Tsuchido, T. Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol. Lett. 2000, 184, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Armenta, S.; Sánchez-Cuapio, Z.; Farrés, A.; Manoutcharian, K.; Hernandez-Santoyo, A.; Sánchez, S.; Rodríguez-Sanoja, R. Data concerning secondary structure and alpha-glucans-binding capacity of the LaCBM26. Data Brief. 2018, 21, 1944–1949. [Google Scholar] [CrossRef]
- Edelmann, D.; Berghoff, B.A. Type I toxin-dependent generation of superoxide affects the persister life cycle of Escherichia coli. Sci. Rep. 2019, 9, 14256. [Google Scholar] [CrossRef] [PubMed]
- Hauf, K.; Kayumov, A.; Gloge, F.; Forchhammer, K. The molecular basis of tnra control by glutamine synthetase in Bacillus subtilis. J. Biol. Chem. 2016, 291, 3483–3495. [Google Scholar] [CrossRef]
- Guérout-Fleury, A.M.; Frandsen, N.; Stragier, P. Plasmids for ectopic integration in Bacillus subtilis. Gene 1996, 180, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Costantino, N.; Court, D.L. A set of recombineering plasmids for gram-negative bacteria. Gene 2006, 379, 109–115. [Google Scholar] [CrossRef] [PubMed]

| Score | emPAI | |||||
|---|---|---|---|---|---|---|
| 4 °C | 30 °C | 42 °C | 4 °C | 30 °C | 42 °C | |
| AlDnaK | 465 | 1508 | 1412 | 0.47 | 0.73 | 0.83 |
| AlClpB | 222 | 532 | 11,703 | 0.09 | 0.31 | 2.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernova, L.S.; Vishnyakov, I.E.; Börner, J.; Bogachev, M.I.; Thormann, K.M.; Kayumov, A.R. The Functionality of IbpA from Acholeplasma laidlawii Is Governed by Dynamic Rearrangement of Its Globular–Fibrillar Quaternary Structure. Int. J. Mol. Sci. 2023, 24, 15445. https://doi.org/10.3390/ijms242015445
Chernova LS, Vishnyakov IE, Börner J, Bogachev MI, Thormann KM, Kayumov AR. The Functionality of IbpA from Acholeplasma laidlawii Is Governed by Dynamic Rearrangement of Its Globular–Fibrillar Quaternary Structure. International Journal of Molecular Sciences. 2023; 24(20):15445. https://doi.org/10.3390/ijms242015445
Chicago/Turabian StyleChernova, Liliya S., Innokentii E. Vishnyakov, Janek Börner, Mikhail I. Bogachev, Kai M. Thormann, and Airat R. Kayumov. 2023. "The Functionality of IbpA from Acholeplasma laidlawii Is Governed by Dynamic Rearrangement of Its Globular–Fibrillar Quaternary Structure" International Journal of Molecular Sciences 24, no. 20: 15445. https://doi.org/10.3390/ijms242015445
APA StyleChernova, L. S., Vishnyakov, I. E., Börner, J., Bogachev, M. I., Thormann, K. M., & Kayumov, A. R. (2023). The Functionality of IbpA from Acholeplasma laidlawii Is Governed by Dynamic Rearrangement of Its Globular–Fibrillar Quaternary Structure. International Journal of Molecular Sciences, 24(20), 15445. https://doi.org/10.3390/ijms242015445







