The Roles of N6-Methyladenosine Modification in Plant–RNA Virus Interactions
Abstract
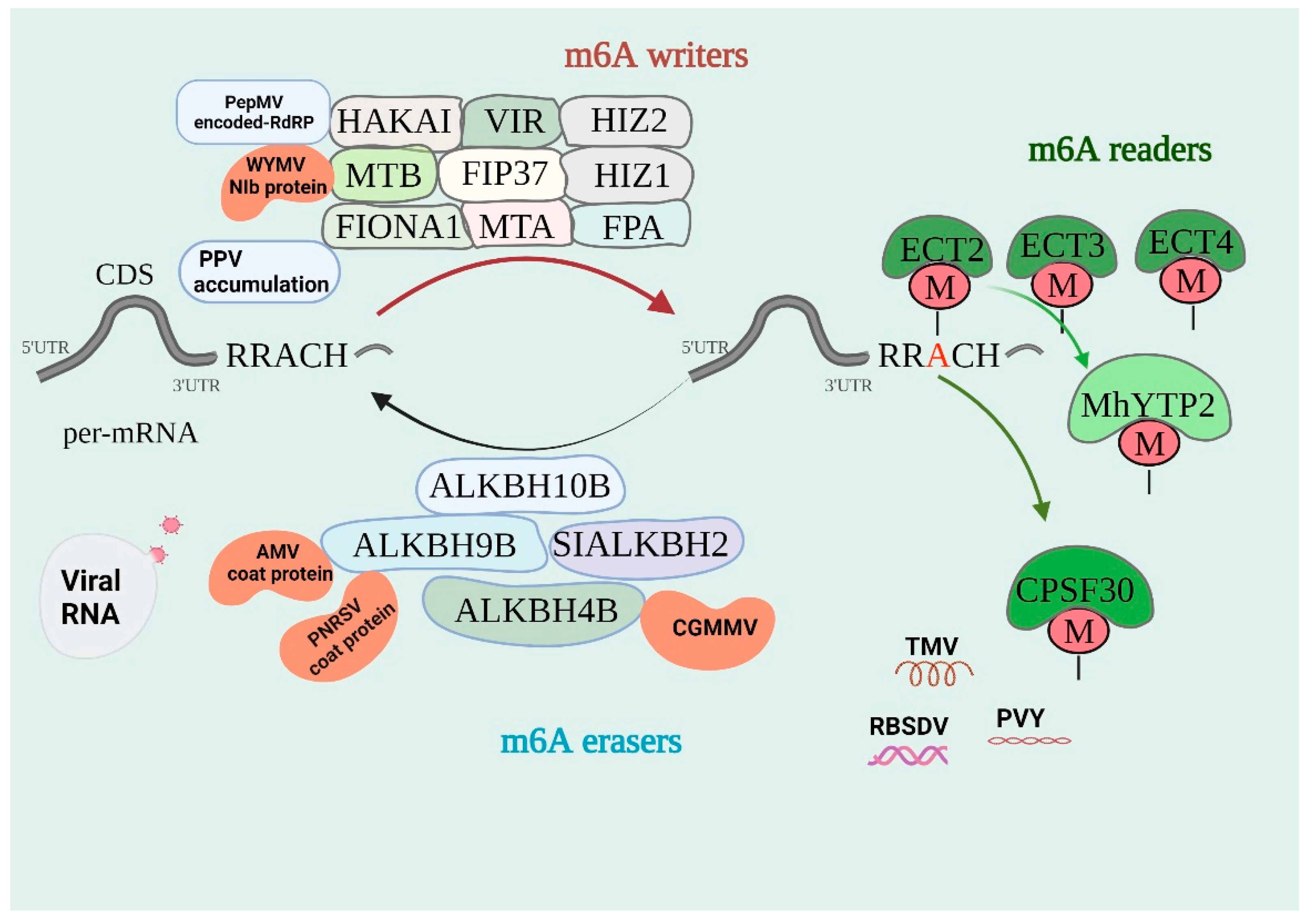
:1. Introduction
2. Molecular Mechanism of m6A Modification
3. Regulation of m6A Methylation in Plant RNA Viruses
| Virus Name | Virus Classification | Mechanisms | Reference |
|---|---|---|---|
| AMV | Bromoviridae I family | AtALKBH9B correlates with the ability to interact with AMV coat proteins. | [19,65,66] |
| PNRSV | Bromoviridae I family | AtALKBH9B was found to interact with the CP of PNRSV (a functionally and phylogenetically closely related AMV). | [67] |
| PVY | Potyviridae family | PVY in Nicotiana benthamiana reduces the level of m6A methylation. | [26] |
| PPV | Potyviridae family | Overexpression of NbMETTL1 and NbMETTL2 caused a decrease in PPV accumulation. | [26] |
| ENMV | Potyviridae family | The P1 of ENMV contains AlkB domains. | [69] |
| WYMV | Potyviridae family | TaMTB binds to the NIb of WYMV and thus promotes viral infection. | [70,71] |
| BlVY | Potyviridae family | BIVY has an AlkB domain of RNA demethylase activity. | [24,69] |
| PepMV | Flexiviridae family | RdRp encoded by PepMV could interact with SlHAKAI and promote its protein degradation. | [72] |
| TMV | Virgaviridae family | The global m6A level was reduced under TMV infection, probably associated with decreased m6A methyltransferase and increased demethylase expression. | [18] |
| CGMMV | Virgaviridae family | ClALKBH4B has a significant induction effect in the early response of resistant watermelon to CGMMV. | [73] |
| RBSDV | Reoviridae family | The m6A methylation is mainly associated with genes that are not actively expressed in virus-infected rice plants. | [74,75] |
| RSV | Phenuiviridae family | [75] |
4. RNA Viruses Affect m6A Methylation
5. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, X.; Xiong, X.; Yi, C. Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat. Methods 2017, 14, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed]
- He, P.C.; He, C. m6A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef]
- Fu, Y.; Jia, G.; Pang, X.; Wang, R.N.; Wang, X.; Li, C.J.; Smemo, S.; Dai, Q.; Bailey, K.A.; Nobrega, M.A. FTO-mediated formation of N6-hydroxymethyladenosine and N 6-formyladenosine in mammalian RNA. Nat. Commun. 2013, 4, 1798. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Canaani, D.; Kahana, C.; Lavi, S.; Groner, Y. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 1979, 6, 2879–2899. [Google Scholar] [CrossRef] [PubMed]
- Beemon, K.; Keith, J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J. Mol. Biol. 1977, 113, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Narayan, P.; Ayers, D.F.; Rottman, F.M.; Maroney, P.A.; Nilsen, T.W. Unequal distribution of N 6-methyladenosine in influenza virus mRNAs. Mol. Cell. Biol. 1987, 7, 1572–1575. [Google Scholar]
- Dang, W.; Xie, Y.; Cao, P.; Xin, S.; Wang, J.; Li, S.; Li, Y.; Lu, J. N6-methyladenosine and viral infection. Front. Microbiol. 2019, 10, 417. [Google Scholar] [CrossRef]
- Williams, G.D.; Gokhale, N.S.; Horner, S.M. Regulation of viral infection by the RNA modification N6-methyladenosine. Annu. Rev. Virol. 2019, 6, 235–253. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Brodersen, P. Occurrence and functions of m6A and other covalent modifications in plant mRNA. Plant Physiol. 2020, 182, 79–96. [Google Scholar] [CrossRef]
- Li, Z.; Shi, J.; Yu, L.; Zhao, X.; Ran, L.; Hu, D.; Song, B. N6-methyl-adenosine level in Nicotiana tabacum is associated with tobacco mosaic virus. Virol. J. 2018, 15, 87. [Google Scholar] [CrossRef]
- Martínez-Pérez, M.; Aparicio, F.; López-Gresa, M.P.; Bellés, J.M.; Sánchez-Navarro, J.A.; Pallás, V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef]
- Roossinck, M.J. Plant RNA virus evolution. Curr. Opin. Microbiol. 2003, 6, 406–409. [Google Scholar] [CrossRef]
- Laliberté, J.-F.; Sanfaçon, H. Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 2010, 48, 69–91. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, X.; Hong, Y.; Liu, Y. Chloroplast in plant-virus interaction. Front. Microbiol. 2016, 7, 1565. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Wei, Y.; Zhao, M. The reversible methylation of m6A is involved in plant virus infection. Biology 2022, 11, 271. [Google Scholar] [CrossRef] [PubMed]
- van den Born, E.; Omelchenko, M.V.; Bekkelund, A.; Leihne, V.; Koonin, E.V.; Dolja, V.V.; Falnes, P.Ø. Viral AlkB proteins repair RNA damage by oxidative demethylation. Nucleic Acids Res. 2008, 36, 5451–5461. [Google Scholar] [CrossRef]
- Bratlie, M.S.; Drabløs, F. Bioinformatic mapping of AlkB homology domains in viruses. BMC Genom. 2005, 6, 1. [Google Scholar] [CrossRef]
- Yue, J.; Wei, Y.; Sun, Z.; Chen, Y.; Wei, X.; Wang, H.; Pasin, F.; Zhao, M. AlkB RNA demethylase homologues and N6-methyladenosine are involved in Potyvirus infection. Mol. Plant Pathol. 2022, 23, 1555–1564. [Google Scholar] [CrossRef]
- Zanardo, L.G.; de Souza, G.B.; Alves, M.S. Transcriptomics of plant–virus interactions: A review. Theor. Exp. Plant Physiol. 2019, 31, 103–125. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Ke, S.; Alemu, E.A.; Mertens, C.; Gantman, E.C.; Fak, J.J.; Mele, A.; Haripal, B.; Zucker-Scharff, I.; Moore, M.J.; Park, C.Y. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015, 29, 2037–2053. [Google Scholar] [CrossRef]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Dierks, D.; Garcia-Campos, M.A.; Uzonyi, A.; Safra, M.; Edelheit, S.; Rossi, A.; Sideri, T.; Varier, R.A.; Brandis, A.; Stelzer, Y. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat. Methods 2021, 18, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, Y.; Dong, S.; Yu, Q.; Jia, G. Antibody-free enzyme-assisted chemical approach for detection of N 6-methyladenosine. Nat. Chem. Biol. 2020, 16, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, S.; Peng, Y.; Ge, R.; Su, R.; Senevirathne, C.; Harada, B.T.; Dai, Q.; Wei, J.; Zhang, L. m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. 2022, 40, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Sun, H.; Yi, Y.; Shen, W.; Li, K.; Xiao, Y.; Li, F.; Li, Y.; Hou, Y.; Lu, B. Absolute quantification of single-base m6A methylation in the mammalian transcriptome using GLORI. Nat. Biotechnol. 2023, 41, 355–366. [Google Scholar] [CrossRef]
- Bokar, J.A.; Rath-Shambaugh, M.E.; Ludwiczak, R.; Narayan, P.; Rottman, F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994, 269, 17697–17704. [Google Scholar] [CrossRef]
- Yang, C.; Hu, Y.; Zhou, B.; Bao, Y.; Li, Z.; Gong, C.; Yang, H.; Wang, S.; Xiao, Y. The role of m6A modification in physiology and disease. Cell Death Dis. 2020, 11, 960. [Google Scholar] [CrossRef]
- Nichols, J. N6-methyladenosine in maize poly (A)-containing RNA. Plant Sci. Lett. 1979, 15, 357–361. [Google Scholar] [CrossRef]
- Kennedy, T.; Lane, B. Wheat embryo ribonucleates. XIII. Methyl-substituted nucleoside constituents and 5′-terminal dinucleotide sequences in bulk poly (A)-rich RNA from imbibing wheat embryos. Can. J. Biochem. 1979, 57, 927–931. [Google Scholar] [CrossRef]
- Nichols, J.; Welder, L. Nucleotides adjacent to N6-methyladenosine in maize poly (A)-containing RNA. Plant Sci. Lett. 1981, 21, 75–81. [Google Scholar] [CrossRef]
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef]
- Růžička, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017, 215, 157–172. [Google Scholar] [CrossRef]
- Vespa, L.; Vachon, G.; Berger, F.; Perazza, D.; Faure, J.-D.; Herzog, M. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 2004, 134, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Ren, H.; Ma, L.; Guo, J.; Mao, D.; Lu, Z.; Lu, L.; Yan, D. Role of Hakai in m6A modification pathway in Drosophila. Nat. Commun. 2021, 12, 2159. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Wu, X.; Wong, C.E.; Fan, S.; Zhang, Y.; Zhang, S.; Liang, Z.; Yu, H.; Shen, L. FIONA1-Mediated m6A Modification Regulates the Floral Transition in Arabidopsis. Adv. Sci. 2022, 9, 2103628. [Google Scholar] [CrossRef]
- Parker, M.T.; Knop, K.; Zacharaki, V.; Sherwood, A.V.; Tome, D.; Yu, X.; Martin, P.G.; Beynon, J.; Michaels, S.D.; Barton, G.J. Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA. Elife 2021, 10, e65537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Bodi, Z.; Mackinnon, K.; Zhong, S.; Archer, N.; Mongan, N.P.; Simpson, G.G.; Fray, R.G. Two zinc finger proteins with functions in m6A writing interact with HAKAI. Nat. Commun. 2022, 13, 1127. [Google Scholar] [CrossRef]
- Alemu, E.A.; He, C.; Klungland, A. ALKBHs-facilitated RNA modifications and de-modifications. DNA Repair 2016, 44, 87–91. [Google Scholar] [CrossRef]
- Fedeles, B.I.; Singh, V.; Delaney, J.C.; Li, D.; Essigmann, J.M. The AlkB family of Fe (II)/α-ketoglutarate-dependent dioxygenases: Repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 2015, 290, 20734–20742. [Google Scholar] [CrossRef]
- Marcinkowski, M.; Pilžys, T.; Garbicz, D.; Steciuk, J.; Zugaj, D.; Mielecki, D.; Sarnowski, T.J.; Grzesiuk, E. Human and Arabidopsis alpha-ketoglutarate-dependent dioxygenase homolog proteins—New players in important regulatory processes. IUBMB Life 2020, 72, 1126–1144. [Google Scholar] [CrossRef]
- Mielecki, D.; Zugaj, D.Ł.; Muszewska, A.; Piwowarski, J.; Chojnacka, A.; Mielecki, M.; Nieminuszczy, J.; Grynberg, M.; Grzesiuk, E. Novel AlkB dioxygenases—Alternative models for in silico and in vivo studies. PLoS ONE 2012, 7, e30588. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.-C.; Wei, L.-H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 2017, 29, 2995–3011. [Google Scholar] [CrossRef] [PubMed]
- Huong, T.T.; Ngoc, L.N.T.; Kang, H. Functional characterization of a putative RNA demethylase ALKBH6 in Arabidopsis growth and abiotic stress responses. Int. J. Mol. Sci. 2020, 21, 6707. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Pickering, B.F.; Jaffrey, S.R. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 2018, 28, 113–127. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef]
- Song, P.; Yang, J.; Wang, C.; Lu, Q.; Shi, L.; Tayier, S.; Jia, G. Arabidopsis N6-methyladenosine reader CPSF30-L recognizes FUE signals to control polyadenylation site choice in liquid-like nuclear bodies. Mol. Plant 2021, 14, 571–587. [Google Scholar] [CrossRef]
- Wei, L.-H.; Song, P.; Wang, Y.; Lu, Z.; Tang, Q.; Yu, Q.; Xiao, Y.; Zhang, X.; Duan, H.-C.; Jia, G. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 2018, 30, 968–985. [Google Scholar] [CrossRef]
- Pontier, D.; Picart, C.; El Baidouri, M.; Roudier, F.; Xu, T.; Lahmy, S.; Llauro, C.; Azevedo, J.; Laudié, M.; Attina, A. The m6A pathway protects the transcriptome integrity by restricting RNA chimera formation in plants. Life Sci. Alliance 2019, 2, e201900393. [Google Scholar] [CrossRef]
- Shen, L.; Liang, Z.; Wong, C.E.; Yu, H. Messenger RNA modifications in plants. Trends Plant Sci. 2019, 24, 328–341. [Google Scholar] [CrossRef]
- Zhang, L.; Hanada, K.; Palukaitis, P. Mapping local and systemic symptom determinants of cucumber mosaic cucumovirus in tobacco. J. Gen. Virol. 1994, 75, 3185–3191. [Google Scholar] [CrossRef]
- Hirata, H.; Lu, X.; Yamaji, Y.; Kagiwada, S.; Ugaki, M.; Namba, S. A single silent substitution in the genome of Apple stem grooving virus causes symptom attenuation. J. Gen. Virol. 2003, 84, 2579–2583. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Borodynko, N.; Jackowiak, P.; Figlerowicz, M.; Pospieszny, H. Single mutation converts mild pathotype of the Pepino mosaic virus into necrotic one. Virus Res. 2011, 159, 57–61. [Google Scholar] [CrossRef]
- Yue, H.; Nie, X.; Yan, Z.; Weining, S. N6-methyladenosine regulatory machinery in plants: Composition, function and evolution. Plant Biotechnol. J. 2019, 17, 1194–1208. [Google Scholar] [CrossRef] [PubMed]
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell 2013, 50, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Marchena, L.; Marquez-Molins, J.; Martinez-Perez, M.; Aparicio, F.; Pallás, V. Mapping of Functional Subdomains in the atALKBH9B m6A-Demethylase Required for Its Binding to the Viral RNA and to the Coat Protein of Alfalfa Mosaic Virus. Front. Plant Sci. 2021, 12, 701683. [Google Scholar] [CrossRef]
- Bujarski, J.; Gallitelli, D.; García-Arenal, F.; Pallás, V.; Palukaitis, P.; Reddy, M.K.; Wang, A.; Consortium, I.R. ICTV virus taxonomy profile: Bromoviridae. J. Gen. Virol. 2019, 100, 1206–1207. [Google Scholar] [CrossRef]
- Aparicio, F.; Sánchez-Navarro, J.; Olsthoorn, R.; Pallás, V.; Bol, J. Recognition of cis-acting sequences in RNA 3 of Prunus necrotic ringspot virus by the replicase of Alfalfa mosaic virus. J. Gen. Virol. 2001, 82, 947–951. [Google Scholar] [CrossRef]
- Sánchez-Navarro, J.; Reusken, C.; Bol, J.; Pallás, V. Replication of alfalfa mosaic virus RNA 3 with movement and coat protein genes replaced by corresponding genes of Prunus necrotic ringspot ilarvirus. J. Gen. Virol. 1997, 78, 3171–3176. [Google Scholar] [CrossRef]
- Desbiez, C.; Schoeny, A.; Maisonneuve, B.; Berthier, K.; Bornard, I.; Chandeysson, C.; Fabre, F.; Girardot, G.; Gognalons, P.; Lecoq, H. Molecular and biological characterization of two potyviruses infecting lettuce in southeastern France. Plant Pathol. 2017, 66, 970–979. [Google Scholar] [CrossRef]
- Zhang, T.-Y.; Wang, Z.-Q.; Hu, H.-C.; Chen, Z.-Q.; Liu, P.; Gao, S.-Q.; Zhang, F.; He, L.; Jin, P.; Xu, M.-Z. Transcriptome-wide N6-methyladenosine (m6A) profiling of susceptible and resistant wheat varieties reveals the involvement of variety-specific m6A modification involved in virus-host interaction pathways. Front. Microbiol. 2021, 12, 656302. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, P.; Zhong, K.; Zhang, F.; Xu, M.; He, L.; Jin, P.; Chen, J.; Yang, J. Wheat yellow mosaic virus NIb interacting with host light induced protein (LIP) facilitates its infection through perturbing the abscisic acid pathway in wheat. Biology 2019, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Ge, L.; Li, Z.; Zhou, X.; Li, F. Pepino mosaic virus antagonizes plant m6A modification by promoting the autophagic degradation of the m6A writer HAKAI. aBIOTECH 2023, 4, 83–96. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, L.; Yao, Y.; Li, Y.; Zhang, H.; Fan, M. Transcriptome-wide N6-methyladenosine (m6A) methylation in watermelon under CGMMV infection. BMC Plant Biol. 2021, 21, 516. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wu, N.; Zhang, L.; Wang, X. RNA N6-methyladenosine modification suppresses replication of rice black streaked dwarf virus and is associated with virus persistence in its insect vector. Mol. Plant Pathol. 2021, 22, 1070–1081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhuang, X.; Dong, Z.; Xu, K.; Chen, X.; Liu, F.; He, Z. The dynamics of N6-methyladenine RNA modification in interactions between rice and plant viruses. Genome Biol. 2021, 22, 189. [Google Scholar] [CrossRef] [PubMed]
- Susaimuthu, J.; Tzanetakis, I.E.; Gergerich, R.C.; Martin, R.R. A member of a new genus in the Potyviridae infects Rubus. Virus Res. 2008, 131, 145–151. [Google Scholar] [CrossRef]
- Yue, J.; Lu, Y.; Sun, Z.; Guo, Y.; San León, D.; Pasin, F.; Zhao, M. Methyltransferase-like (METTL) homologues participate in Nicotiana benthamiana antiviral responses. Plant Signal. Behav. 2023, 18, 2214760. [Google Scholar] [CrossRef]
- Brocard, M.; Ruggieri, A.; Locker, N. m6A RNA methylation, a new hallmark in virus-host interactions. J. Gen. Virol. 2017, 98, 2207–2214. [Google Scholar] [CrossRef]
- Zhang, T.; Shi, C.; Hu, H.; Zhang, Z.; Wang, Z.; Chen, Z.; Feng, H.; Liu, P.; Guo, J.; Lu, Q. N6-methyladenosine RNA modification promotes viral genomic RNA stability and infection. Nat. Commun. 2022, 13, 6576. [Google Scholar] [CrossRef]
- Guo, T.; Liu, C.; Meng, F.; Hu, L.; Fu, X.; Yang, Z.; Wang, N.; Jiang, Q.; Zhang, X.; Ma, F. The m6A reader MhYTP2 regulates MdMLO19 mRNA stability and antioxidant genes translation efficiency conferring powdery mildew resistance in apple. Plant Biotechnol. J. 2022, 20, 511–525. [Google Scholar] [CrossRef]
- Ren, Z.; Tang, B.; Xing, J.; Liu, C.; Cai, X.; Hendy, A.; Kamran, M.; Liu, H.; Zheng, L.; Huang, J. MTA1-mediated RNA m6A modification regulates autophagy and is required for infection of the rice blast fungus. New Phytol. 2022, 235, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Steinwand, M.A.; Ronald, P.C. Crop biotechnology and the future of food. Nat. Food 2020, 1, 273–283. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, S.; Yu, L.; Xiao, Y.; Zhang, S.; Wang, X.; Xu, Y.; Yu, H.; Li, Y.; Yang, J. RNA demethylation increases the yield and biomass of rice and potato plants in field trials. Nat. Biotechnol. 2021, 39, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, M.; Li, Z.; Xie, X. The Roles of N6-Methyladenosine Modification in Plant–RNA Virus Interactions. Int. J. Mol. Sci. 2023, 24, 15608. https://doi.org/10.3390/ijms242115608
He M, Li Z, Xie X. The Roles of N6-Methyladenosine Modification in Plant–RNA Virus Interactions. International Journal of Molecular Sciences. 2023; 24(21):15608. https://doi.org/10.3390/ijms242115608
Chicago/Turabian StyleHe, Min, Zhiqiang Li, and Xin Xie. 2023. "The Roles of N6-Methyladenosine Modification in Plant–RNA Virus Interactions" International Journal of Molecular Sciences 24, no. 21: 15608. https://doi.org/10.3390/ijms242115608
APA StyleHe, M., Li, Z., & Xie, X. (2023). The Roles of N6-Methyladenosine Modification in Plant–RNA Virus Interactions. International Journal of Molecular Sciences, 24(21), 15608. https://doi.org/10.3390/ijms242115608






