Optimizing Eco-Friendly Degradation of Polyvinyl Chloride (PVC) Plastic Using Environmental Strains of Malassezia Species and Aspergillus fumigatus
Abstract
:1. Introduction
2. Results
2.1. Isolation and Screening of Plastic Degrading Fungi
2.2. Depolymerase and Lipase Enzyme Activities (Quantitative Method)
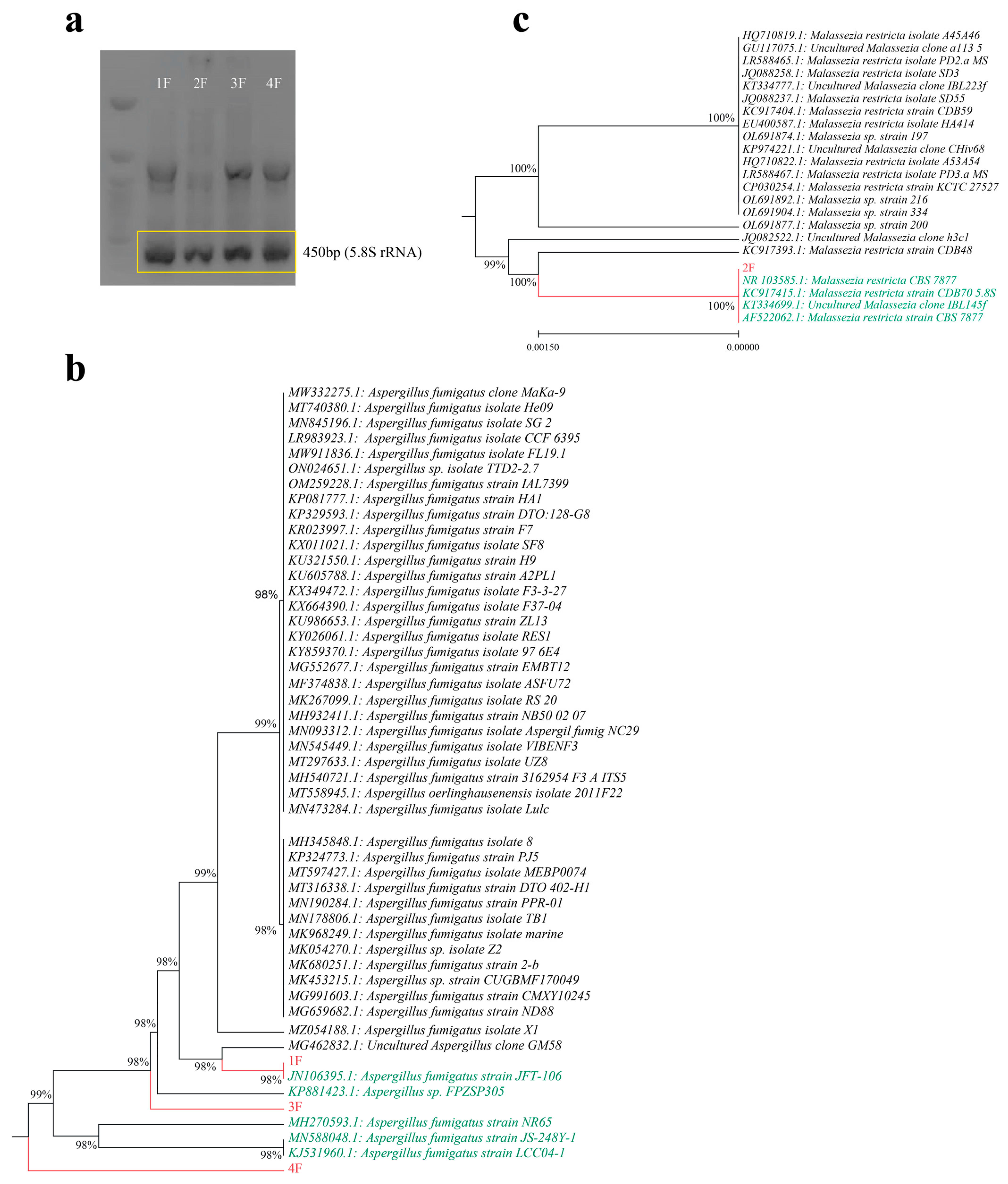
2.3. Phenotypic Identification, Molecular Identification and Phylogenic Analysis
2.4. Hydrophobicity Assay
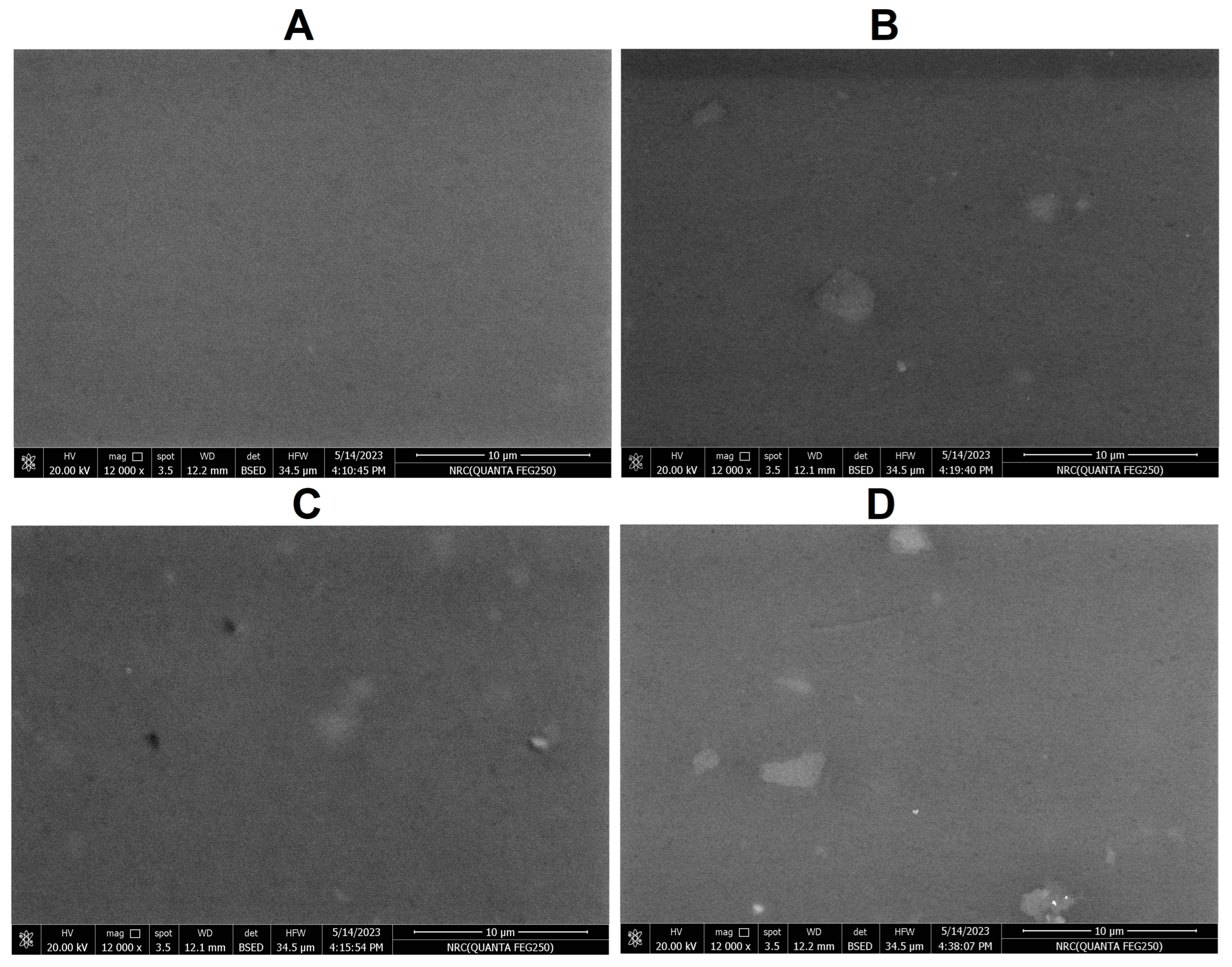
2.5. Weight Reduction Analysis and SEM Analysis of PVC Strips
2.6. Effect of pH and Temperature on Fungal Growth
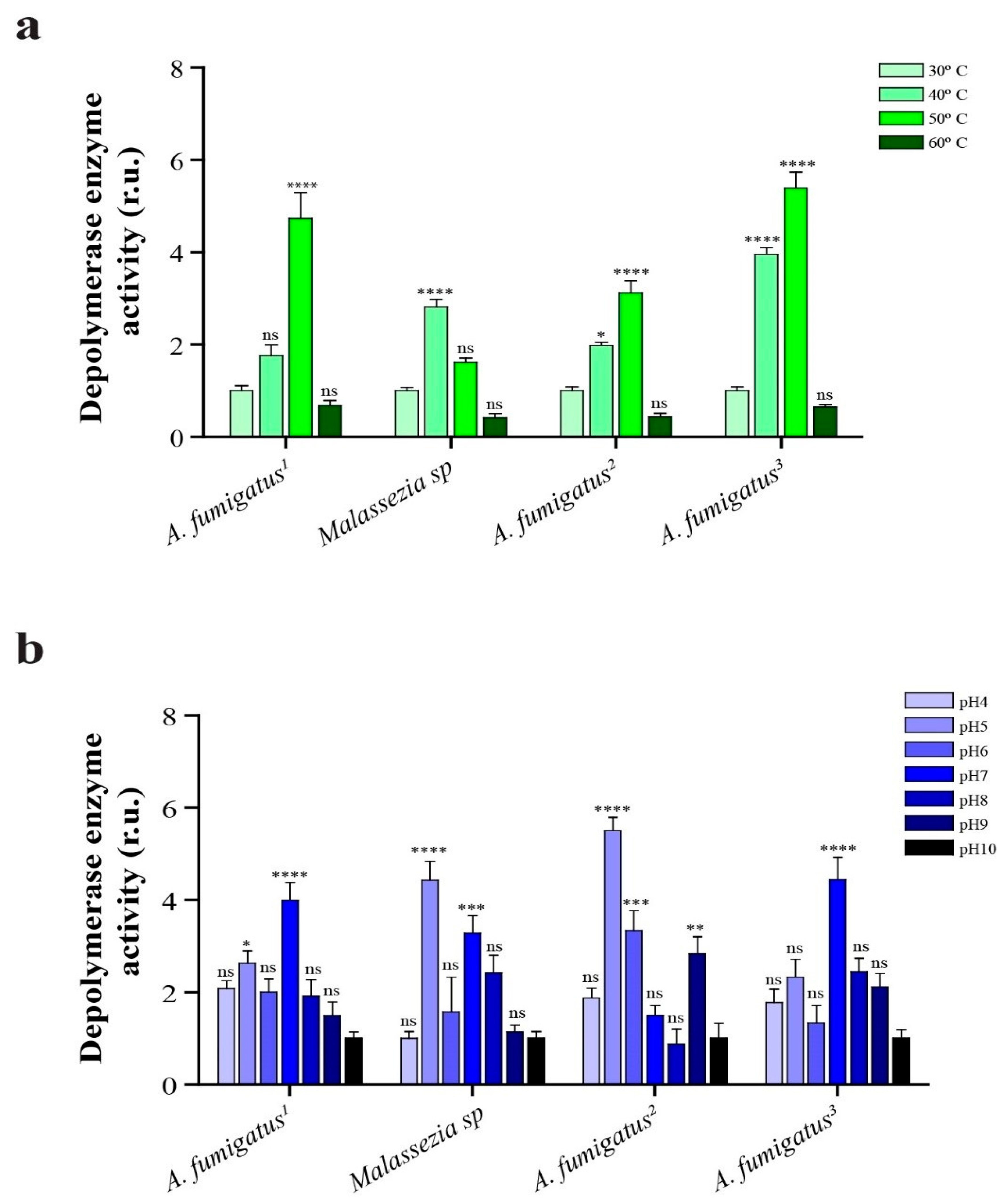
2.7. Depolymerase Enzyme Activity at Different Temperatures and pH
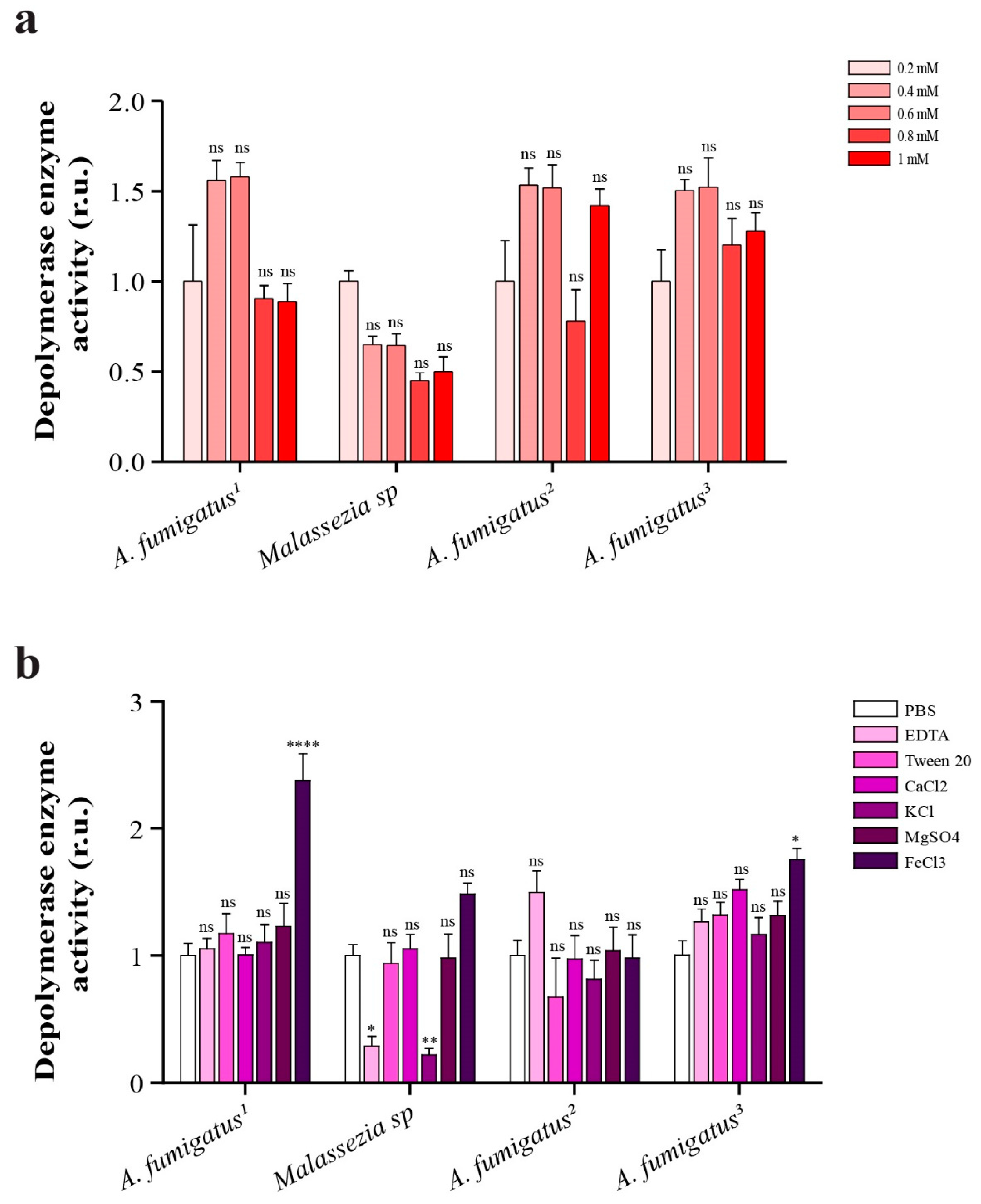
2.8. Depolymerase Enzyme Activity in Presence of Different Concentrations of EDTA
2.9. Comparison of Depolymerase Enzyme Activity in Presence of 1 mM (EDTA, Ca2+, K1+, Mg2+, Fe3+) and 1% v/v Tween 80
3. Discussion
4. Materials and Methods
4.1. Polymer Preparation
4.2. Plastic Degrading Isolates
4.3. Qualitative Assay of Plastic Biodegradation (Zone of Clearance Method)
4.4. Fungal Spore Hydrophobicity Assay
4.5. Quantitative Determination of Depolymerase Enzyme Activity
4.6. Quantitative Determination of Esterase and Lipase Enzyme Activities
4.7. Molecular Identification of Isolates via 5.8S-rRNA Sequencing and Phylogenetic Analysis
4.8. Analysis of Plastic Biodegradation via the Weight Reduction Method and Scanning Electron Microscope (SEM)
4.9. Effect of pH and Temperature on Fungal Growth
4.10. Effect of Temperature and pH on Depolymerase Enzyme Activity
4.11. Effect of Different Concentrations of EDTA, Different Ions and Tween 80 on Depolymerase Enzyme Activity
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andrady, A.L.; Neal, M.A. Applications and societal benefits of plastics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 1977–1984. [Google Scholar] [CrossRef]
- Ghaffar, I.; Rashid, M.; Akmal, M.; Hussain, A. Plastics in the environment as potential threat to life: An overview. Environ. Sci. Pollut. Res. 2022, 29, 56928–56947. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Vinoda, K.S.; Papireddy, M.; Gowda, A.N.S. Toxic pollutants from plastic waste-a review. Procedia Environ. Sci. 2016, 35, 701–708. [Google Scholar] [CrossRef]
- Gu, J.D. On environmental biotechnology of bioremediation. Appl. Environ. Biotechnol. 2020, 5, 3–8. [Google Scholar] [CrossRef]
- Kotova, I.B.; Taktarova, Y.V.; Tsavkelova, E.A.; Egorova, M.A.; Bubnov, I.A.; Malakhova, D.V.; Shirinkina, L.I.; Sokolova, T.G. Bonch-Osmolovskaya, E.A. Microbial degradation of plastics and approaches to make it more efficient. Microbiology 2021, 90, 671–701. [Google Scholar] [CrossRef]
- Temporiti, M.E.E.; Nicola, L.; Nielsen, E.; Tosi, S. Fungal Enzymes Involved in Plastics Biodegradation. Microorganisms 2022, 10, 1180. [Google Scholar] [CrossRef] [PubMed]
- Chiellini, F.; Ferri, M.; Morelli, A.; Dipaola, L.; Latini, G. Perspectives on alternatives to phthalate plasticized poly (vinyl chloride) in medical devices applications. Prog. Polym. Sci. 2013, 38, 1067–1088. [Google Scholar] [CrossRef]
- Khandare, S.D.; Chaudhary, D.R.; Jha, B. Bioremediation of polyvinyl chloride (PVC) films by marine bacteria. Mar. Pollut. Bull. 2021, 169, 112566. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.Y.; Chen, Z.; Chen, J.; Yu, H.; Zhou, X.; Criddle, C.S.; Wu, W.M.; Zhang, Y. Biodegradation of polyvinyl chloride (PVC) in Tenebrio molitor (Coleoptera: Tenebrionidae) larvae. Environ. Int. 2020, 145, 106106. [Google Scholar] [CrossRef] [PubMed]
- Hosney, H.; Nadiem, B.; Ashour, I.; Mustafa, I.; El-Shibiny, A. Epoxidized vegetable oil and bio-based materials as PVC plasticizer. J. Appl. Polym. Sci. 2018, 135, 46270. [Google Scholar] [CrossRef]
- Dovjak, M.; Kristl, Ž. Health concerns of PVC materials in the built environment. Int. J. Sanit. Eng. Res. 2011, 5, 4–26. [Google Scholar]
- Ali, M.I.; Ahmed, S.; Robson, G.; Javed, I.; Ali, N.; Atiq, N.; Hameed, A. Isolation and molecular characterization of polyvinyl chloride (PVC) plastic degrading fungal isolates. J. Basic Microbial. 2014, 54, 18–27. [Google Scholar] [CrossRef]
- Pardo-Rodríguez, M.L.; Zorro-Mateus, P.J.P. Biodegradation of polyvinyl chloride by Mucor s.p. and Penicillium s.p. isolated from soil. Rev. Investig. Desarro. Innov. 2021, 11, 387–399. [Google Scholar] [CrossRef]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011, 9, 177–192. [Google Scholar] [CrossRef]
- Sánchez, C. Fungal potential for the degradation of petroleum-based polymers: An overview of macro-and microplastics biodegradation. Biotechnol. Adv. 2020, 40, 107501. [Google Scholar] [CrossRef]
- Sang, B.I.; Hori, K.; Tanji, Y.; Unno, H. Fungal contribution to in situ biodegradation of poly (3-hydroxybutyrate-co-3-hydroxyvalerate) film in soil. Appl. Microbiol. Biotechnol. 2002, 58, 241–247. [Google Scholar] [CrossRef]
- Barratt, S.R.; Ennos, A.R.; Greenhalgh, M.; Robson, G.D.; Handley, P.S. Fungi are the predominant micro-organisms responsible for degradation of soil-buried polyester polyurethane over a range of soil water holding capacities. J. Appl. Microbiol. 2003, 95, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Zeghal, E.; Vaksmaa, A.; Vielfaure, H.; Boekhout, T.; Niemann, H. The potential role of marine fungi in plastic degradation—A review. Front. Mar. Sci. 2021, 8, 738877. [Google Scholar] [CrossRef]
- Srikanth, M.; Sandeep, T.S.R.S.; Sucharitha, K.; Godi, S. Biodegradation of plastic polymers by fungi: A brief review. Bioresour. Bioprocess. 2022, 9, 42. [Google Scholar] [CrossRef]
- Kaushal, J.; Khatri, M.; Arya, S.K. Recent insight into enzymatic degradation of plastics prevalent in the environment: A mini-review. Clean. Eng. Technol. 2021, 2, 100083. [Google Scholar] [CrossRef]
- Hu, X.; Gao, Z.; Wang, Z.; Su, T.; Yang, L.; Li, P. Enzymatic degradation of poly (butylene succinate) by cutinase cloned from Fusarium solani. Polym. Degrad. Stabil. 2016, 134, 211–219. [Google Scholar] [CrossRef]
- El-Morsy, E.M.; Hassan, H.M.; Ahmed, E. Biodegradative activities of fungal isolates from plastic contaminated soils. Mycosphere 2017, 8, 1071–1087. [Google Scholar] [CrossRef]
- Jung, H.W.; Yang, M.K.; Su, R.C. Purification, characterization, and gene cloning of an Aspergillus fumigatus polyhydroxybutyrate depolymerase used for degradation of polyhydroxybutyrate, polyethylene succinate, and polybutylene succinate. Polym. Degrad. Stabil. 2018, 154, 186–194. [Google Scholar] [CrossRef]
- Jabeen, N.; Majid, I.; Nayik, G.A. Bioplastics and food packaging: A review. Cogent Food Agric. 2015, 1, 1117749. [Google Scholar] [CrossRef]
- Krueger, M.C.; Harms, H.; Schlosser, D. Prospects for microbiological solutions to environmental pollution with plastics. Appl. Microbial. Biotechnol. 2015, 99, 8857–8874. [Google Scholar] [CrossRef]
- Amobonye, A.; Bhagwat, P.; Singh, S.; Pillai, S. Plastic biodegradation: Frontline microbes and their enzymes. Sci. Total Environ. 2021, 759, 143536. [Google Scholar] [CrossRef]
- Ali, S.S.; Elsamahy, T.; Al-Tohamy, R.; Zhu, D.; Mahmoud, Y.A.; Koutra, E.; Metwally, M.A.; Kornaros, M.; Sun, J. Plastic wastes biodegradation: Mechanisms, challenges and future prospects. Sci. Total Environ. 2021, 780, 146590. [Google Scholar] [CrossRef]
- Liauw, C.M.; Slate, A.J.; Butler, J.A.; Wilson-Nieuwenhuis, J.S.; Deisenroth, T.; Preuss, A.; Verran, J.; Whitehead, K.A. The effect of surface hydrophobicity on the attachment of fungal conidia to substrates of polyvinyl acetate and polyvinyl alcohol. J. Polym. Environ. 2020, 28, 1450–1464. [Google Scholar] [CrossRef]
- Jensen, H.L. The fungus flora of the soil. Soil Sci. 1931, 31, 123–158. [Google Scholar] [CrossRef]
- Scherer, T.M. Biological and Enzymatic Mechanisms of Polyester Biodegradation by Fungi. Ph.D. Thesis, University of Massachusetts Amherst, Amherst, MA, USA, 1996. [Google Scholar]
- Aly, M.M.; Jaar, T.A.M.; Bokhari, F.M. Poly-β-hydroxybutyrate degradation by Aspergillus fumigates isolated from soil samples collected from Jeddah, Saudi Arabia. IOSR J. Pharm. Biol. Sci. 2017, 12, 53–61. [Google Scholar] [CrossRef]
- Theelen, B.; Cafarchia, C.; Gaitanis, G.; Bassukas, I.D.; Boekhout, T.; Dawson, T.L., Jr. Malassezia ecology, pathophysiology, and treatment. Med. Mycol. 2018, 56 (Suppl. 1), S10–S25. [Google Scholar] [CrossRef]
- Amend, A. From dandruff to deep-sea vents: Malassezia-like fungi are ecologically hyper-diverse. PLoS Pathog. 2014, 10, e1004277. [Google Scholar] [CrossRef]
- Zeinali, E.; Sadeghi, G.; Yazdinia, F.; Shams-Ghahfarokhi, M.; Razzaghi-Abyaneh, M. Clinical and epidemiological features of the genus Malassezia in Iran. Iran. J. Microbiol. 2014, 6, 354–360. [Google Scholar]
- Hađina, S.; Bruvo Mađarić, B.; Kazazić, S.; Paradžik, T.; Reljić, S.; Pinter, L.; Huber, Đ.; Vujaklija, D. Malassezia pachydermatis from brown bear: A comprehensive analysis reveals novel genotypes and distribution of all detected variants in domestic and wild animals. Front. Microbiol. 2023, 14, 1151107. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Goh, B.N.; Teh, W.K.; Jiang, Z.; Goh, J.P.Z.; Goh, A.; Wu, G.; Hoon, S.S.; Raida, M.; Camattari, A.; et al. Skin commensal Malassezia globosa secreted protease attenuates Staphylococcus aureus biofilm formation. J. Investig. Dermatol. 2018, 138, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Jayasinghe-Arachchige, V.M.; Prabhakar, R. Degradation of a main plastic pollutant polyethylene terephthalate by two distinct proteases (neprilysin and cutinase-like enzyme). J. Chem. Inf. Model. 2021, 61, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Krasowska, A.; Sigler, K. How microorganisms use hydrophobicity and what does this mean for human needs? Front. Cell. Infect. Microbiol. 2014, 4, 112. [Google Scholar] [CrossRef]
- Kwon-Chung, K.J.; Sugui, J.A. Aspergillus fumigatus—What makes the species a ubiquitous human fungal pathogen? PLoS Pathog. 2013, 9, e1003743. [Google Scholar] [CrossRef]
- Al Hosni, A.S.; Pittman, J.K.; Robson, G.D. Microbial degradation of four biodegradable polymers in soil and compost demonstrating polycaprolactone as an ideal compostable plastic. Waste Manag. 2019, 97, 105–114. [Google Scholar] [CrossRef]
- Maurya, A.; Bhattacharya, A.; Khare, S.K. Enzymatic remediation of polyethylene terephthalate (PET)-based polymers for effective management of plastic wastes: An overview. Front. Bioeng. Biotechnol. 2020, 8, 602325. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.; Shah, R.; Sharma, A.; Jendrossek, D.; Desai, A. Purification of Aspergillus fumigatus (Pdf1) poly (β-hydroxybutyrate) (PHB) depolymerase using a new, single-step substrate affinity chromatography method: Characterization of the PHB depolymerase exhibiting novel self-aggregation behavior. J. Environ. Polym. Degrad. 2000, 8, 197–203. [Google Scholar] [CrossRef]
- Saeed, S.; Iqbal, A.; Deeba, F. Biodegradation study of Polyethylene and PVC using naturally occurring plastic degrading microbes. Arch. Microbiol. 2022, 204, 497. [Google Scholar] [CrossRef]
- Malachová, K.; Novotný, Č.; Adamus, G.; Lotti, N.; Rybková, Z.; Soccio, M.; Šlosarčíková, P.; Verney, V.; Fava, F. Ability of Trichoderma hamatum isolated from plastics-polluted environments to attack petroleum-based, synthetic polymer films. Processes 2020, 8, 467. [Google Scholar] [CrossRef]
- Webb, J.S.; Nixon, M.; Eastwood, I.M.; Greenhalgh, M.; Robson, G.D.; Handley, P.S. Fungal colonization and biodeterioration of plasticized polyvinyl chloride. Appl. Environ. Microbiol. 2000, 66, 3194–3200. [Google Scholar] [CrossRef] [PubMed]
- Dsouza, G.C.; Sheriff, R.S.; Ullanat, V.; Shrikrishna, A.; Joshi, A.V.; Hiremath, L.; Entoori, K. Fungal biodegradation of low-density polyethylene using consortium of Aspergillus species under controlled conditions. Heliyon 2021, 7, e07008. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Jin, L.; Yu, X.; Wang, B.; Hu, S.; Ruan, H.; Sung, Y.-J.; Lee, H.-G.; Jin, F. Biodegradation of Low Density Polyethylene by the Fungus Cladosporium sp. Recovered from a Landfill Site. J. Fungi 2023, 9, 605. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, H.; Chen, S.; Liu, D.; Xia, H. Purification and properties of a poly (β-hydroxybutyrate) depolymerase from Penicillium sp. J. Polym. Environ. 2006, 14, 419–426. [Google Scholar] [CrossRef]
- Ekanayaka, A.H.; Tibpromma, S.; Dai, D.; Xu, R.; Suwannarach, N.; Stephenson, S.L.; Dao, C.; Karunarathna, S.C. A Review of the fungi that degrade plastic. J. Fungi 2022, 8, 772. [Google Scholar] [CrossRef] [PubMed]
- Raaman, N.; Rajitha, N.; Jayshree, A.; Jegadeesh, R. Biodegradation of plastic by Aspergillus spp. isolated from polythene polluted sites around Chennai. J. Acad. Indus Res. 2012, 1, 313–316. [Google Scholar]
- Verma, N.; Gupta, S. Assessment of LDPE degrading potential Aspergillus species isolated from municipal landfill sites of Agra. SN Appl. Sci. 2019, 1, 701. [Google Scholar] [CrossRef]
- Mathur, G.; Mathur, A.; Prasad, R. Colonization and degradation of thermally oxidized high-density polyethylene by Aspergillus niger (ITCC No. 6052) isolated from plastic waste dumpsite. Bioremediation J. 2011, 15, 69–76. [Google Scholar] [CrossRef]
- Devi, R.S.; Kannan, V.R.; Nivas, D.; Kannan, K.; Chandru, S.; Antony, A.R. Biodegradation of HDPE by Aspergillus spp. from marine ecosystem of Gulf of Mannar, India. Mar. Pollut. Bull. 2015, 96, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Osman, M.; Satti, S.M.; Luqman, A.; Hasan, F.; Shah, Z.; Shah, A.A. Degradation of polyester polyurethane by Aspergillus sp. strain S45 isolated from soil. J. Polym. Environ. 2018, 26, 301–310. [Google Scholar] [CrossRef]
- Chien, H.L.; Tsai, Y.T.; Tseng, W.S.; Wu, J.A.; Kuo, S.L.; Chang, S.L.; Huang, S.J.; Liu, C.T. Biodegradation of PBSA films by Elite Aspergillus isolates and farmland soil. Polymers 2022, 14, 1320. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Rank, C.; Nielsen, K.F.; Larsen, T.O. Metabolomics of Aspergillus fumigatus. Med. Mycol. 2009, 47 (Suppl. 1), S53–S71. [Google Scholar] [CrossRef]
- Urbanek, A.K.; Rymowicz, W.; Strzelecki, M.C.; Kociuba, W.; Franczak, Ł.; Mirończuk, A.M. Isolation and characterization of Arctic microorganisms decomposing bioplastics. AMB Express 2017, 7, 148. [Google Scholar] [CrossRef]
- Usha, R.; Sangeetha, T.; Palaniswamy, M. Screening of polyethylene degrading microorganisms from garbage soil. Libyan Agric. Res. Cent. J. Int. 2011, 2, 200–204. [Google Scholar]
- Kaminskyj, S.G. Fundamentals of growth, storage, genetics and microscopy of Aspergillus nidulans. Fungal Genet. Rep. 2001, 48, 25–31. [Google Scholar] [CrossRef]
- Kadry, A.A.; El-Ganiny, A.M.; Mosbah, R.A.; Kaminskyj, S.G. Deletion of Aspergillus nidulans GDP-mannose transporters affects hyphal morphometry, cell wall architecture, spore surface character, cell adhesion, and biofilm formation. Med. Mycol. 2018, 56, 621–630. [Google Scholar] [CrossRef]
- Kumar, D.; Kumar, L.; Nagar, S.; Raina, C.; Parshad, R.; Gupta, V.K. Screening, isolation and production of lipase/esterase producing Bacillus sp. strain DVL2 and its potential evaluation in esterification and resolution reactions. Arch. Appl. Sci. Res. 2012, 4, 1763–1770. [Google Scholar]
- Kobayashi, T.; Sugiyama, A.; Kawase, Y.; Saito, T.; Mergaert, J.; Swings, J. Biochemical and genetic characterization of an Extracellular Poly (3-Hydroxybutyrate) Depolymerase from Acidovorax sp. Strain TP4. J. Environ. Polym. Degrad. 1999, 7, 9–18. [Google Scholar] [CrossRef]
- Ansari, F.N.; Amirul, A.A. Preparation and characterization of polyhydroxyalkanoates macroporous scaffold through enzyme-mediated modifications. Appl. Biochem. Biotechnol. 2013, 170, 690–709. [Google Scholar] [CrossRef] [PubMed]
- Ramnath, L.; Sithole, B.; Govinden, R. Identification of lipolytic enzymes isolated from bacteria indigenous to Eucalyptus wood species for application in the pulping industry. Biotechnol. Rep. 2017, 15, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.K.; El-Ganiny, A.M.; Afroz, S.; Sanders, D.A.; Liu, J.; Kaminskyj, S.G. Aspergillus nidulans galactofuranose biosynthesis affects antifungal drug sensitivity. Fungal Genet. Boil. 2012, 49, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Op De Beeck, M.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS ONE 2014, 9, e97629. [Google Scholar] [CrossRef]
- Yahya, G.; Ebada, A.; Khalaf, E.M.; Mansour, B.; Nouh, N.A.; Mosbah, R.A.; Saber, S.; Moustafa, M.; Negm, S.; El-Sokkary, M.M.A.; et al. Soil-Associated Bacillus Species: A Reservoir of Bioactive Compounds with Potential Therapeutic Activity against Human Pathogens. Microorganisms 2021, 9, 1131. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Halim, M.S.; Askoura, M.; Mansour, B.; Yahya, G.; El-Ganiny, A.M. In vitro activity of celastrol in combination with thymol against carbapenem-resistant Klebsiella pneumoniae isolates. J. Antibiot. 2022, 75, 679–690. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Youssef, C.R.B.; Kadry, A.A.; El-Ganiny, A.M. The alarming coincidence of toxin genes with staphylococcal cassette Chromosome mec (SCCmec) in clinical MRSA isolates. Saudi J. Biol. Sci. 2022. (in press) [Google Scholar] [CrossRef]
- Matousek, J.L.; Campbell, K.L.; Kakoma, I.; Solter, P.F.; Schaeffer, D.J. Evaluation of the effect of pH on in vitro growth of Malassezia pachydermatis. Can. J. Vet. Res. 2003, 67, 56–59. [Google Scholar]
- Yousef, N.E.; Abdelatif, H.K.; Serry, F.M.; Cameron, J.A. Microbial degradation of microbial polyester copolymer polyhydroxy-butyrate/valerate. Zagazig J. Pharm. Sci. 1998, 7, 33–45. [Google Scholar] [CrossRef]
- Yahya, G.; Wu, Y.; Peplowska, K.; Röhrl, J.; Soh, Y.M.; Bürmann, F.; Gruber, S.; Storchova, Z. Phospho-regulation of the Shugoshin—Condensin interaction at the centromere in budding yeast. PLoS Genet. 2020, 16, e1008569. [Google Scholar] [CrossRef]
| Isolate No. | Zone of Clearance (cm) on BHA | Depolymerase Activity (U/mL/min) | Lipase Activity (U/mL/min) | Percentage of Hydrophobicity | Weight Loss of PVC Strips |
|---|---|---|---|---|---|
| F1 | 4 | 0.032 | 0.02 | 41.86% | 0.718 ± 0.1 |
| F2 | 3.5 | 0.014 | 0 | 68.82% | 1.46 ± 0.7 |
| F3 | 4 | 0.032 | 0 | 58.12% | 1.92 ± 0.51 |
| F4 | 3.2 | 0.024 | 0.017 | 82.29% | 2.15 ± 0.42% |
| A. fumigatus1 | Malassezia sp. | A. fumigatus2 | A. fumigatus3 | |
|---|---|---|---|---|
| OD620 nm after 24 h incubation at different pH | ||||
| pH 4 | 1.11 (0.035) a | 5.84 (0.181) a | 2.35 (0.068) a | 1.76 (0.197) a |
| pH 5 | 0.28 (0.097) ns | 1.76 (0.041) a | 1.62 (0.036) a | 2.33 (0.042) a |
| pH 6 | 1.23 (0.034) a | 3.58 (0.102) a | 1.64 (0.055) a | 2.04 (0.058) a |
| pH 7 | 0.67 (0.081) b | 0.55 (0.044) c | 1.40 (0.045) a | 3.42 (0.169) a |
| pH 8 | 3.48 (0.282) a | 1.66 (0.032) a | 1.79 (0.036) a | 3.56 (0.053) a |
| pH 9 | 2.13 (0.025) a | 1.83 (0.046) a | 1.61 (0.030) a | 5.08 (0.070) a |
| pH 10 * | 0.12 (0.014) | 0.11 (0.032) | 0.17 (0.043) | 0.11 (0.023) |
| OD620 nm after 24 h incubation at different temperatures (optimum pH for each isolate) | ||||
| 30 °C | 1.05 (0.012) a | 1.06 (0.013) a | 2.87 (0.044) a | 1.37 (0.031) a |
| 40 °C | 2.74 (0.073) a | 5.89 (0.058) a | 2.08 (0.109) a | 4.75 (0.066) a |
| 50 °C | 0.08 (0.014) ns | 0.27 (0.048) ns | 0.34 (0.047) ns | 0.20 (0.033) ns |
| 60 °C * | 0.14 (0.022) | 0.17 (0.022) | 0.31 (0.035) | 0.17 (0.025) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Dash, H.A.; Yousef, N.E.; Aboelazm, A.A.; Awan, Z.A.; Yahya, G.; El-Ganiny, A.M. Optimizing Eco-Friendly Degradation of Polyvinyl Chloride (PVC) Plastic Using Environmental Strains of Malassezia Species and Aspergillus fumigatus. Int. J. Mol. Sci. 2023, 24, 15452. https://doi.org/10.3390/ijms242015452
El-Dash HA, Yousef NE, Aboelazm AA, Awan ZA, Yahya G, El-Ganiny AM. Optimizing Eco-Friendly Degradation of Polyvinyl Chloride (PVC) Plastic Using Environmental Strains of Malassezia Species and Aspergillus fumigatus. International Journal of Molecular Sciences. 2023; 24(20):15452. https://doi.org/10.3390/ijms242015452
Chicago/Turabian StyleEl-Dash, Heba A., Nehal E. Yousef, Abeer A. Aboelazm, Zuhier A. Awan, Galal Yahya, and Amira M. El-Ganiny. 2023. "Optimizing Eco-Friendly Degradation of Polyvinyl Chloride (PVC) Plastic Using Environmental Strains of Malassezia Species and Aspergillus fumigatus" International Journal of Molecular Sciences 24, no. 20: 15452. https://doi.org/10.3390/ijms242015452
APA StyleEl-Dash, H. A., Yousef, N. E., Aboelazm, A. A., Awan, Z. A., Yahya, G., & El-Ganiny, A. M. (2023). Optimizing Eco-Friendly Degradation of Polyvinyl Chloride (PVC) Plastic Using Environmental Strains of Malassezia Species and Aspergillus fumigatus. International Journal of Molecular Sciences, 24(20), 15452. https://doi.org/10.3390/ijms242015452






