Photosynthesis Response and Transcriptional Analysis: Dissecting the Role of SlHB8 in Regulating Drought Resistance in Tomato Plants
Abstract
:1. Introduction
2. Results
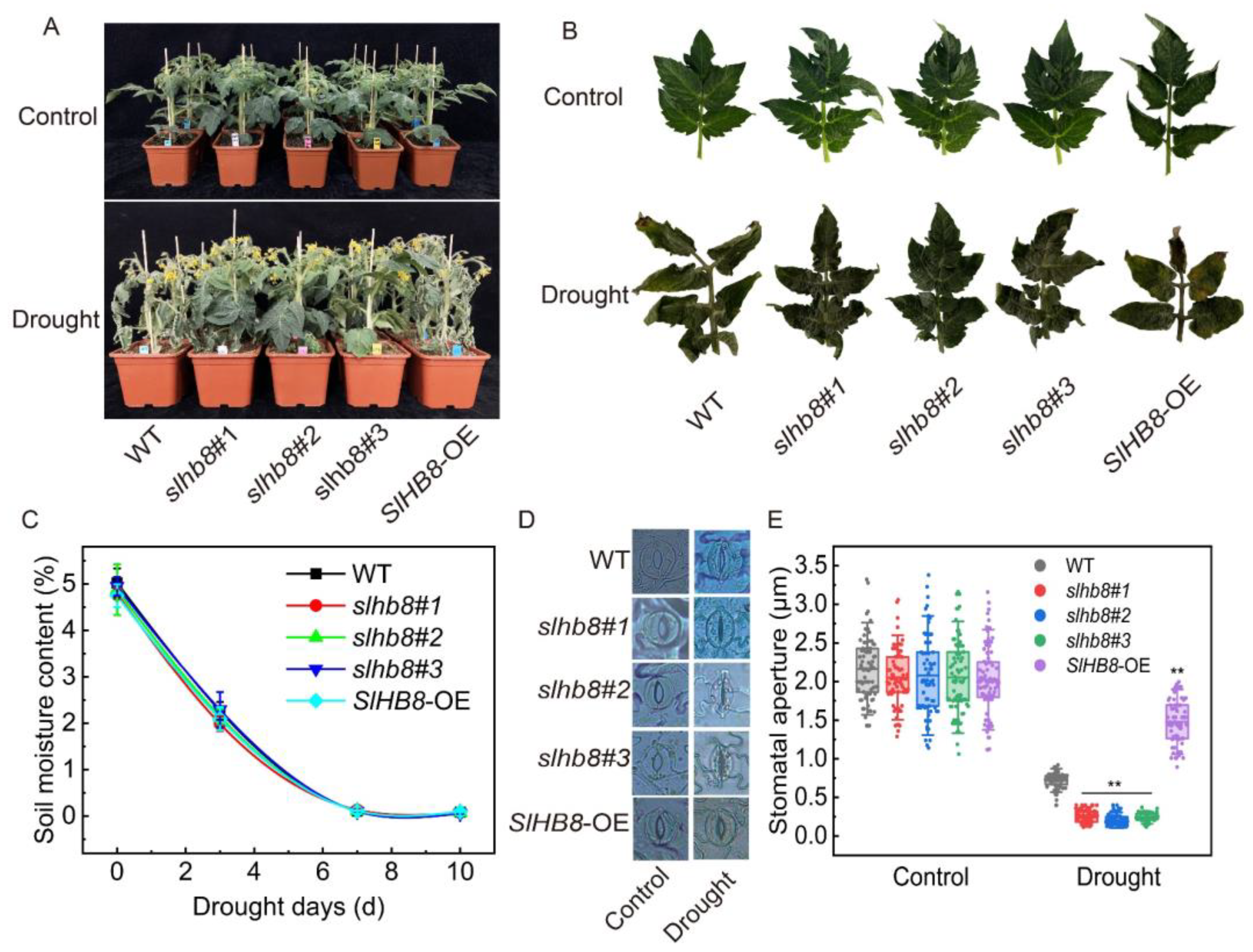
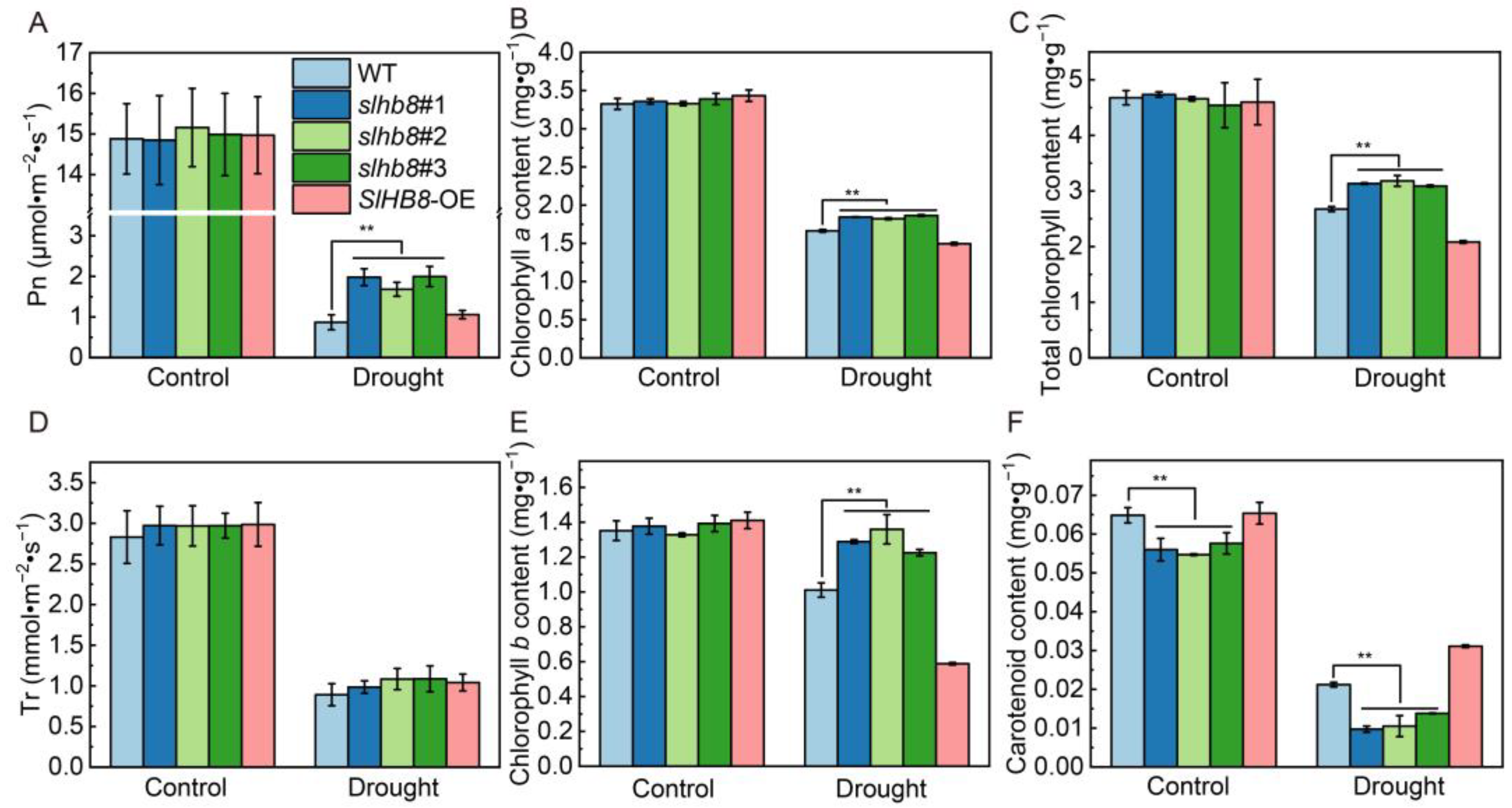
2.1. Effects of SlHB8 on Stomatal Aperture and Photosynthetic Response of Tomato Leaves under Drought Stress
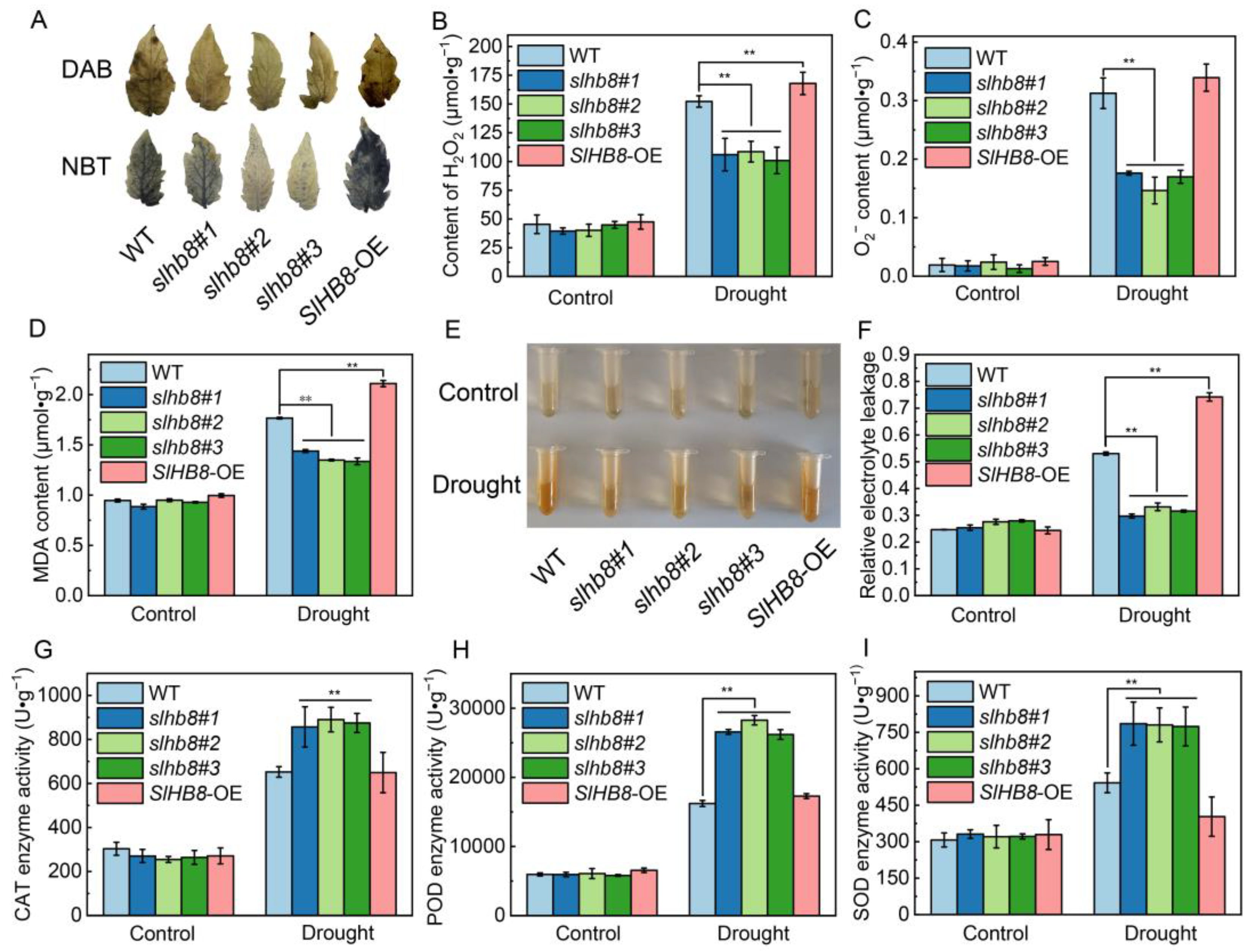
2.2. Effects of SlHB8 on Oxidative Stress and Antioxidant Enzyme Activity in Tomato Leaves under Drought Stress
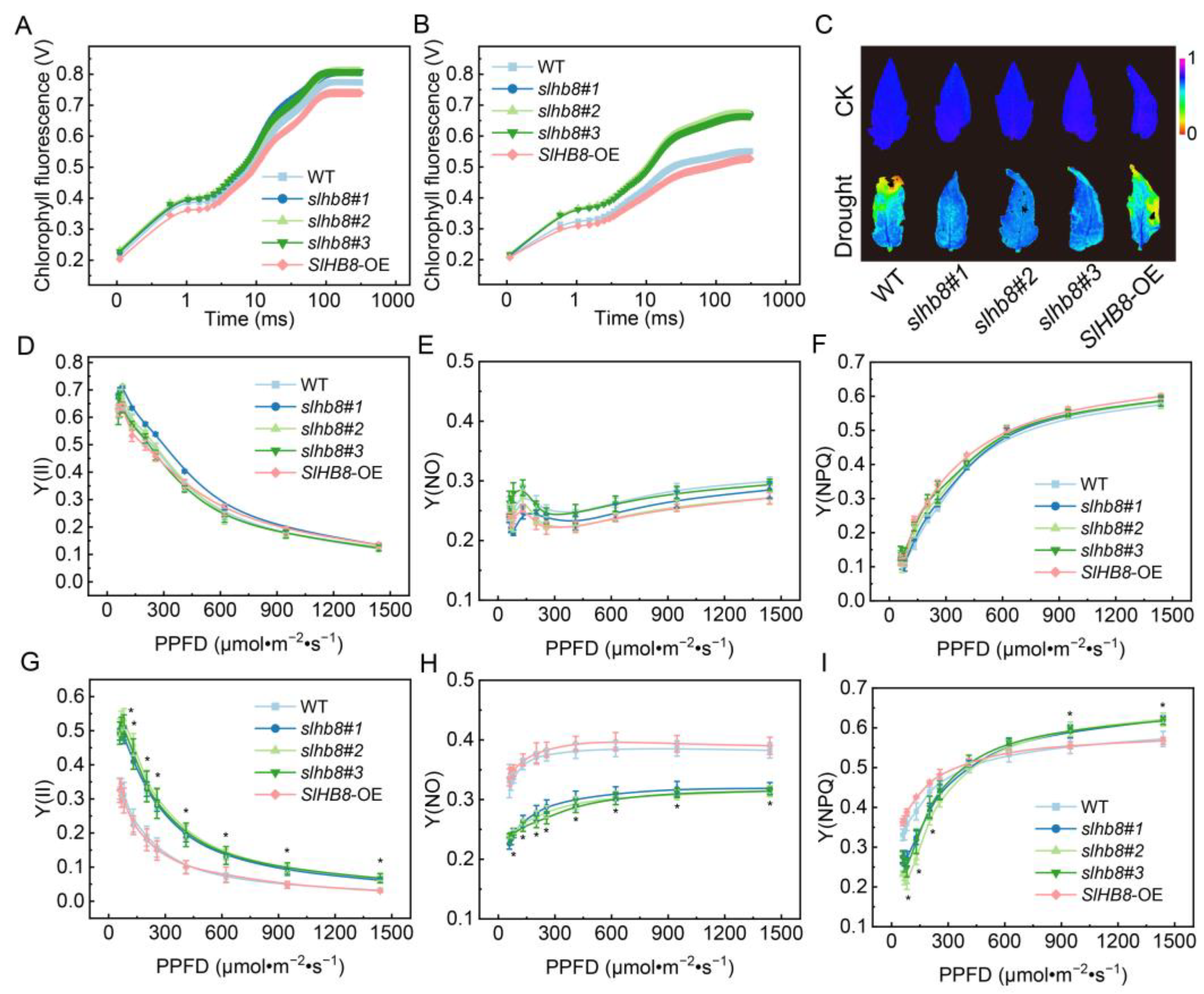
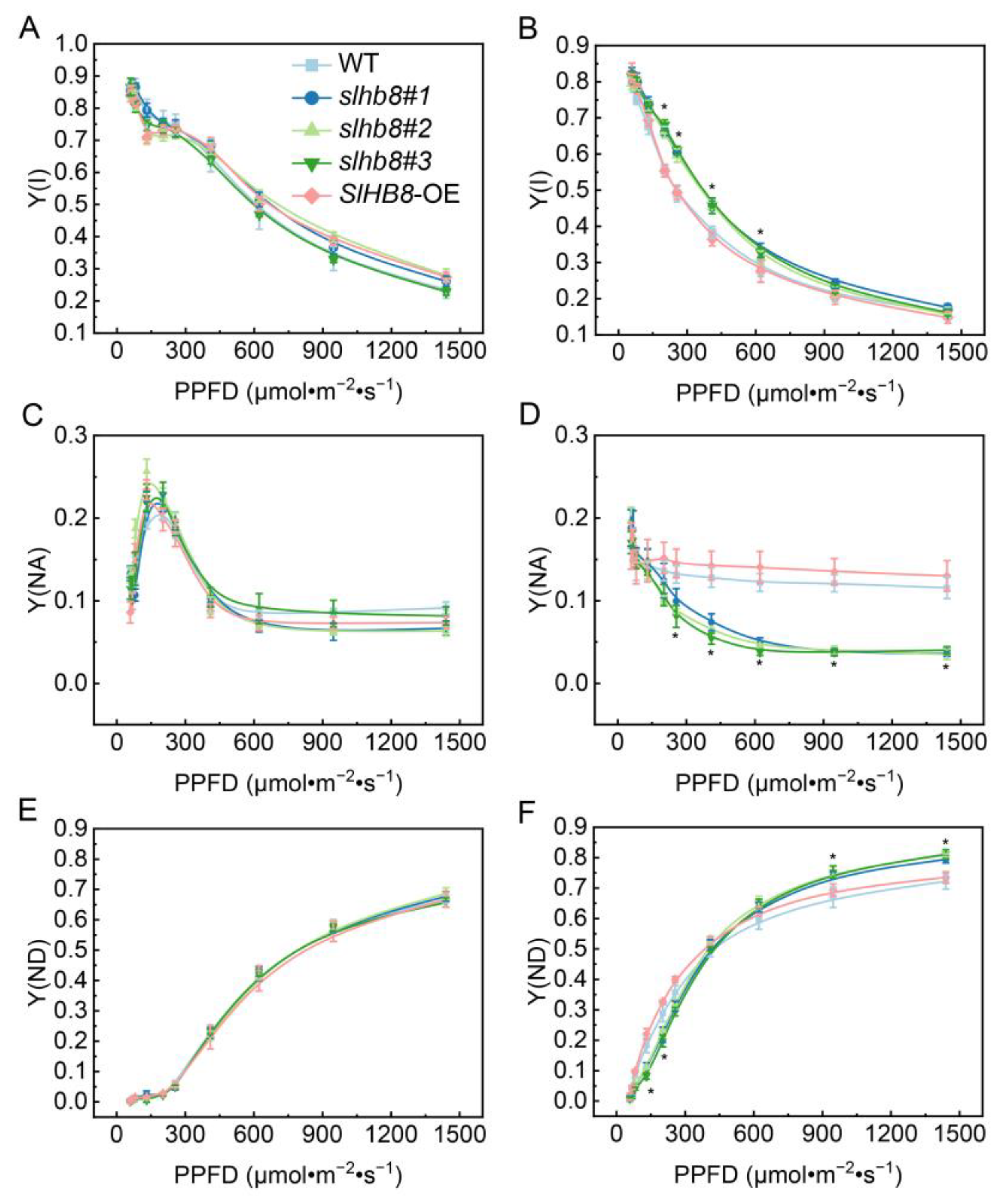
2.3. Effects of SlHB8 on Photochemical Reaction Efficiency of Photosystems in Tomato Leaves under Drought
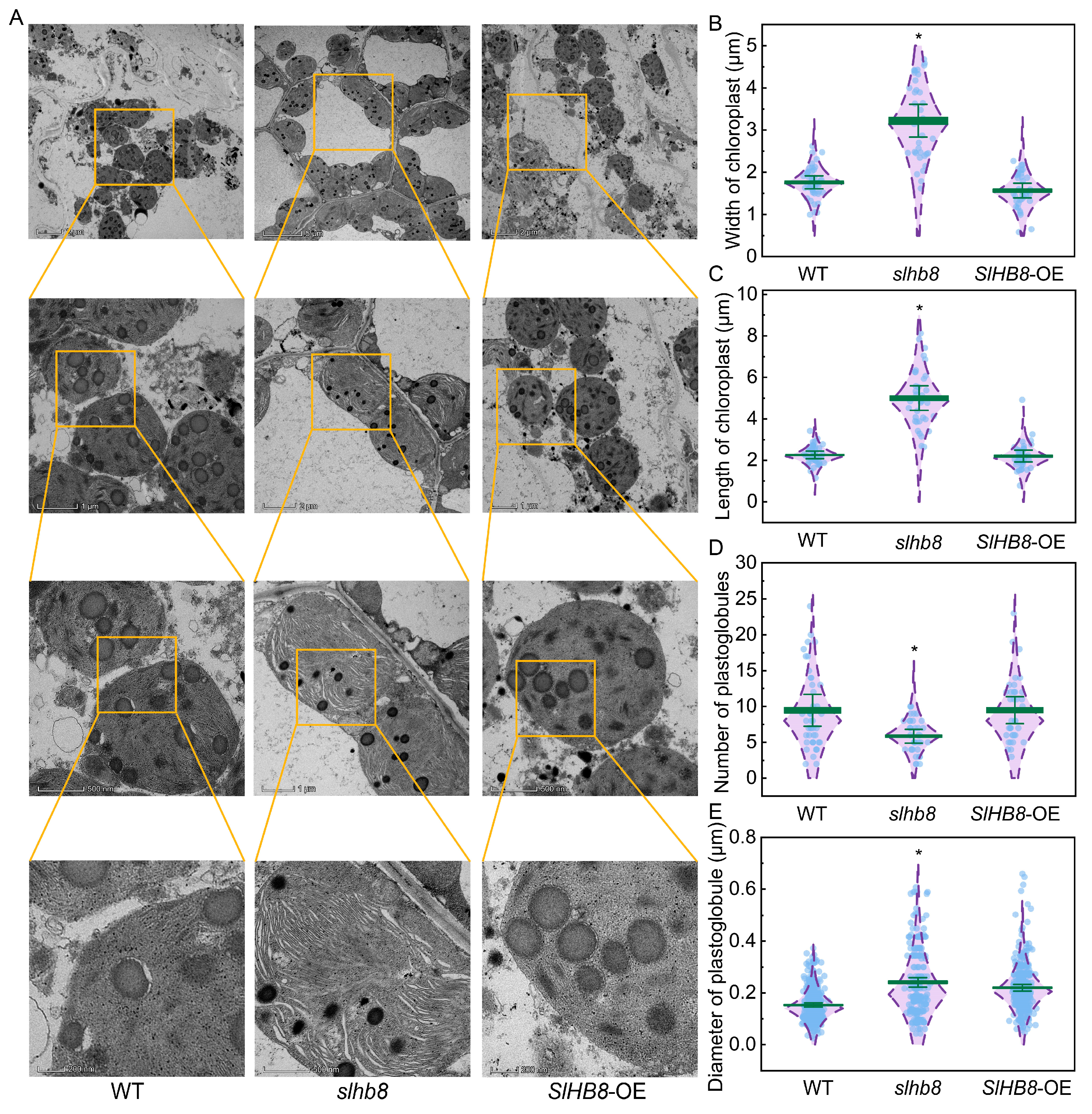
2.4. Effects of SlHB8 on Chloroplast Ultrastructure in Tomato Leaves under Drought Stress
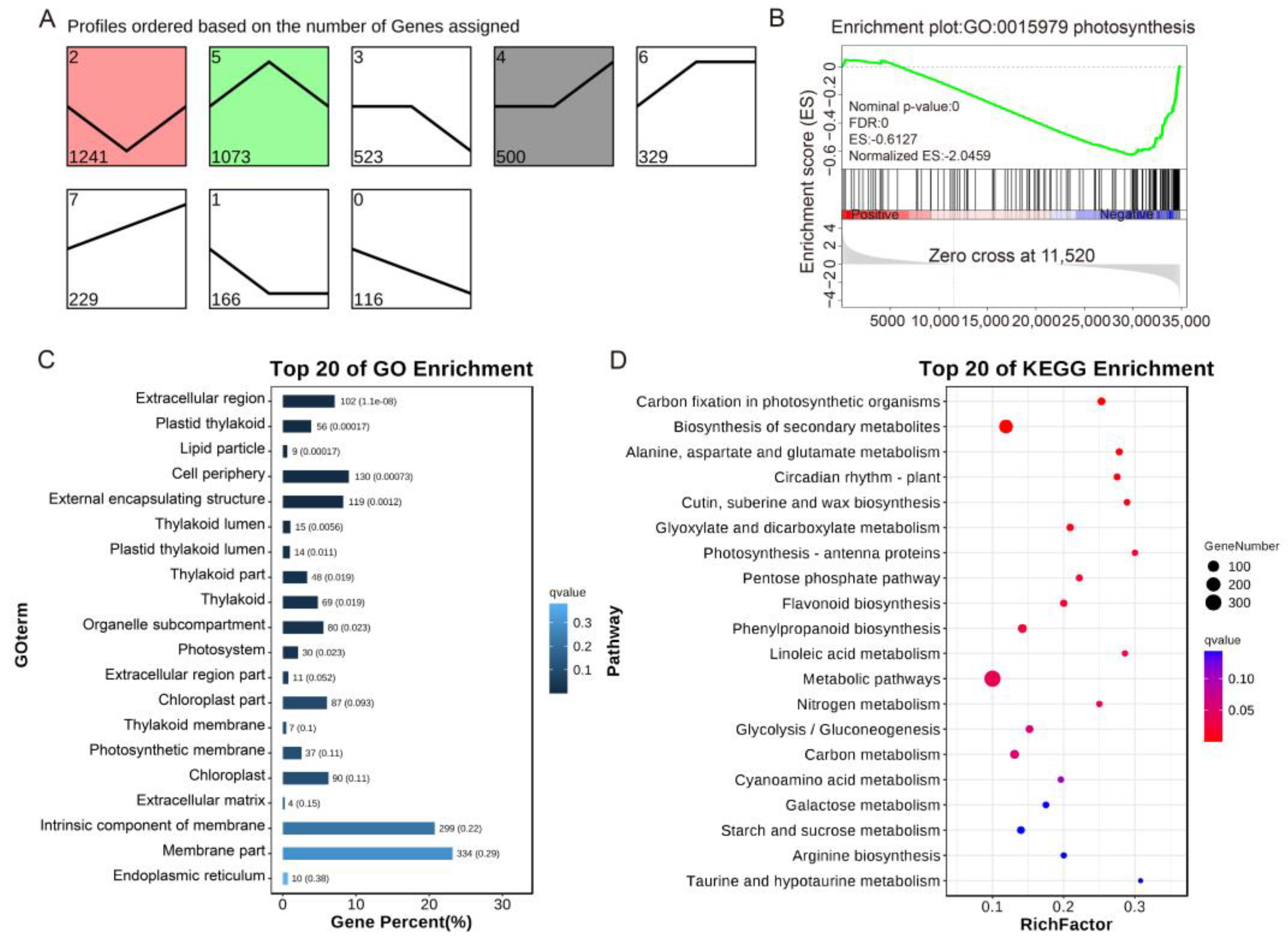
2.5. RNA-seq Analysis the Effects of SlHB8 on Gene Expression in Tomato Leaves under Drought Stress
2.6. Analysis of Gene Expression Related to Photosynthesis and Drought Resistance
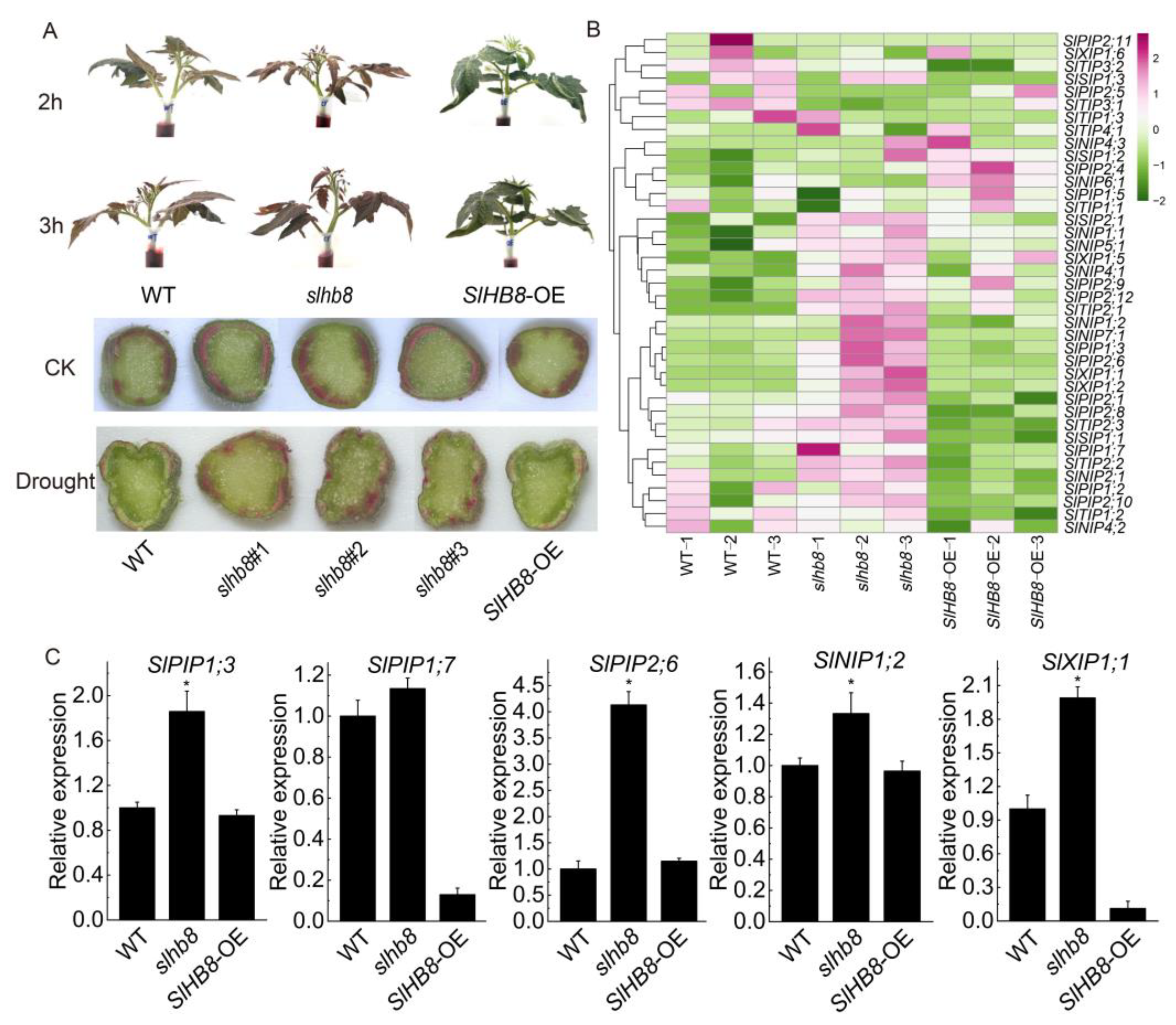
2.7. Analysis of Water Absorption Capacity of Plants and Expression Analysis of Aquaporin Genes
3. Discussion
3.1. The Role of HD-ZIP III Proteins in Plant Development and Stress Resistance
3.2. The Loss of Function of SlHB8 Can Effectively Alleviate Oxidative Damage Caused by Drought Stress in Tomato Plants
3.3. The Role of SlHB8 in Mediating Drought Resistance Involves the Regulation of Photosynthesis and the Maintenance of Chloroplast Quality
3.4. Expression of Drought-Related Genes and Water Absorption Capacity Regulated by SlHB8 in Drought Resistance
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Observation of Stomatal Aperture
4.3. Determination of Photosynthetic Pigment Content
4.4. Determination of Oxidative Stress and Antioxidant Enzyme Activity
4.5. Measurement of Photosynthetic Gas Exchange Parameters
4.6. Measurement of Photosystem Activity
4.7. Observation of Chloroplast Structure by Transmission Electron Microscopy
4.8. Transcriptomic Analysis
4.9. Quantitative Real-Time Polymerase Chain Reaction Assays
4.10. Assessment of Red Ink Absorption Capacity of Tomato Plants
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Muhammad Aslam, M.; Waseem, M.; Jakada, B.H.; Okal, E.J.; Lei, Z.; Saqib, H.S.A.; Yuan, W.; Xu, W.; Zhang, Q. Mechanisms of abscisic acid-mediated drought stress responses in plants. Int. J. Mol. Sci. 2022, 23, 1084. [Google Scholar] [CrossRef] [PubMed]
- Mukarram, M.; Choudhary, S.; Kurjak, D.; Petek, A.; Khan, M.M.A. Drought: Sensing, signalling, effects and tolerance in higher plants. Physiol. Plant. 2021, 172, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Dubey, R.; Pandey, B.K.; Sawant, S.V.; Shirke, P.A. Drought stress inhibits stomatal development to improve water use efficiency in cotton. Acta Physiol. Plant. 2023, 45, 30. [Google Scholar] [CrossRef]
- Zargar, S.M.; Gupta, N.; Nazir, M.; Mahajan, R.; Malik, F.A.; Sofi, N.R.; Shikari, A.B.; Salgotra, R.K. Impact of drought on photosynthesis: Molecular perspective. Plant Gene 2017, 11, 154–159. [Google Scholar] [CrossRef]
- Zahra, N.; Hafeez, M.B.; Kausar, A.; Al Zeidi, M.; Asekova, S.; Siddique, K.H.; Farooq, M. Plant photosynthetic responses under drought stress: Effects and management. J. Agron. Crop Sci. 2023, 209, 651–672. [Google Scholar] [CrossRef]
- Nosalewicz, A.; Okoń, K.; Skorupka, M. Non-photochemical quenching under drought and fluctuating light. Int. J. Mol. Sci. 2022, 23, 5182. [Google Scholar] [CrossRef]
- Arellano, J.B. Non-photochemical quenching of photosystem I as an adaptive response to prolonged drought. J. Exp. Bot. 2023, 74, 16–18. [Google Scholar] [CrossRef]
- Sapeta, H.; Yokono, M.; Takabayashi, A.; Ueno, Y.; Cordeiro, A.M.; Hara, T.; Tanaka, A.; Akimoto, S.; Oliveira, M.M.; Tanaka, R.; et al. Reversible down-regulation of photosystems I and II leads to fast photosynthesis recovery after long-term drought in Jatropha curcas. J. Exp. Bot. 2023, 74, 336–351. [Google Scholar] [CrossRef]
- Razi, K.; Muneer, S. Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Crit. Rev. Biotechnol. 2021, 41, 669–691. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; Chen, H.; Huang, J.; Zhang, Y.; Qi, M.; Liu, Y.; Li, T. Photosynthetic response mechanism of soil salinity-induced cross-tolerance to subsequent drought stress in tomato plants. Plants 2020, 9, 363. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Song, S.; Zhang, H.; Li, Y.; Niu, L.; Zhang, J.; Wang, W. Signaling transduction of ABA, ROS, and Ca2+ in plant stomatal closure in response to drought. Int. J. Mol. Sci. 2022, 23, 14824. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, C.; Su, D.; Yang, Y.; Xian, Z.; Yu, C.; Li, Z.; Hao, Y.; Chen, R. The SlHB8 acts as a negative regulator in stem development and lignin biosynthesis. Int. J. Mol. Sci. 2021, 22, 13343. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lin, Y.; Wu, C.; Yang, Y.; Su, D.; Xian, Z.; Zhu, Y.; Yu, C.; Hu, G.; Deng, W.; et al. The SlARF4-SlHB8 regulatory module mediates leaf rolling in tomato. Plant Sci. 2023, 335, 111790. [Google Scholar] [CrossRef]
- Wu, C.; Yang, Y.; Su, D.; Yu, C.; Xian, Z.; Pan, Z.; Guan, H.; Hu, G.; Chen, D.; Li, Z.; et al. The SlHB8 acts as a negative regulator in tapetum development and pollen wall formation in tomato. Hortic. Res. 2022, 9, uhac185. [Google Scholar] [CrossRef]
- Yadav, A.; Kumar, S.; Verma, R.; Lata, C.; Sanyal, I.; Rai, S.P. microRNA 166: An evolutionarily conserved stress biomarker in land plants targeting HD-ZIP family. Physiol. Mol. Biol. Plants 2021, 27, 2471–2485. [Google Scholar] [CrossRef]
- Hu, G.; Fan, J.; Xian, Z.; Huang, W.; Lin, D.; Li, Z. Overexpression of SlREV alters the development of the flower pedicel abscission zone and fruit formation in tomato. Plant Sci. 2014, 229, 86–95. [Google Scholar] [CrossRef]
- Clepet, C.; Devani, R.S.; Boumlik, R.; Hao, Y.; Morin, H.; Marcel, F.; Verdenaud, M.; Mania, B.; Brisou, G.; Citerne, S.; et al. The miR166–SlHB15A regulatory module controls ovule development and parthenocarpic fruit set under adverse temperatures in tomato. Mol. Plant 2021, 14, 1185–1198. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Z.; Zhang, Y.; Guo, J.; Liu, L.; Wang, C.; Wang, B.; Han, G. The roles of HD-ZIP proteins in plant abiotic stress tolerance. Front. Plant Sci. 2022, 13, 1027071. [Google Scholar] [CrossRef]
- Shen, W.; Li, H.; Teng, R.; Wang, Y.; Wang, W.; Zhuang, J. Genomic and transcriptomic analyses of HD-Zip family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis). Genomics 2019, 111, 1142–1151. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, Y.; Feng, M.; Cui, Y.; Li, S.; Liu, L.; Wang, Y.; Xu, W.; Li, F. Genome-wide identification of the HD-ZIP III subfamily in upland cotton reveals the involvement of GhHB8-5D in the biosynthesis of secondary wall in fiber and drought resistance. Front. Plant Sci. 2022, 12, 806195. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Han, X.; Liang, Z.; Yan, J.; Cong, P.; Zhang, C. Genome-wide identification, classification, and expression analysis of the HD-zip transcription factor family in apple (Malus domestica Borkh.). Int. J. Mol. Sci. 2022, 23, 2632. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Iwamoto, K.; Yamagami, A.; Nakano, T.; Fukuda, H. HD-ZIP III-dependent local promotion of brassinosteroid synthesis suppresses vascular cell division in Arabidopsis root apical meristem. Proc. Nat. Acad. Sci. USA 2023, 120, e2216632120. [Google Scholar] [CrossRef] [PubMed]
- Andrade Galan, A.G.; Doll, J.; Saile, S.C.; Wünsch, M.; Roepenack-Lahaye, E.V.; Pauwels, L.; Goossens, A.; Bresson, J.; Zentgraf, U. The Non-JAZ TIFY protein TIFY8 of Arabidopsis thaliana interacts with the HD-ZIP III transcription factor REVOLUTA and regulates leaf senescence. Int. J. Mol. Sci. 2023, 24, 3079. [Google Scholar] [CrossRef]
- Sharif, R.; Raza, A.; Chen, P.; Li, Y.; El-Ballat, E.M.; Rauf, A.; Hano, C.; El-Esawi, M.A. HD-ZIP gene family: Potential roles in improving plant growth and regulating stress-responsive mechanisms in plants. Genes 2021, 12, 1256. [Google Scholar] [CrossRef]
- Yan, X.; Yue, Z.; Pan, X.; Si, F.; Li, J.; Chen, X.; Li, X.; Luan, F.; Yang, J.; Zhang, X.; et al. The HD-ZIP gene family in watermelon: Genome-wide identification and expression analysis under abiotic stresses. Genes 2022, 13, 2242. [Google Scholar] [CrossRef]
- Qiu, X.; Wang, G.; Abou-Elwafa, S.F.; Fu, J.; Liu, Z.; Zhang, P.; Xie, X.; Ku, L.; Ma, Y.; Guan, X.; et al. Genome-wide identification of HD-ZIP transcription factors in maize and their regulatory roles in promoting drought tolerance. Physiol. Mol. Biol. Plants 2022, 28, 425–437. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Shi, Y.; Chang, Y.L.; Wu, H.T.; Shalmani, A.; Liu, W.T.; Li, W.Q.; Xu, J.W.; Chen, K.M. OsRbohB-mediated ROS production plays a crucial role in drought stress tolerance of rice. Plant. Cell Rep. 2020, 39, 1767–1784. [Google Scholar] [CrossRef]
- Zhao, S.; Gao, H.; Jia, X.; Wang, H.; Ke, M.; Ma, F. The HD-Zip I transcription factor MdHB-7 regulates drought tolerance in transgenic apple (Malus domestica). Environ. Exp. Bot. 2020, 180, 104246. [Google Scholar] [CrossRef]
- Sun, H.; Shi, Q.; Liu, N.Y.; Zhang, S.B.; Huang, W. Drought stress delays photosynthetic induction and accelerates photoinhibition under short-term fluctuating light in tomato. Plant Physiol. Biochem. 2023, 196, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Elias, E.; Nawrocki, W.J.; Croce, R. Drought affects both photosystems in Arabidopsis thaliana. New Phytol. 2023, 240, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Mohapatra, S.; Dogra, V. Improving photosynthetic efficiency by modulating non-photochemical quenching. Trends Plant Sci. 2023, 28, 264–266. [Google Scholar] [CrossRef] [PubMed]
- Miyake, C. Molecular mechanism of oxidation of P700 and suppression of ROS production in photosystem I in response to electron-sink limitations in C3 plants. Antioxidants 2020, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Krieger-Liszkay, A.; Shimakawa, G. Regulation of the generation of reactive oxygen species during photosynthetic electron transport. Biochem. Soc. Trans. 2022, 50, 1025–1034. [Google Scholar] [CrossRef]
- van Wijk, K.J.; Kessler, F. Plastoglobuli: Plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu. Rev. Plant Biol. 2017, 68, 253–289. [Google Scholar] [CrossRef]
- Choi, H.; Yi, T.; Ha, S.H. Diversity of plastid types and their interconversions. Front. Plant Sci. 2021, 12, 692024. [Google Scholar] [CrossRef]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.Y.; Li, J.; Wang, P.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Shivaraj, S.M.; Sharma, Y.; Chaudhary, J.; Rajora, N.; Sharma, S.; Thakral, V.; Ram, H.; Sonah, H.; Singla-Pareek, S.L.; Sharma, T.R.; et al. Dynamic role of aquaporin transport system under drought stress in plants. Environ. Exp. Bot. 2021, 184, 104367. [Google Scholar] [CrossRef]
- Ahmed, S.; Kouser, S.; Asgher, M.; Gandhi, S.G. Plant aquaporins: A frontward to make crop plants drought resistant. Physiol. Plant. 2021, 172, 1089–1105. [Google Scholar] [CrossRef]
- Patel, J.; Mishra, A. Plant aquaporins alleviate drought tolerance in plants by modulating cellular biochemistry, root-architecture, and photosynthesis. Physiol. Plant. 2021, 172, 1030–1044. [Google Scholar] [CrossRef]
- Wu, L.; Chang, Y.; Wang, L.; Wang, S.; Wu, J. The aquaporin gene PvXIP1; 2 conferring drought resistance identified by GWAS at seedling stage in common bean. Theor. Appl. Genet. 2021, 135, 485–500. [Google Scholar] [CrossRef]
- Bai, J.; Wang, X.; Yao, X.; Chen, X.; Lu, K.; Hu, Y.; Wang, Z.; Mu, Y.; Zhang, L.; Dong, H. Rice aquaporin OsPIP2; 2 is a water-transporting facilitator in relevance to drought-tolerant responses. Plant Direct 2021, 5, e338. [Google Scholar] [CrossRef]
- Liu, M.; Wang, C.; Ji, Z.; Lu, J.; Zhang, L.; Li, C.; Huang, J.; Yang, G.; Yan, K.; Zhang, S.; et al. Regulation of drought tolerance in Arabidopsis involves the PLATZ4-mediated transcriptional repression of plasma membrane aquaporin PIP2; 8. Plant J. 2023, 115, 434–451. [Google Scholar] [CrossRef]
- Chen, X.; Tao, H.; Wu, Y.; Xu, X. Effects of Cadmium on metabolism of photosynthetic pigment and photosynthetic system in Lactuca sativa L. revealed by physiological and proteomics analysis. Sci. Hortic. 2022, 305, 111371. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Zhang, X.; Zhong, Q.; Liu, X.; Guan, H.; Chen, R.; Hao, Y.; Yang, X. Photosynthesis Response and Transcriptional Analysis: Dissecting the Role of SlHB8 in Regulating Drought Resistance in Tomato Plants. Int. J. Mol. Sci. 2023, 24, 15498. https://doi.org/10.3390/ijms242015498
Yang Y, Zhang X, Zhong Q, Liu X, Guan H, Chen R, Hao Y, Yang X. Photosynthesis Response and Transcriptional Analysis: Dissecting the Role of SlHB8 in Regulating Drought Resistance in Tomato Plants. International Journal of Molecular Sciences. 2023; 24(20):15498. https://doi.org/10.3390/ijms242015498
Chicago/Turabian StyleYang, Yinghua, Xinyue Zhang, Qiuxiang Zhong, Xiaojuan Liu, Hongling Guan, Riyuan Chen, Yanwei Hao, and Xiaolong Yang. 2023. "Photosynthesis Response and Transcriptional Analysis: Dissecting the Role of SlHB8 in Regulating Drought Resistance in Tomato Plants" International Journal of Molecular Sciences 24, no. 20: 15498. https://doi.org/10.3390/ijms242015498
APA StyleYang, Y., Zhang, X., Zhong, Q., Liu, X., Guan, H., Chen, R., Hao, Y., & Yang, X. (2023). Photosynthesis Response and Transcriptional Analysis: Dissecting the Role of SlHB8 in Regulating Drought Resistance in Tomato Plants. International Journal of Molecular Sciences, 24(20), 15498. https://doi.org/10.3390/ijms242015498





