Abstract
Papain-like cysteine proteases are composed of 11 human cysteine cathepsins, originally located in the lysosomes. They exhibit broad specificity and act as endopeptidases and/or exopeptidases. Among them, only cathepsins B, H, C, and X/Z exhibit exopeptidase activity. Recently, cysteine cathepsins have been found to be present outside the lysosomes and often participate in various pathological processes. Hence, they have been considered key signalling molecules. Their potentially hazardous proteolytic activities are tightly regulated. This review aims to discuss recent advances in understanding the structural aspects of these four cathepsins, mechanisms of their zymogen activation, regulation of their activities, and functional aspects of these enzymes in neurodegeneration and cancer. Neurodegenerative effects have been evaluated, particularly in Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis, multiple sclerosis, and neuropsychiatric disorders. Cysteine cathepsins also participate in tumour progression and metastasis through the overexpression and secretion of proteases, which trigger extracellular matrix degradation. To our knowledge, this is the first review to provide an in-depth analysis regarding the roles of cysteine cathepsins B, H, C, and X in neurodegenerative diseases and cancer. Further advances in understanding the functions of cysteine cathepsins in these conditions will result in the development of novel, targeted therapeutic strategies.
1. Cysteine Cathepsins: Structural and Functional Aspects
The discovery of lysosomes by Christian de Duve has been crucial for understanding intracellular degradation processes [1]. The release of enzymes from injured lysosomes results in the destruction of their own cells and cell death [2,3].
Lysosomes are present in almost all eukaryotic cells and contain more than 50 acid hydrolases. The primary function of lysosomes, which can degrade and digest proteins, is not to destroy cells but rather to maintain cellular homeostasis and recycle cell constituents, as has been demonstrated in several physiological processes (reviewed in [4,5,6,7,8,9]).
This classical view has changed more recently after the discovery of cysteine cathepsins in the nucleus, mitochondria, cytoplasm, and extracellular space [9,10,11,12,13].
Recent developments in quantitative proteomics and in vivo imaging have elucidated protease specificity profiling and identified physiological substrates [14], resulting from the concept of proteases, including cysteine cathepsins, as degrading enzymes and proteases as key signalling molecules [15,16].
A typical example of signalling is the activation of the pro-apoptotic protein Bid, a member of the Bcl-2 family, which initiates apoptosis [17,18,19,20].
When released from the lysosomes, cathepsins are potentially hazardous and frequently associated with various human pathologies, including cancer [21,22,23,24], cardiovascular diseases [25,26], neurodegeneration [24,27,28], bone disorders and inflammatory diseases [29,30,31], coronavirus disease caused by SARS-CoV-2 [32,33], and, although less investigated, diseases with a genetic deficiency of cysteine cathepsins F, K, C, and H and lysosomal storage diseases [34].
Proteases catalyse irreversible hydrolytic reactions; therefore, their proteolytic activity must be strictly regulated. This can be achieved at multiple levels by various mechanisms, such as gene expression, post-translational modification, autocatalytic activation of their inactive zymogens, or by other proteases targeting specific compartments, intracellular protein processing and degradation, oxidants and endogenous protein inhibitors, or exogenous inhibitors [16,35,36]. Once activated, mature enzymes are proteolytically active and must be regulated by pH, temperature, oxidation, glycosaminoglycans, and endogenous protein inhibitors.
Inhibitors can be classified into emergency and regulatory inhibitors based on their localisation. Emergency inhibitors are normally localised in different cellular compartments than the enzyme, cystatins being a typical example, whereas regulatory inhibitors are often co-localised with their target [16,37,38].
The most well-studied are cystatins (family I25), which are divided into stefins (I25A), cystatins (I25B), and kininogens (I25C) subfamilies [39]. They are competitive, reversible, tight-binding inhibitors that can discriminate between endo- and exopeptidases (more in reviews by [37,38,40,41,42]).
The crystal structure of chicken cystatin has served as the foundation for elucidating the new mechanism of interaction between cystatins and papain-like enzymes [43], which is thus confirmed by the crystal structure of the human stefin B-papain complex [44].
The chicken cystatin nuclear magnetic resonance structure exhibits the same overall fold but also notable differences in some segments of the polypeptide chain that are more similar to those of human stefin B [45].
Recently, thyropins, which are novel protein inhibitors structurally different from cystatins, have been identified. They belong to family I31 of clan IX [39]. The physiologically most important inhibitor of this family is the p41 fragment of the invariant chain, which inhibits several cathepsins and is involved in regulating major histocompatibility complex-II antigen presentation [46,47].
There are numerous synthetic inhibitors, among which epoxysuccinate derivatives were the first identified inhibitors of cysteine cathepsins [48]. One of these, the irreversible inhibitor CA030 (ethyl ester of epoxysuccinyl-Ile-Pro-OH), has been crystallised in complex with cathepsin B. Notably, the Ile-Pro-OH region of CA030 mimics the P1′ and P2′ residues in the substrate. Therefore, this structure initially revealed a substrate-like interaction with the S1′ and S2′ residues of papain-like enzymes [49].
Among lysosomal hydrolases, proteases (also termed peptidases) play an important role. There are 15 cathepsins in humans, which are classified as follows, according to their catalytic type: serine proteases (cathepsins A and G), aspartic proteases (cathepsins D and E), and cysteine proteases (cathepsins B, C, F, H, K, L, O, S, V, W, and X/Z) (Table 1 and Table S1). For consistency, the name cathepsin X has been used throughout the manuscript when referring to cathepsin X or Z, since they refer to the same enzyme that has been simultaneously reported by two independent groups using different names [50,51].

Table 1.
Background information on all 15 human cathepsins (11 cysteine, 2 serine, and 2 aspartic proteases).
These 11 lysosomal cysteine cathepsins are members of the papain family (C1A) from the cysteine peptidases clan (CA). They are predominantly endopeptidases, except for cathepsins C and X, which are strictly exopeptidases. Moreover, cathepsin B is a carboxydipeptidase, and cathepsin H is an aminopeptidase. Both of these cathepsins are predominantly exopeptidases, exhibiting limited access to their active sites. Cathepsin B contains an insertion of approximately 20 amino acid residues, termed the occluding loop, which blocks the active-site cleft and consecutively removes two amino acids [53]. With increasing pH, the loop becomes flexible, allowing cathepsin B to function as an endopeptidase [54]. An irreversible loss of cathepsin B activity accompanied by structural changes has been observed at neutral or alkaline pH [55]. Meanwhile, the exopeptidase activity of cathepsin B is limited to an acidic pH. Deletion of the occluding loop results in cathepsin B with endopeptidase activity only [56]. Therefore, the pH-dependence of the propeptide binding can be explained by competitive binding of the occluding loop and the propeptides [57]. The crystal structures of the cathepsin B-stefin A complex [58] and the cathepsin B-chagasin complex [59] displace the occluding loop, thus inhibiting the cathepsin B endopeptidase activity.
In vitro studies have demonstrated that only cathepsin L cleaves and activates procathepsin H [60]. Upon activation, the mature enzyme primarily acts as an aminopeptidase, thus cleaving a single N-terminal residue from the polypeptide chain. An octapeptide known as the “mini-chain,” which is disulphide-linked to the main enzyme structure in the narrow active-site cleft in the substrate-binding direction, is responsible for the strong aminopeptidase activity. The mini-chain is positioned in the active-site cleft via carbohydrate residues attached to the enzyme structure [61]. From the 38-residue propeptide, the mini-chain EPQNCSAT octapeptide contains non-primed substrate-binding sites starting at the S2 position. The positioning of the mini-chain and substrate, which is based on the displacement of residues within the active-site cleft, allows cathepsin H to exhibit exopeptidase activity. The crystal structure of stefin A-cathepsin H reveals structural changes along its interaction surface [62]. Recombinant cathepsin H lacking a mini-chain exhibits only endopeptidase activity, confirming that the mini-chain is responsible for the enzyme’s aminopeptidase activity [63].
All amino acid sequences were determined and confirmed by bioinformatics analysis of the human genome draft sequence [64]. While the majority of the above-described cysteine cathepsins are ubiquitously expressed, the other four cathepsins, K, S, V, and W (also named lymphopain), show a more restricted cell- or tissue-specific distribution, suggesting their specific cellular functions [65].
Cysteine cathepsins are optimally active at acidic pH values (pH 3.5–6.0) and in a reducing environment and are mostly unstable and inactivated at neutral pH values, except cathepsin S, which is stable and active at neutral or slightly alkaline pH values [66]. Heparin-like glycosaminoglycans can potentiate the endopeptidase activity of cathepsin B at alkaline pH values by interacting with heparin and heparan sulphate in the occluding loop of the enzyme [67]. Recently, dual activities, namely dipeptidyl carboxypeptidase and endopeptidase activities, of cathepsin B under both acidic and neutral pH conditions have been reported [68]. Furthermore, these researchers also developed a novel synthetic tripeptide substrate that is highly specific for monitoring high cathepsin B activity at acidic to neutral pH values [69].
Lysosomal cathepsins are synthesised as preproenzymes (Figure 1, Figure 2, Figure 3 and Figure 4). After removal of the N-terminal signal peptide in the endoplasmic reticulum, the resulting inactive proenzymes are transported to late endosomes or lysosomes, where the prodomain (propeptide) is removed by limited proteolytic processing to obtain active mature enzymes (Figure 1, Figure 2, Figure 3 and Figure 4). This activation process occurs autocatalytically at acidic pH values as a combination of unimolecular and bimolecular processes [70]. We further proposed a model for the autocatalytic activation of cysteine cathepsins. This involved the low catalytic activity of procathepsin B in dissociating the propeptide from the active-site cleft as the first unimolecular step during zymogen activation. The second step is the bimolecular proteolytic removal of the propeptide [71]. This activation is facilitated by glycosaminoglycans [72,73]. In contrast, procathepsin C is activated to its mature form by cathepsin L and S but not by autocatalytic processing [74]. Similarly, procathepsin X is incapable of autocatalytic processing but can be processed in vitro using cathepsin L under reducing conditions [75].
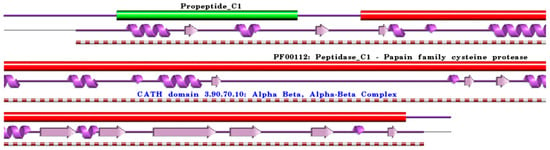
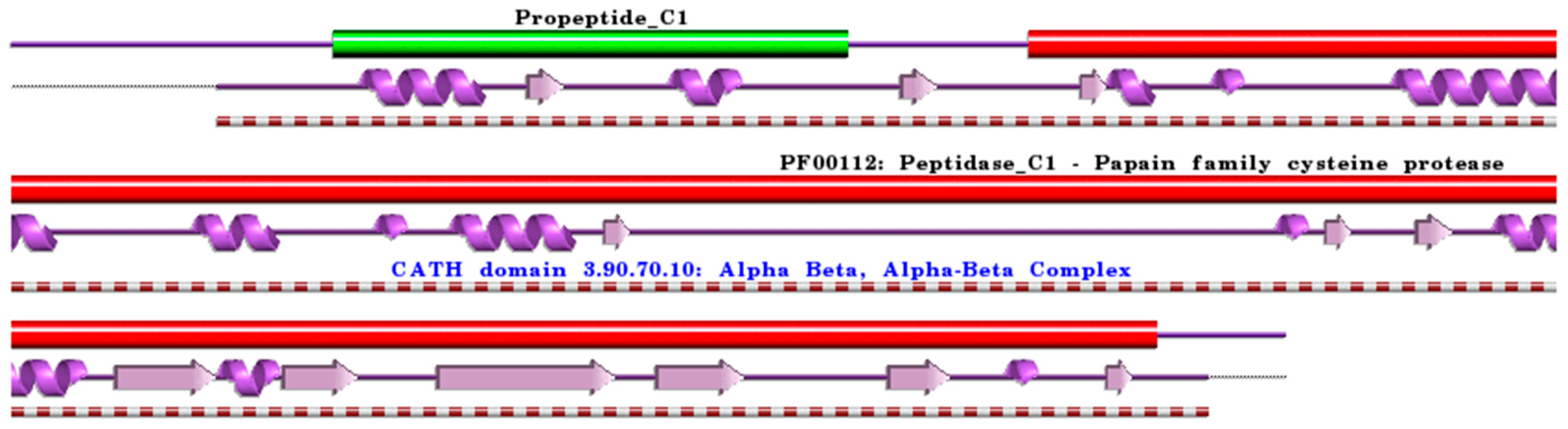
Figure 1.
Schematic representation of aligned PDB and UniProt sequences of human procathepsin B. The upper panel shows the domain composition of human procathepsin B [76] (UniProtKB ID P07858) [52], namely the “Activation peptide” (PfamA domain: Propeptide_C1) (green) and the “Cathepsin B chain” (PfamA domain: PF00112: Peptidase_C1) (red). In the lower panels, a schematic “wiring diagram” of human procathepsin B (PDB ID: 3pbh:A, [77]) 2D structure highlights the helices (purple springs) and strands (pink arrows), along with the CATH structural hierarchy classification of protein domain structures, which clusters proteins at four major levels, namely Class (C), Architecture (A), Topology (T), and Homologous superfamily (H). The figure was generated using the PDBsum web server [78]. The sequence alignment is shown in Supplementary Materials (File S1). Details regarding molecular processing are provided in the main text and Table 1.
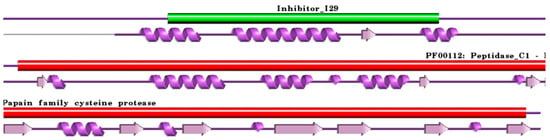
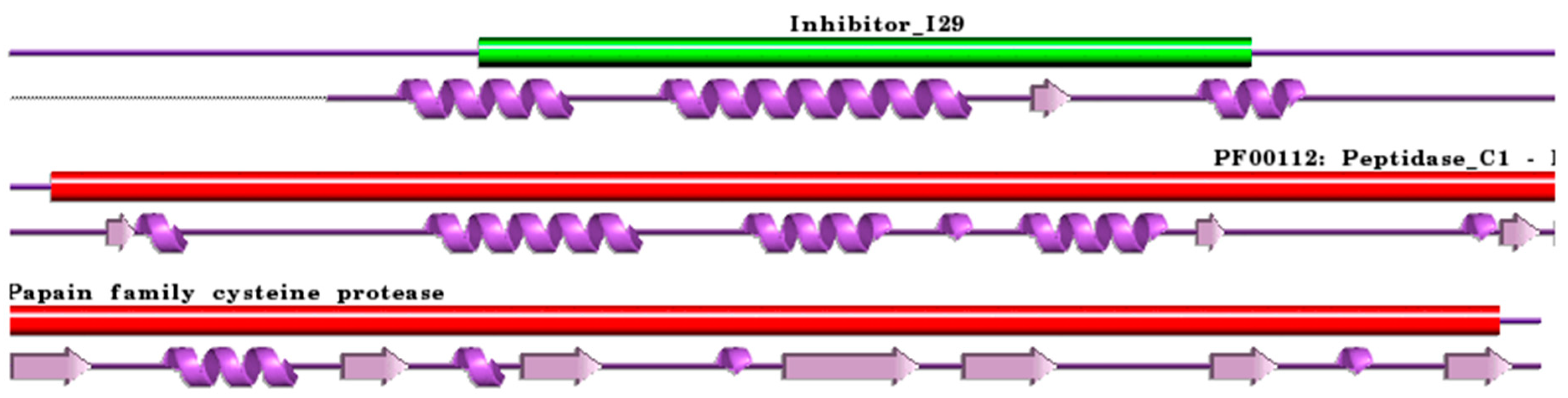
Figure 2.
Schematic representation aligned PDB and UniProt sequences of human procathepsin H. The upper panel shows the domain composition of human procathepsin H [79] (UniProtKB ID P09668) [52], namely the “propeptide region” (PfamA domain: Inhibitor_I29) (green) and the “Cathepsin H chain” (PfamA domain: PF00112: Peptidase_C1) (red). In the lower panel, a schematic “wiring diagram” of human procathepsin H (PDB ID: 6czk:A, [60]) 2D structure highlights the helices (purple springs) and strands (pink arrows). The figure was generated using the PDBsum web server [78]. The sequence alignment is shown in Supplementary Materials (File S2). Details regarding molecular processing are provided in the main text and Table 1.
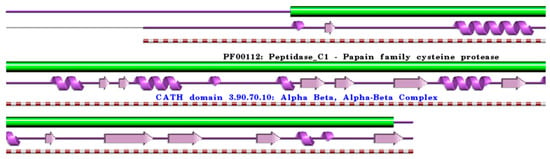
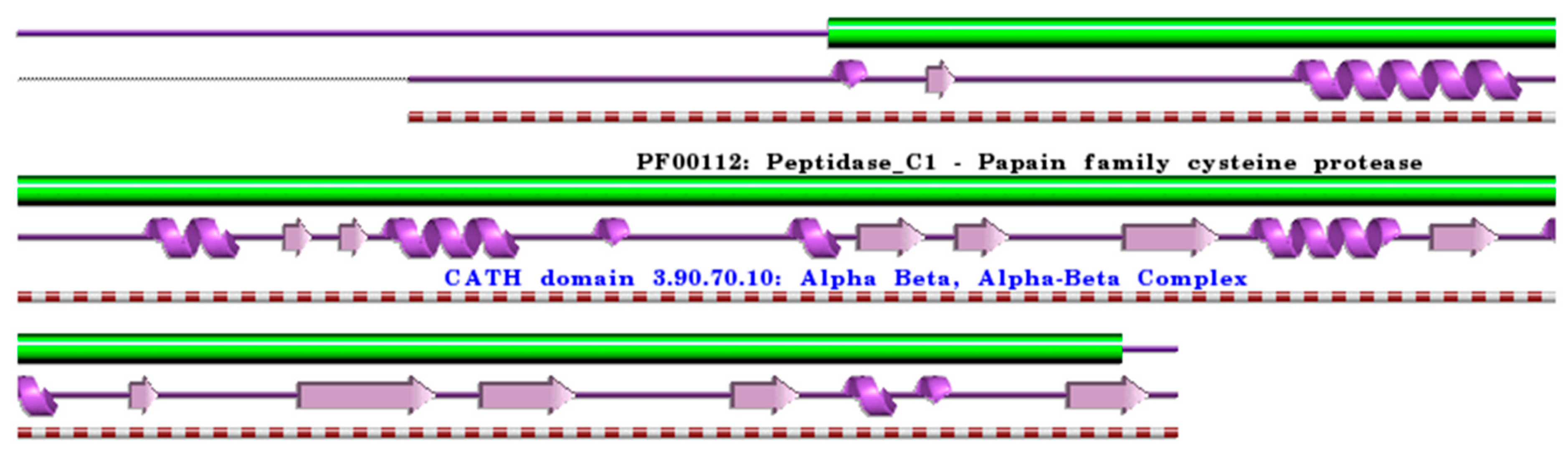
Figure 3.
Schematic representation of the aligned PDB and UniProt sequences of human procathepsin X. The upper panel shows the domain composition of human procathepsin X [50,51] (UniProtKB ID Q9UBR2) [52], namely the “Cathepsin X chain” (PfamA domain: PF00112: Peptidase_C1) (green). In the lower panels, a schematic “wiring diagram” of human procathepsin X (PDB ID: 1deu:A, [75]) 2D structure highlights the helices (purple springs) and strands (pink arrows), along with the CATH structural hierarchy classification. The figure was generated using the PDBsum web server [78]. The sequence alignment is shown in Supplementary Materials (File S3). Details regarding molecular processing are provided in the main text and Table 1.
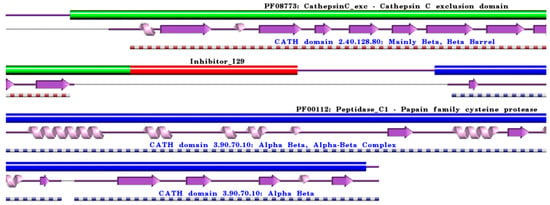
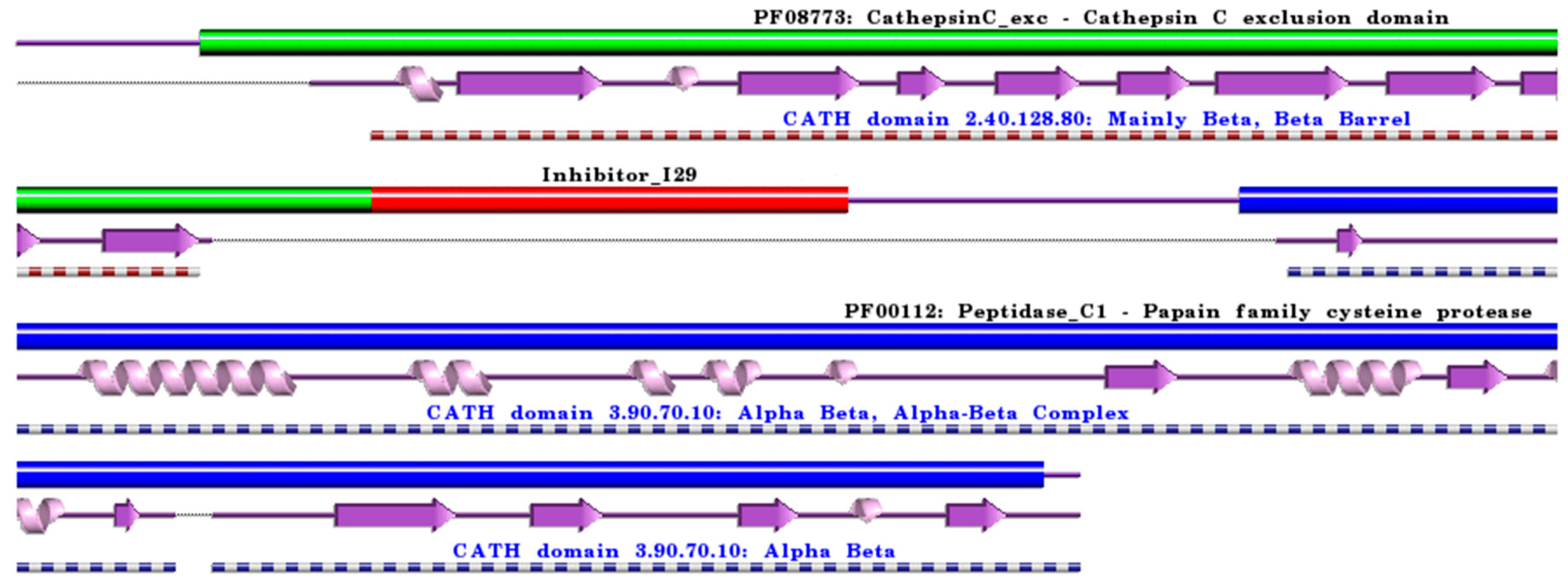
Figure 4.
Schematic representation of aligned PDB and UniProt sequences of human procathepsin C. The upper panel highlights the domain composition of human procathepsin C [80] (UniProtKB ID P53634) [52], comprising the mature form of the enzyme, namely the “Cathepsin C exclusion domain chain” (PfamA domain: PF08773: CathepsinC_exc) (green), the “Cathepsin C chain” (PfamA domain: PF00112: Peptidase_C1) (blue), and the “propeptide” (PfamA domain: Inhibitor_I29) (red). In the lower panels, a schematic “wiring diagram” of human cathepsin C (PDB ID: 3pdf:A, [81]) 2D structure highlights the helices (purple springs) and strands (pink arrows), along with the CATH structural hierarchy classification. The figure was generated using the PDBsum web server [78]. The sequence alignment is shown in Supplementary Materials (File S4), where it is evident that the propeptide (Inhibitor_I29) is absent in the 3D structure. Details regarding molecular processing are provided in the main text and Table 1.
The crystal structures of human procathepsins, including those of procathepsin B [77,82], H [60], X [75], and L [83], have revealed that these propeptides share the same fold despite differences in amino acid sequences and lengths (Figure S1).
Most propeptides contain approximately 100 amino acid residues; the shortest propeptide of cathepsin X contains only 38 residues [50,75], whereas the longest are cathepsin C, with 206 amino acid residues [80], and cathepsin F, which has 251 residues and contains a cystatin-like domain unique to cysteine cathepsin zymogens [84]. Propeptides fold on the enzyme surface, covering the catalytic site and acting as inhibitors, suggesting that this mode of inhibition is common to all enzymes of the papain superfamily [83,85]. The propeptides unfold at an acidic pH, thus exposing the active site of the enzyme and suggesting a mechanism of acidic zymogen activation [86].
During activation, propeptides from endopeptidases, including cathepsin B, dissociate from the enzyme surface, whereas exopeptidases, such as cathepsin C and cathepsin H, show different activation processes. The crystal structure of cathepsin C, which is unique among papain-like enzymes, reveals that the mature enzyme is a tetramer composed of four identical papain-like endopeptidases and four exposed active sites [87,88]. The proenzyme is a dimer [74], which oligomerizes into a tetramer. The additional domain, termed the “exclusion” domain, with no homology to papain-like enzymes, contributes to the tetrameric structure and is extended to the active site cleft, thus limiting access to the polypeptide apart from the N-terminus. The crystal structure shows that the mature enzyme contains 119 residues in the exclusion domain (from Asp 1 to 119) and 233 residues in the two papain-like domains (from Leu207 to Leu439). The 87-residue propeptide is cleaved off (from Thr120 to His206) during activation of the proenzyme by cathepsin L and cathepsin S [74]. It blocks not only the active site of the enzyme but also prevents oligomerization [88]. The active site is blocked beyond the S2 binding site by the exclusion domain (more details are provided in [88]). Cathepsin C, also called dipeptidyl peptidase I, sequentially cleaves dipeptides and is the only cathepsin that requires halide ions for its activity. Recombinant cathepsin C, which lacks its exclusion domain, is a monomer with endopeptidase activity [89].
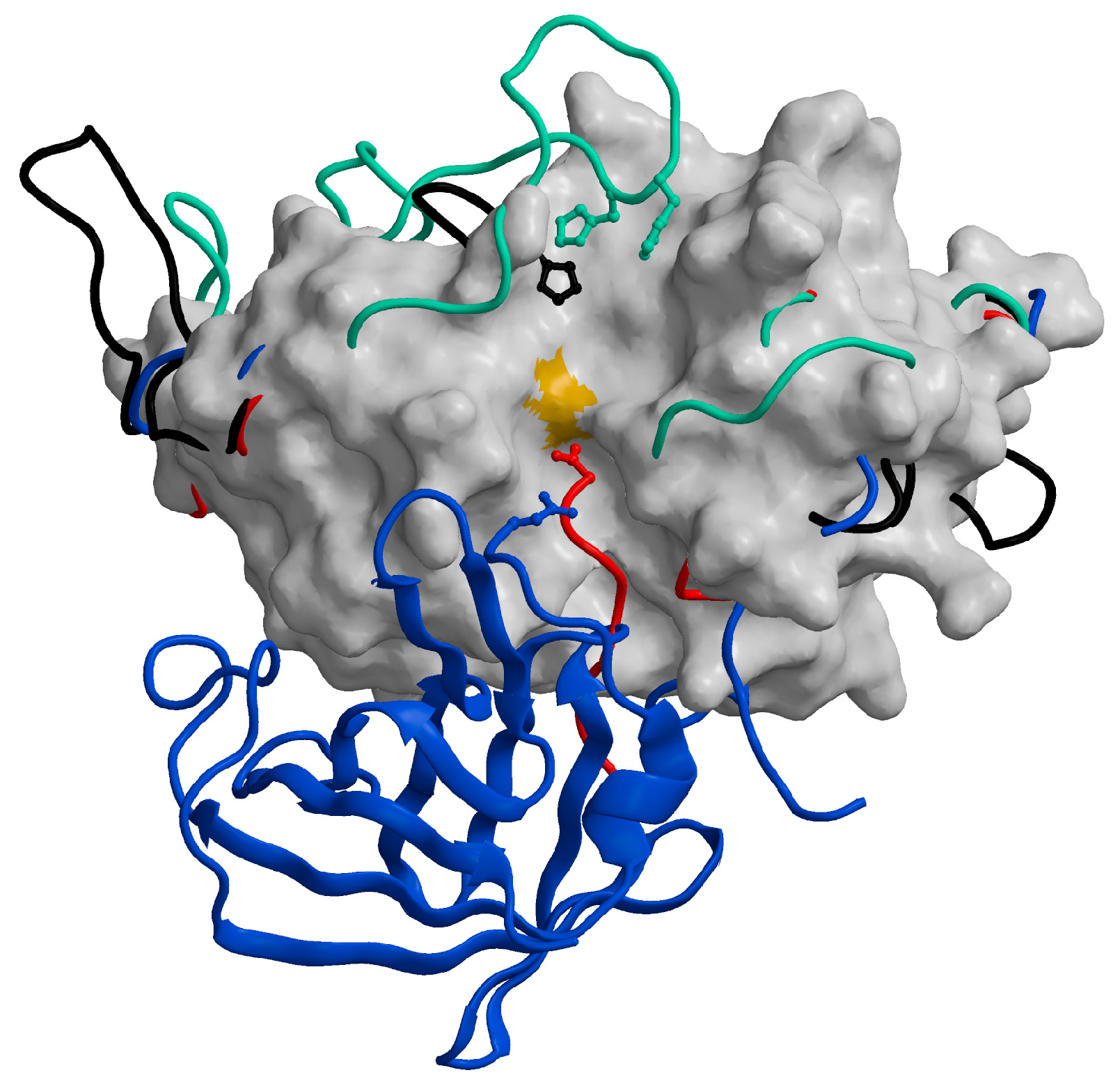
The mature forms of all cysteine cathepsins share similar sequences and a typical papain-like fold, which consists of two domains forming a “V” active site cleft with a catalytic dyad of Cys25 and His159 on opposite sides of the domains, forming a thiolate-imidazolium ion pair responsible for enzyme activity [90]. All cysteine cathepsins are monomers with molecular weights (MWs) of approximately 30 kDa, with the exception of tetrameric cathepsin C (200 kDa) [87] and the active homodimer of cathepsin X (55 kDa) [91]. From the crystal structures of the exopeptidases cathepsin B [53], cathepsin H [61], cathepsin C [88], and cathepsin X [92], it is evident that their exopeptidase activities result from additional structural elements such as loops (cathepsins B and X) and propeptide regions (cathepsins C and H) (Figure 5). Cathepsin X has later been reported to be a carboxymonopeptidase [93]. Detailed information is available in the original structural papers and reviews [65,90].
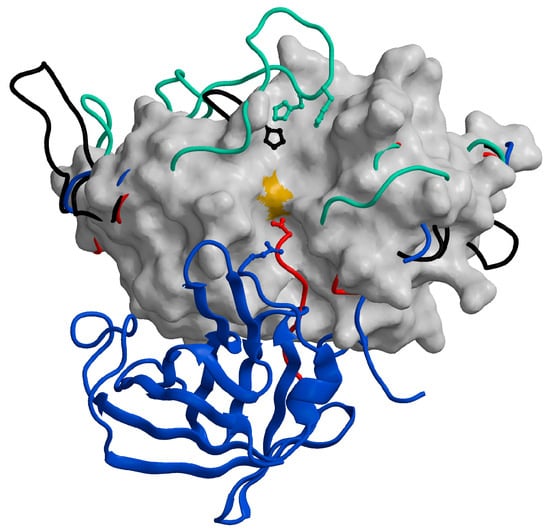
Figure 5.
Distinctive features of cysteine cathepsins with exopeptidase activity. The chain traces of the structural elements responsible for the exopeptidase activity of cathepsins B (1huc; [53]; green), C (1k3b; [88]; blue), H (8pch; [61]; red), and X (1ef7; [92]; black) are highlighted over the surface of the endopeptidase cathepsin L (1icf) [94]. The figure has been generated using the MAIN programme [95] and modified from [65].
Regarding the following sections, to our knowledge, this is the first review to provide an in-depth analysis regarding the roles of cysteine cathepsins B, H, C, and X in neurodegenerative diseases and cancer; notably, these cathepsins exhibit exopeptidase activity. Nevertheless, some recent reviews have focused on some of these cathepsins in either neurodegeneration [24,27,28,96,97,98,99] or cancer [23,24].
2. Cathepsins B, H, C, and X in Neurodegenerative and Neuropsychiatric Disorders
Age-related neurodegenerative disorders are often termed ‘proteinopathies’ due to the presence of misfolded and aggregated proteins that lose their physiological roles and acquire neurotoxic properties [100,101]. Notably, most neurodegenerative disorders share an endolysosomal dysfunction due to the accumulation and spread of oligomeric forms of neurotoxic proteins [100,102], where cathepsins play an important role [27,96,97,103,104,105,106,107,108,109]. Several proteins associated with neurodegenerative diseases have been identified as cathepsin substrates [98]. Recently, cysteine cathepsins were also found to be involved in neuroinflammation [24,110,111,112,113,114,115,116,117,118,119,120], a process closely linked to synaptic dysfunction and neurodegeneration [121,122,123]. Neuroinflammatory processes considerably impact the pathology of neuropsychiatric disorders [124]. Therefore, the role of cathepsins in these conditions has attracted increasing interest [125,126]. Noteworthy, cathepsin B [127,128,129,130] and cathepsin X [116,117], which have been linked to neuronal damage under a variety of pathological conditions [128,129,131], are released by activated microglia.
α-Synuclein (α-Syn) aggregation, which is clinically found in the inclusion bodies of post-mortem brain tissues from patients with Parkinson’s disease (PD) [132], activates microglia [133,134,135]. Overall, neuroinflammation in activated microglia is presumably neurotoxic [116,127,135,136,137].
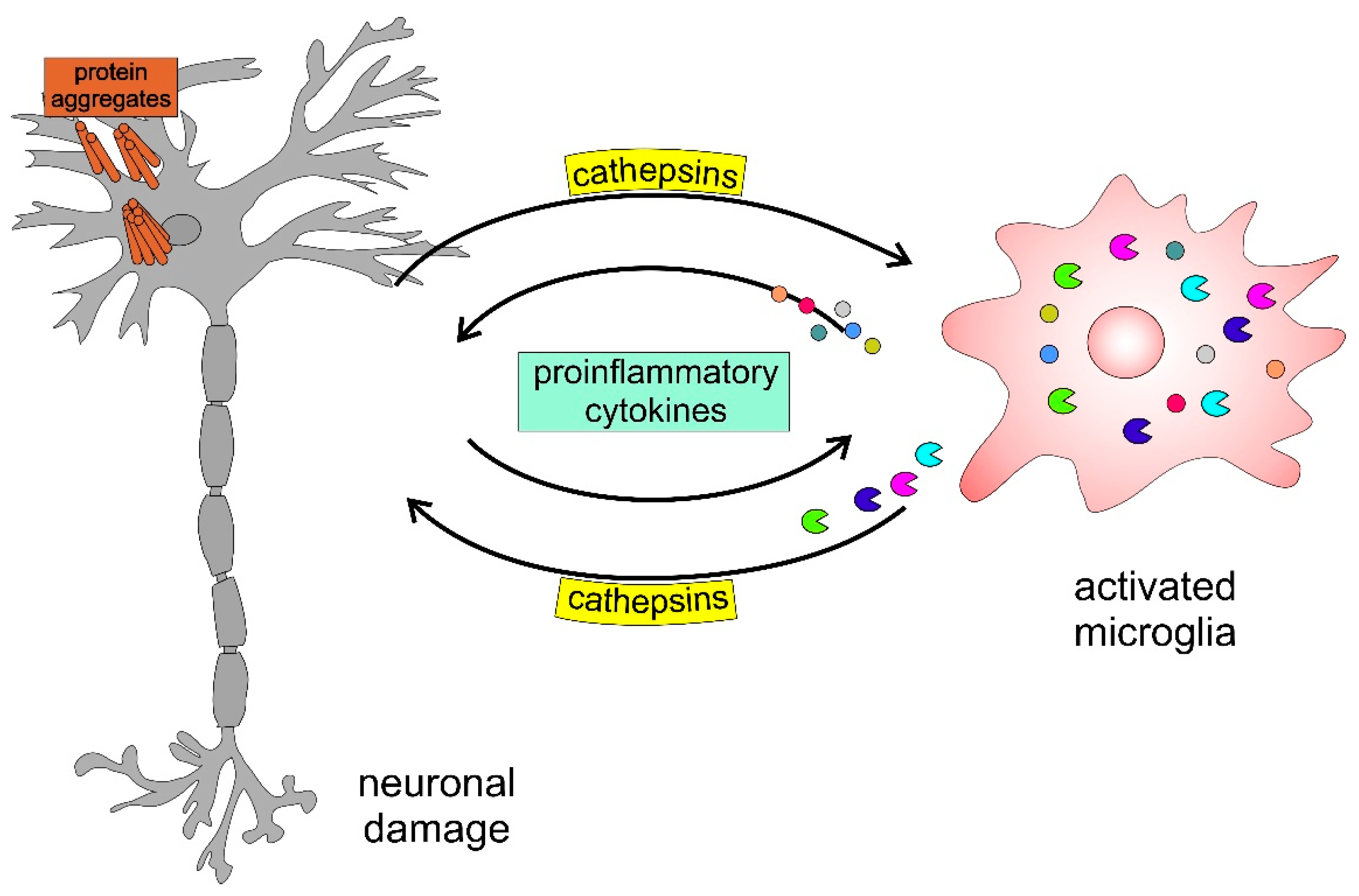
Figure 6 presents a schematic model highlighting the involvement of cathepsins in neurodegenerative disorders. The accumulation of protein aggregates, such as amyloid-β (Aβ), α-Syn, and mutated huntingtin, activates microglia, inducing the activation and release of cysteine cathepsins (i.e., B, H, C, X) and proinflammatory cytokines, including interleukin-1β (IL-1β) and tumour necrosis factor-α (TNF-α), which further enhance this self-propelling neurotoxicity, leading to neurodegeneration.
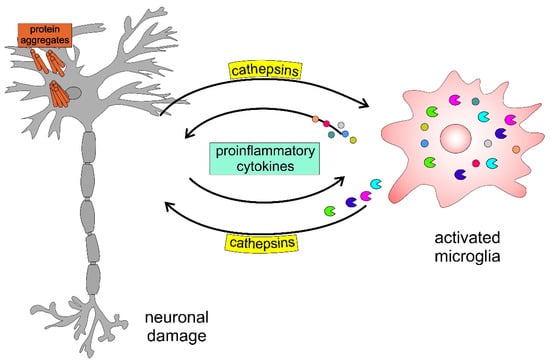
Figure 6.
Schematic model of neuronal-microglial crosstalk with emphasis on the role of cathepsins in neurodegenerative diseases.
2.1. Roles of Cathepsins B and X in Alzheimer’s Disease
Alzheimer’s disease (AD) is a progressive neurodegenerative disease frequently associated with memory deficits and cognitive decline [138,139]. Extracellular Aβ and amyloid precursor protein (APP) deposits, intracellular neurofibrillary tangles, dystrophic neuritis, and amyloid angiopathy are the neuropathological markers of AD [140].
The “amyloid cascade hypothesis”, presented by Hardy and Higgins in the 1990s, claims that the pathology of AD is caused by the deposition of Aβ, the primary component of plaques, consequently causing neurofibrillary tangles, cell death, vascular damage, and dementia [141]. Although this concept has influenced and guided much academic and pharmaceutical research, Aβ is necessary but insufficient to cause AD [142].
In contrast, Cataldo and Nixon proposed that APP within senile plaques is processed by lysosomal proteases principally derived from degenerating neurons [143]. Ten years later, Nixon proposed the “protease activation cascade”, which is pertinent to the pathogenesis of sporadic AD and entails the early and progressive activation of proteolytic systems such as the calpain-calpastatin and endolysosomal systems, but not exclusively [105]. Using thioflavin T fluorescence, liquid chromatography, and mass spectrometry, Lambeth and Julian recently investigated the proteolysis of Aβ by cathepsins B, H, L, and D and demonstrated that all Aβ fibril morphologies are resistant to cathepsin digestion [144].
Since the development of therapeutic agents for AD based on the amyloid cascade hypothesis was unsuccessful, considerable attention was given to the “amyloid cascade-inflammatory hypothesis” [145]. AD presumably results from an inflammatory response induced by extracellular Aβ deposits, which are later enhanced by tau aggregates. The inflammatory response driven by activated microglia increases with disease progression [145]. Importantly, cathepsins play a crucial role in the activation of microglia during chronic neuroinflammation [120,130].
On the one hand, cathepsins and other lysosomal hydrolases accumulate within senile plaques in the brains of patients with AD [105,146]. On the other hand, cathepsins are involved in the initiation and mediation of apoptosis and other forms of cell death [128]. Thus, dysfunction of the lysosomal system is a potential pathogenic mechanism in AD-related neurodegeneration. Cathepsin X is also associated with plaques in patients with AD [147] and AD transgenic mouse models APP/PS1 [147] and Tg2576 [148].
Moreover, Sun et al. proposed the “cystatin C-cathepsin B axis” and showed that cystatin C regulates soluble Aβ and Aβ-associated neuronal deficits by inhibiting cathepsin B-induced Aβ degradation [149]. Bernstein and Keilhoff recently reviewed the putative roles of cathepsin B in AD pathology and highlighted that it shows a neuroprotective effect by lowering Aβ levels and improving neuronal dysfunction; in contrast, it may also contribute to AD pathology by acting as a β-secretase and generating pyroglutamate Aβ [150].
Recently, Nixon proposed a multifactorial disease model wherein β-amyloidogenesis and the endolysosomal network have been identified as essential for the cause and progression of AD [151].
Bai et al. showed that oxidative stress activates the NLRP3 inflammasome by upregulating cathepsin B activity, thus highlighting the role of cathepsin B in neuroinflammation and as a potential target in AD therapy [152]. Recently, Nakanishi reviewed how intracellular and extracellular proteolytic mechanisms of microglial cathepsin B contribute to inflammatory brain disorders and brain aging [130]. Nuclear factor-κB (NF-κB) is activated by proteolytic degradation of κBα inhibitor (IκBα), an endogenous inhibitor of NF-κB, and subsequent nuclear translocation of NF-κB. The signalling-induced degradation of IκBα is mediated by the ubiquitin-proteasome system [153]. However, autophagy machinery may also be involved in IκBα degradation [154]. In activated microglia, cathepsin B induces the autophagic degradation of IκBα, leading to chronic neuroinflammation [155]. Various studies have demonstrated the involvement of microglial cathepsin B in cell death and Aβ clearance [128,129,130]. Therefore, the phagocytic clearance of Aβ by microglia may potentially resolve chronic neuroinflammation in AD [130]. Recently, Ni and Wu reviewed the molecular mechanisms governing the crosstalk between systemic inflammation and neuroinflammation. They suggested that disseminating inflammation indicates a negative spiral between systemic diseases and AD and proposed that inhibition of cathepsin B or S may delay the onset of AD and enable early intervention [155]. A systematic review of human post-mortem immunohistochemical studies and bioinformatics analyses revealed the complexity of AD reactive astrogliosis, which involves cathepsins [156]. In addition, Thygesen et al. demonstrated the involvement of cathepsin X expressed in myeloid cells of the central nervous system (CNS) in AD [157].
Since compromised synapses and cognition are improved by safely increasing protein clearance through modulated cathepsin B, Hwang et al.’s research supports the idea that early cathepsin B upregulation is a disease-modifying therapy that may also retard the progression of mild cognitive impairment to dementia [158].
The neuropathology of AD, traumatic brain injury, and other related brain disorders have all been linked to cathepsin B, according to extensive research from Hook’s lab [96,159,160]. Cathepsin B is possibly redistributed from the lysosomes to the cytosol, where it initiates cell death and inflammatory processes linked to neurodegeneration [96]. Therefore, cathepsin B has been proposed as a potential target for AD prevention and therapy [161,162,163,164].
Dunlop and Carney reported that L-serine selectively induces the activity of autophagic-lysosomal enzymes, cathepsins B and L, but not proteasome-hydrolysing activities, thus contributing to its neuroprotective effect [164]. Moreover, Cecarini et al. demonstrated that metabolites such as phenyl-γ-valerolactones exert neuroprotective activity by regulating intracellular proteolysis and confirmed the role of cathepsin B in autophagy [162]. However, the repertoire of potent small molecules that act as potential cathepsin B inhibitors is expanding. This includes E64d [161], pyridine, acetamide, and benzohydrazide compounds [163], and various natural and synthetic heterocyclic scaffolds [165].
Very recently, Cheng et al. reviewed the use of nanomedicines targeting AD lesions as a more suitable strategy than conventional therapy for AD treatment ([166] and references therein).
2.2. Roles of Cathepsins B and X in Parkinson’s Disease
Parkinson’s disease (PD) is the second most common age-associated neurodegenerative disorder and is characterised by the loss of dopaminergic neurons and the presence of α-Syn-containing aggregates in the substantia nigra pars compacta. Chronic neuroinflammation is a hallmark of PD pathophysiology [167], and microglial cathepsin B has been proposed as a key driver of inflammatory brain diseases and brain ageing [130].
To investigate the mechanisms underlying astrocyte ATP13A2-regulated lysosomal function and neuroinflammation following 1-methyl-4-phenylpyridinium treatment, Qiao et al. used a PD model of cultured primary neurons and astrocytes from the mouse midbrain [168]. The authors showed that the lack of ATP13A2 increases lysosomal membrane permeabilization and cathepsin B release, which in turn exacerbates activation of the NLRP3 inflammasome to produce excess IL-1β from astrocytes, thus suggesting a direct link between astrocyte lysosomes and neuroinflammation [168].
Codolo et al. demonstrated that although the monomeric and fibrillar α-Syn forms can promote pro-IL-1β expression, following the engagement of Toll-like receptor (TLR) 2, secretion of the mature cytokine is specific to the fibrillated protein, a process involving NLRP3 inflammasome activation [169]. This relies on the phagocytosis of fibrillar α-Syn, followed by the increased production of reactive oxygen species (ROS) and the release of cathepsin B into the cytosol [169]. In addition, Freeman et al. reported that α-Syn aggregates can induce lysosome rupture following endocytosis in neuronal cell lines via a mechanism that induces a cathepsin B-dependent ROS increase in target cells [170]. They also observed that α-Syn aggregates induce inflammasome activation in THP-1 cells [170]. NLRP3 inflammasome activation by α-Syn upon microglial endocytosis and subsequent lysosomal cathepsin B release has also been confirmed in the midbrain of PD model mice and in the serum of patients with PD [171]. Thus, fibrillar α-Syn released during neuronal degeneration endogenously triggers the cathepsin B-mediated inflammatory response in PD, which likely precedes neurodegeneration [170,171].
In contrast, cysteine cathepsin activity is essential for the lysosomal degradation of α-Syn [172] and C-terminal α-Syn truncations in PD [173]. Hu et al. showed that α-Syn is primarily degraded in the lysosomes, whereas the Leucine-rich repeat serine/threonine-protein kinase 2 (LRRK2) G2019S mutation, which is the most common genetic cause of PD, inhibits α-Syn degradation and promotes its aggregation. Moreover, LRRK2 G2019S decreases the activity of lysosomal enzymes, including cathepsins B and L, indicating that the inhibitory effect of LRRK2 G2019S on α-Syn degradation could underlie the pathogenesis of aberrant α-Syn aggregation in PD with LRRK2 mutation [174]. In addition, α-Syn fibril-induced intracellular aggregate formation requires lysosomal function, which is dependent on cathepsin B and not aspartic cathepsin D [175].
Recently, Blauwendraat et al. demonstrated a decrease in active cathepsin B protein levels in iPSC-derived neurones among glucosylceramidase β1 (GBA) variant carriers compared to those in non-carriers, suggesting a further reduction in lysosomal protease function in these cases. Moreover, α-Syn levels remain unaltered in the forebrain neurones carrying the GBA variant, suggesting that the overall reduction in lysosomal proteases allows for a faster accumulation of α-Syn aggregates as neurones age [176].
In contrast, Nelson et al. revealed that cathepsin D activity significantly decreases in the temporal cortex of patients with late-stage PD in the absence of cathepsin B as well as glucocerebrosidase (GCase) activity [177]. Moreover, a significant correlation exists between a decrease in GCase activity and an increase in p129S-α-Syn, whereas cathepsin D or cathepsin B do not correlate significantly with α-Gal A activity or levels [177].
More recently, Kim et al. demonstrated that ceramide activates cathepsin B and identified a novel role for cathepsin B in mediating prosaposin cleavage to form saposin C, the lysosomal coactivator of GCase [178]. Senkevich et al. reported that genetic modifiers such as LRRK2, endosomal/lysosomal proton channel TMEM175, α-Syn→(SNCA), and cathepsin B (CTSB) can either affect GCase activity or modulate the risk and age at the onset of GBA-associated PD [179]. In addition, Kim et al. suggested that loss of GBA1, Sphingomyelin phosphodiesterase (SMPD1), or Galactocerebrosidase (GALC) function in PD causes lysosomal ceramide deficiency; reduced ceramide-mediated cathepsin B activation in the lysosomes subsequently impairs the processing of prosaposin to saposin C, ultimately impairing GCase activity [178]. Kim et al. were the first to report a mechanistic link between ceramide and cathepsin B in regulating GCase activity and suggested that targeting lysosomal ceramide or cathepsin B is an important therapeutic strategy for activating GCase in PD and related disorders [178].
Recently, Pišlar et al. reported the upregulation of cathepsin X in the 6-hydroxydopamine (6-OHDA) model of PD and suggested cathepsin X as an important factor leading to the progressive loss of dopaminergic neurones and a potential therapeutic target for PD intervention [108]. Moreover, dopamine neuron cell death on treatment with 6-OHDA induces the loss of tyrosine hydroxylase, caspase-3 activation, intracellular ROS generation, and mitochondrial dysfunction, including the release of cytochrome c and an imbalanced Bax/Bcl-2 ratio [109]. This process is prevented by the cathepsin X inhibitor AMS36, which interferes with NF-κB activation by blocking IκBα degradation and preventing NF-κB nuclear translocation [109]. In addition, Lee et al. showed that PC12 cells exposed to 6-OHDA exhibit lysosomal dysregulation, caspase activation, and cell death, which are attenuated by the inhibitors pepstatin A and DEVD-Cho, whereas the cathepsin B inhibitor, CA-074Me, fails to protect cells [180]. In contrast, Wu et al. reported that the autophagy/lysosomal pathway is involved in the 6-OHDA-induced death of PC12 cells. The authors showed that overactive autophagy due to mitochondrial disability increases cathepsin B expression and diminishes Bcl-2 expression, whereas necrostatin-1 exerts a protective effect against injury in dopaminergic neurones [181].
Recently, Milanowski et al. identified CTSB p.Gly284Val as a rare variant in PD pathogenesis, suggesting that the CTSB locus harbours variants with varying penetrance that determine disease risk [182]. This finding expands the known repertoire of PD-linked genes (PARK1-21) [97,183,184] and the comprehensive genetic database for PD (Gene4PD) [185].
2.3. Roles of Cathepsins B, H, and X in Huntington’s Disease
Huntington’s disease (HD) is a progressive, fatal, autosomal dominant neurodegenerative disorder characterised by uncontrolled excessive motor movements and cognitive and emotional deficits [186,187,188,189].
Early studies by Mantle et al. reported a significant increase in protease activity, particularly of cathepsins H and D, in the brain tissue of patients with HD [104]. Nagata et al. provided direct evidence of abnormalities in HD tissues outside the brain under basal conditions by examining patient lymphoblasts. The authors reported pronounced vacuole formation with huntingtin remnants and cathepsin B staining, suggesting autophagy [190]. Later, Zhang et al. used an HD mouse model to demonstrate involvement of the p53 pathway in signalling, both autophagy and apoptosis, a process involving active cathepsins B and D [191].
Moreover, Kegel et al. identified the endolysosomal pathway as the main pathway for the removal of excess huntingtin, and lysosomal activity may regulate the cleavage of N-terminal fragments, which later aggregate in the nuclear and cytoplasmic inclusions of HD neurones [192]. Several proteases, including cathepsins B, L, X, and D, caspases, calpain, metalloproteases, and proteasomes, contribute to the N-terminal proteolysis of mutant huntingtin [107,192,193,194,195,196].
Interestingly, using cathepsin-deficient cells and pharmacological inhibitors, cathepsins L and X were found to degrade polyQ proteins and peptides but not other aggregation-prone proteins, suggesting that they may have a crucial role in host defence against the toxic accumulation of polyQ proteins [197]. Lai et al. reported that scyllo-inositol promotes the robust degradation of mutant huntingtin protein mediated by lysosomes and proteasomes but not autophagosomes. The rescue of degradation pathways is due to a reduction in mutant polyQ-huntingtin protein levels and is not a direct result of the compound on the lysosome or proteasome [198].
2.4. Roles of Cathepsins B, H, and X in Amyotrophic Lateral Sclerosis
Amyotrophic lateral sclerosis (ALS) is a degenerative motor neuron disease with a complex aetiology involving protein misfolding. This feature is shared by other neurodegenerative diseases, although there is a distinct common thread among ALS genes, associating them with the autophagy cascade [199].
To clarify the possible association of ALS neurodegeneration with the endolysosomal system, Kikuchi et al. examined the pathological expression of cysteine cathepsins B, H, and L and aspartic cathepsin D in the anterior horns of 15 ALS cases and five controls [106]. Consequently, only cathepsin B expression was upregulated, suggesting that it may play an important role in motor neuron degeneration in ALS [106]. Recently, Mori et al. showed that autophagy is a common degradation pathway for Bunina Bodies and TAR DNA-binding protein 43 (TDP-43) inclusions, which may explain the frequent coexistence of these inclusions in anterior horn cells in sporadic ALS [200].
Lee et al. demonstrated that proteasome inhibitors, but not cathepsin B inhibitors, increase superoxide dismutase [Cu-Zn] (SOD1) aggregate formation but do not promote cell death, indicating the absence of an association between SOD1 aggregates and cell death in familial ALS [201].
cDNA microarray analysis of post-mortem spinal cord specimens from four patients with sporadic ALS compared to four age-matched non-neurological controls revealed 60 differentially expressed genes, including an increase in the expressions of cathepsins B and D, apolipoprotein E, epidermal growth factor receptor, ferritin, and lysosomal trafficking regulator [202]. Since the findings from patients with sporadic ALS corroborate those of the SOD1 transgenic mouse model, the examined genes are suggested to play a specific role in the pathogenesis of ALS [202].
In addition, Boutahar et al. evaluated the effect of oxidative or excitotoxic stress on the transcriptional profile of ALS-linked mutant SOD1-cultured neurones and observed that both the ubiquitin-proteasome and endolysosomal systems are upregulated in transgenic neuron cultures [203]. Moreover, a meta-analysis of gene expression profiling in ALS consistently confirmed that the differential expressions of cathepsins B and D, GFAP, and SERPINA3 are significant in both the mouse model and patients with ALS [204].
Fukada et al. analysed gene expressions in the spinal cord of SOD1 (L126delTT) Tg mice using a cDNA microarray and identified four genes (Crym, Hspb1/Hsp27, CtsH, and Paip1) potentially related to the pathogenesis of familial ALS, including the progression of reactive astrocytes and the inflammatory response of microglial cells. In particular, cathepsin H was present in reactive astrocytes and microglial cells, suggesting that its overexpression might be associated with a reaction against misfolded proteins due to failure of the ubiquitin–proteasome system [205].
Gene profiling of skeletal muscles in an ALS mouse model showed that before the onset of overt clinical symptoms and motor neuron death, early changes affect genes involved in detoxification, regeneration, tissue degradation, and cell death. Notably, cathepsin X, metallothionein-1 and -2, ATF3, and galectin-3 genes appear to be regulated in both the skeletal muscle and spinal motor neurons of paralysed ALS mice [206].
In addition, Wendt et al. showed that cathepsin X is critical in degenerative processes during normal aging and under pathological conditions, as it has been found to be upregulated in numerous glial cells in the degenerating brain regions of a transgenic ALS mouse model [147].
Moreover, a neurodegeneration-specific gene expression signature of acutely isolated microglia from an ALS mouse model has revealed co-regulated genes in the lysosome pathway, which include several cathepsins (A, B, D, L, S, X, and E), a host of lysosome enzymes (HexA), membrane markers (Cd68, Cd63, and Lamp1), and components of lysosomal ATPase (Atp6v0d1) [207]. Therefore, cathepsins may be involved in the removal of mutant SOD1 aggregates and neuronal debris in ALS mice [207]. Conversely, Ulbrich et al. reported evidence for a reciprocal influence of SOD1 and stefin B/cystatin B genes and a direct interaction between the two proteins [208].
Watanabe et al. demonstrated that cystatin C, the main component of Bunina bodies in ALS, is an endogenous neuroprotective factor that functions via the coordinated activation of two distinct neuroprotective pathways, namely, induction of autophagy and inhibition of aberrant cathepsin B activity [209].
2.5. Roles of Cathepsins B, H, C, and X in Multiple Sclerosis
Multiple sclerosis (MS) affects the CNS and is characterised by inflammation, demyelination, and neurodegeneration [210,211]. Increased cathepsin B levels have been reported in monocytes and macrophages, cells known to be activated in the peripheral blood of patients with MS and implicated as effectors of demyelination [212].
Moreover, biochemical analysis of MS brain tissue suggests that monocytes, macrophages, and reactive astrocytes are potential sources of increased cathepsin B levels [213]. Since proteasomal dysfunction is observed in the white and grey matter of patients with MS, an increase in cathepsin B activity may represent a compensatory mechanism for intracellular protein degradation [214].
To identify the proteases involved in MS pathogenesis, cDNA microarray analysis was performed on the brains of transgenic (plptg/−) mice, an animal model that closely mimics the failure of remyelination in MS [215]. Cathepsins B, H, and L are upregulated in the microglia and macrophages of the brain white matter, whereas elevated cystatin C expression is found in astrocytes, suggesting that the imbalance between cathepsins and their inhibitors may be cytotoxic to neurones (axons) and oligodendrocytes [215].
Using an animal model of MS, Allan and Yates demonstrated that cathepsin L−/− attenuates myelin oligodendrocyte glycoprotein (MOG) antigen presentation and the development of experimental autoimmune encephalomyelitis (EAE) [216]. In contrast, neither cathepsin B−/− nor cathepsin S−/− showed any effect, whereas their double-mutant mice showed attenuated MOG antigen presentation and EAE development [216]. Moreover, Okada et al. showed that cathepsin H deficiency impairs the TLR3-mediated activation of the interferon regulatory factor 3 (IRF3) and interferon-β (IFN-β) secretion from dendritic cells, thus enhancing Th1 cell differentiation and resulting in early-onset EAE in an animal MS model [217]. Therefore, functional redundancy among cathepsins B, L, and S in EAE suggests that the inhibition of multiple cysteine cathepsins may improve autoimmune disorders, such as MS. In contrast, the inhibition of cathepsin H may have an adverse effect on MS.
Recently, Liang et al. demonstrated that the absence of the cystatin F gene and the resulting disinhibition of cathepsin C aggravate demyelination. This finding may be related to increased expression of the glia-derived chemokine CXCL2, which may attract inflammatory cells to sites of myelin sheath damage, an effect that is reversed by knockdown of the cathepsin C gene [113]. Shimizu et al. showed that the balance between cathepsin C and cystatin F controls remyelination in the brain of Plp1-overexpressing mice, a model of chronic demyelinating disease [218]. From the same group, Durose et al. confirmed that cathepsin C and cystatin F are strongly associated with inflammatory demyelination; they demonstrated that the severity of EAE is reduced in the absence of cathepsin C. In contrast, increased microglial cathepsin C expression enhances clinical severity, suggesting that the interaction between cathepsin C and cystatin F plays an essential role in the pathogenesis of inflammatory demyelination in EAE [219].
In addition, cathepsin X propagates IL-1β-driven neuroinflammation, thus providing mechanistic support for the epigenetic risk factors in MS [114]. Haves-Zburof et al. evaluated whether the expression levels of cathepsins B and S and their inhibitors, cystatins B and C, are affected by the MS disease state and therapies (IFN-β and methylprednisolone) and whether they are associated with the IFN-β response phenotype. The authors demonstrated that cathepsin S expression levels are aberrantly elevated in patients with MS, in contrast to cathepsin B. However, the value of cathepsin S and cystatin C as predictive biomarkers for disease type, response to therapy, and the development of new targeted therapies for immune-mediated disorders, such as MS, requires further validation [220].
2.6. Roles of Cathepsins B and C in Neuropsychiatric Disorders
Transcriptome analysis of inbred mouse lines selected for low or high anxiety-related behaviour with depression-like behaviour revealed that cathepsin B is responsible for low anxiety in female mice [221]. Assessment of anxiety-related and depression-like behaviours in cathepsin B-deficient mice revealed an increase in depression-like behaviours in females. In contrast, cathepsin C aggravates neuroinflammation involved in behavioural and neurochemical disturbances in acute and chronic stress-induced murine models of depression [222]. In contrast, cathepsin C knockdown partially prevents inflammation, which may help alleviate the symptoms of depression in mice.
The “monoamine hypothesis of depression” postulates that the underlying pathophysiologic basis of depression is the decreased levels of 5-hydroxytriotamin, noradrenalin, and/or dopamine in the CNS. More recently, the “neuroplasticity hypothesis of depression” identified dysfunctional neural plasticity as the pathophysiological basis of depression [223]. The role of cathepsin C in promoting anxiety- and depression-like behaviours may be due to the involvement of cathepsin C in neuroinflammation induced by activated microglia [111,119], because depression-like behaviour induced by cathepsin C overexpression is associated with increased neuroinflammation and decreased 5-hydroxytryptamine levels [222]. In contrast, cathepsin B induces neuroinflammation by activated microglia [120,130] and protects against anxiety- and depressive-like behaviours. Therefore, the mechanisms underlying the protective effect of cathepsin B against these disorders may stem from its role in activity-dependent neuronal plasticity by activating matrix metalloprotease-9 [224,225]. Nevertheless, the specific pathophysiological roles of cathepsins in neuropsychiatric disorders should be elucidated in future studies.
3. Cathepsins B, H, C, and X in Cancer
Cysteine proteases play prominent roles in multiple molecular pathways involved in tumour progression and metastasis [21,24,226]. Cathepsin B, the most abundant and ubiquitously expressed exopeptidase of the papain family, is associated with tumour progression in numerous cancer types, including colorectal, breast, lung, pancreatic, and gastric cancer [227,228,229,230,231,232,233]. Cathepsin B expression correlates with increased malignancy and poor prognosis; thus, it has been proposed as a predictive biomarker for oral squamous cell carcinoma [234], cervical cancer [235], endometrial cancer [236], and colorectal cancer [232]. Another carboxypeptidase of the clan CA/C1 cysteine protease family, cathepsin X, has been primarily implicated in the development of gastrointestinal cancers, including colorectal [237,238], gastric [239], liver [240], and pancreatic cancers [241].
Moreover, Wang et al. demonstrated the involvement of cathepsin X in regulating the epithelial-to-mesenchymal transition and invasion in hepatocellular carcinoma [240]. Aminopeptidases H and C are overexpressed in various cancers and are involved in malignant transformations [242,243,244,245]. Cathepsin H regulates the processing of talin, a large focal adhesion protein, thereby promoting PC3 prostate cancer cell progression by modulating integrin activation and adhesion strength [246].
Endopeptidase cathepsin L is implicated in tumourigenesis and the malignant progression of different tumour types [24,247,248]. It may act as a tumour promoter by interfering with the tumour suppressor CDK2-AP1, leading to the progression of breast cancer [249]. Tumour-secreted cytokines, which are closely associated with malignant progression, significantly enhance the transcriptional upregulation of cathepsin L. Moreover, increased promoter activity and cathepsin L synthesis by vascular endothelial growth factor A (VEGF) have been demonstrated in glioblastoma cells [250].
Although lysosomal cysteine cathepsins are predominantly intracellular, they are secreted into the extracellular space under multiple physiological and pathological conditions [10,21]. The secretion of cysteine cathepsins is often accompanied by acidification of the extracellular milieu [251], which is a characteristic feature of the tumour microenvironment (TME). Notably, the slightly acidic pH of tumours provides a favourable environment for extracellular cathepsin activity, thereby promoting the execution of their function.
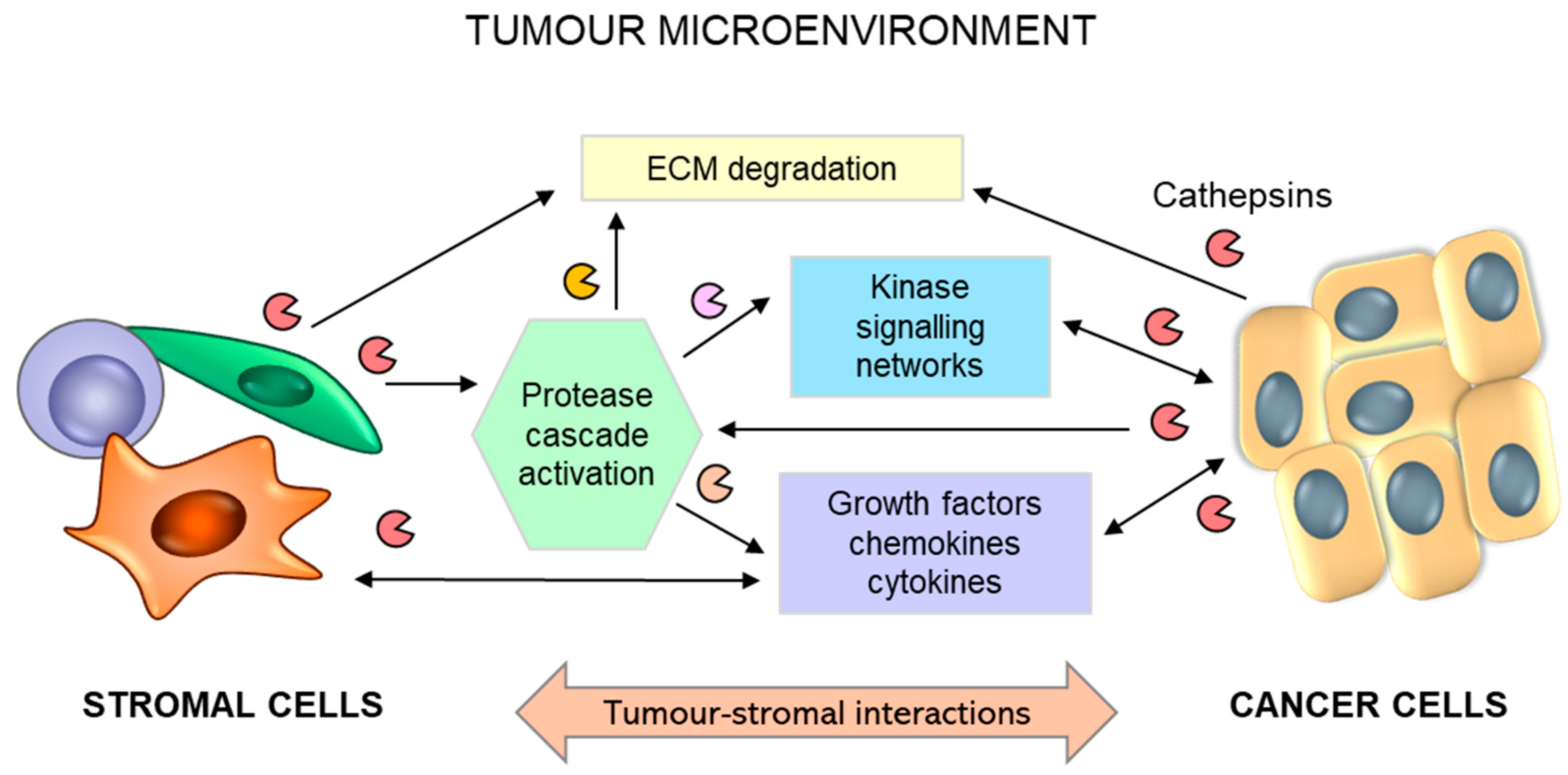
In addition to tumour cells secreting substantial levels of cathepsins, tumour stromal cells, such as endothelial cells, mast cells, tumour-associated macrophages, and fibroblasts, are important contributors to the increased levels of cysteine cathepsins in the TME [11,252] (Figure 7). The bulk of cathepsin B and X activity in several cancer types emanates from immune cells of the myeloid lineage [253], such as peritumoral macrophages [254,255,256,257] and myeloid-derived suppressor cells [258]. Secreted cysteine cathepsins can participate in the degradation of extracellular matrix (ECM) proteins, such as E-cadherin [259], collagen IV [260,261], or tenascin-C [262].
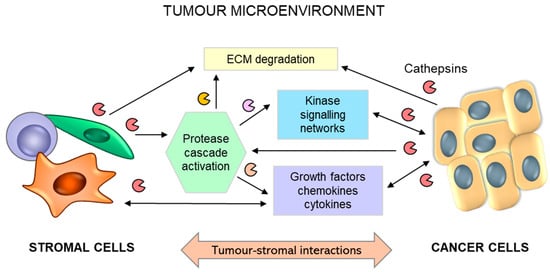
Figure 7.
Schematic representation for the role of reciprocal interactions between tumour and stromal cells in promoting tumour progression. Tumour-stromal crosstalk leads to activation of the stroma and overexpression and secretion of proteolytic enzymes, including cathepsins (B, H, C, and X), triggering extracellular matrix (ECM) degradation and the release of soluble factors. Activated stromal cells (macrophages, fibroblasts, and mast cells) secrete additional growth factors, cytokines, and chemokines, which regulate numerous interrelated events leading to tumour progression and metastasis.
Nevertheless, more specific roles of cysteine cathepsins have recently been discovered in modulating extra- and intracellular signal transduction pathways, which can also be executed through the shedding of receptors and adhesion molecules or the processing of respective cytokines and growth factors [11]. It was recently demonstrated that cathepsin C can promote proliferation and metastasis in hepatocellular carcinoma through activation of the TNF-α/MAPK (p38) signalling pathway [263]. Moreover, cathepsin B expression is implicated in regulating TGF-β1 signalling [264] and MAP and PI3 kinase pathways in malignant meningiomas [265].
Genetically engineered mouse models, in combination with genetic ablation or the overexpression of specific proteases, are valuable research tools for elucidating the multiple roles of cathepsins in tumorigenesis and cancer progression. Critical roles of cathepsins B and X in the carcinogenesis, progression, and metastasis of breast cancer have been discovered using a transgenic MMTV-PymT model of metastasising breast cancer [254,257,266]. Furthermore, the impact of cathepsin B on tumour formation and progression has been confirmed in multiple models, including the pancreatic cancer RIP1-Tag2 model [259] and the renal cell carcinoma xenograft model [267]. Although no data are available on the role of cathepsin H in MMTV-PymT breast cancer progression, cathepsin H depletion significantly impairs the establishment and maintenance of tumour vasculature and reduces the tumour burden in the RIP1-Tag2 model of pancreatic islet carcinogenesis [268]. Notably, a cathepsin C tumour-promoting effect has been demonstrated in a squamous cell carcinoma K14-HPV16 model [269], but not in RIP1-Tag2 [259] or MMTV-PymT [269] transgenic mouse models. In summary, the functions of individual proteases may be hardwired into a specific tissue paradigm and thus depend on the cancer type and biology of the primary and metastatic lesion host tissues.
Cathepsin activity exists within a larger integrated network of protease activities known as the protease web [270]. Through interactions with other proteases and their inhibitors, cathepsins can alter the general proteolytic activity within the TME. In addition to directly regulating multiple processes involved in tumour progression and metastasis, many proteases can indirectly impact the activation of multiple cascades of enzymatic activities [271,272]. This can be illustrated by cathepsin B processing of the urokinase-type plasminogen activator (pro-uPA) pro-form, thus converting plasminogen into plasmin [273], which may activate zymogens of matrix metalloproteinases, and thus, together with the precursor proteases of this proteolytic activation cascade, execute the numerous functions associated with tumour progression and metastasis [274]. Notably, these proteolytic webs or networks can interact with other important signalling pathways in tumour biology, including cytokines, chemokines, and kinases [275].
In summary, as essential elements of the proteolytic network balance, cysteine cathepsins B, X, C, and H are involved in multiple steps of cancer development and progression. Therefore, elucidating their roles in tumour biology and regulating relevant signalling pathways can be utilised as novel targeted anticancer therapeutic approaches.
4. Conclusions
Since the discovery of lysosomes and lysosomal cathepsins, the understanding of intracellular proteolysis and its role in normal biology and disease states has advanced rapidly. Major progress in this area particularly occurred with the determination of the crystal structures of cathepsins, including B, H, and C, and the last discovered cathepsin, X. Endogenous protein inhibitors and synthetic inhibitors are crucial for understanding the mechanism of interaction with their target enzymes and structure-function relationships, both of which are of crucial importance for the treatment of various diseases, including neurodegeneration and cancer. Currently, research is focused on channelling the existing knowledge to detect proteases in various diseases and treat their overexpression. Activity-based probes are under development, but the identification of physiological substrates remains unexplored, which is expected to be addressed using mass spectroscopy. The finding that cathepsins act as signalling molecules requires an understanding of their signalling pathways and their regulation. Therefore, further basic research is required to develop new therapeutic approaches in the near future. Advances in understanding the function of cysteine cathepsins in neurodegeneration, cancer, and other diseases will result in the development of novel, targeted therapeutic strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms242115613/s1, Table S1: Classification of human cathepsins according to the MEROPS database of proteolytic enzymes; File S1: Sequence alignment of the UniProt and PDB sequences of human procathepsin B; File S2: Sequence alignment of the UniProt and PDB sequences of human procathepsin H; File S3: Sequence alignment of the UniProt and PDB sequences of human procathepsin X; File S4: Sequence alignment of the UniProt and PDB sequences of human procathepsin C; Figure S1: Schematic representation of the multiple sequence alignment of human procathepsins B, C, H and X.
Author Contributions
Conceptualisation, V.S. and V.T.; data curation, V.S.; formal analysis, V.S.; writing—original draft preparation, V.S., O.V., H.N. and V.T.; writing—review and editing, V.S. and V.T.; project administration, V.S.; funding acquisition, V.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Slovenian Research Agency, grant numbers J1-2473 (V.S.) and P1-0140.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this review article were obtained from published articles and publicly accessible databases. All sources and references were appropriately cited within the article. All data are provided within the manuscript and the Supplementary Materials accordingly.
Acknowledgments
The authors are grateful to Dušan Turk for Figure 5 preparation and Iztok Dolenc for assistance with reference formatting.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, the collection, analysis, or interpretation of data, the writing of the manuscript, or the decision to publish the results.
Abbreviations
| 6-OHDA | 6-hydroxydopamine |
| α-Syn | α-Synuclein |
| AD | Alzheimer’s disease |
| ALS | Amyotrophic Lateral Sclerosis |
| CATH | Class (C), Architecture (A), Topology (T), and Homologous superfamily (H) |
| CNS | Central Nervous System |
| EAE | Experimental Autoimmune Encephalomyelitis |
| ECM | Extracellular matrix |
| GBA | Glucosylceramidase β1 |
| GCase | Glucocerebrosidase |
| HD | Huntington’s disease |
| IFN-β | Interferon-β |
| IκBα | κBα inhibitor |
| LRRK2 | Leucine-rich repeat serine/threonine-protein kinase 2 |
| MS | Multiple sclerosis |
| NF-κB | Nuclear factor-κB |
| PD | Parkinson’s disease |
| PDB | Protein Data Bank |
| ROS | Reactive Oxygen Species |
| SOD1 | Superoxide dismutase [Cu-Zn] |
| Tg | Transgenic mouse |
| TLR | Toll-like receptor |
| TME | Tumour microenvironment |
| TNFα | Tumour necrosis factor α |
| UniProt | Universal Protein Knowledgebase |
References
- De Duve, C.; Pressman, B.C.; Gianetto, R.; Wattiaux, R.; Appelmans, F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem. J. 1955, 60, 604–617. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C. Lysosomes revisited. Eur. J. Biochem. 1983, 137, 391–397. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C. The lysosome turns fifty. Nat. Cell Biol. 2005, 7, 847–849. [Google Scholar] [CrossRef] [PubMed]
- Amaral, O.; Martins, M.; Oliveira, A.R.; Duarte, A.J.; Mondragão-Rodrigues, I.; Macedo, M.F. The biology of lysosomes: From order to disorder. Biomedicines 2023, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Brix, K.; Dunkhorst, A.; Mayer, K.; Jordans, S. Cysteine cathepsins: Cellular roadmap to different functions. Biochimie 2008, 90, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.; Turk, V. Lysosomes as “suicide bags” in cell death: Myth or reality? J. Biol. Chem. 2009, 284, 21783–21787. [Google Scholar] [CrossRef] [PubMed]
- Repnik, U.; Stoka, V.; Turk, V.; Turk, B. Lysosomes and lysosomal cathepsins in cell death. Biochim. Biophys. Acta 2012, 1824, 22–33. [Google Scholar] [CrossRef]
- Brix, K.; McInnes, J.; Al-Hashimi, A.; Rehders, M.; Tamhane, T.; Haugen, M.H. Proteolysis mediated by cysteine cathepsins and legumain-recent advances and cell biological challenges. Protoplasma 2015, 252, 755–774. [Google Scholar] [CrossRef]
- Biasizzo, M.; Javoršek, U.; Vidak, E.; Zarić, M.; Turk, B. Cysteine cathepsins: A long and winding road towards clinics. Mol. Aspects Med. 2022, 88, 101150. [Google Scholar] [CrossRef]
- Vizovišek, M.; Fonović, M.; Turk, B. Cysteine cathepsins in extracellular matrix remodeling: Extracellular matrix degradation and beyond. Matrix Biol. 2019, 75–76, 141–159. [Google Scholar] [CrossRef]
- Vidak, E.; Javoršek, U.; Vizovišek, M.; Turk, B. Cysteine cathepsins and their extracellular roles: Shaping the microenvironment. Cells 2019, 8, 264. [Google Scholar] [CrossRef] [PubMed]
- Yadati, T.; Houben, T.; Bitorina, A.; Shiri-Sverdlov, R. The ins and outs of cathepsins: Physiological function and role in disease management. Cells 2020, 9, 1679. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Inoue, A.; Lei, Y.; Wu, H.; Hong, L.; Cheng, X.W. Cathepsins in the extracellular space: Focusing on non-lysosomal proteolytic functions with clinical implications. Cell Signal. 2023, 103, 110531. [Google Scholar] [CrossRef] [PubMed]
- Vizovišek, M.; Vidmar, R.; Drag, M.; Fonović, M.; Salvesen, G.S.; Turk, B. Protease specificity: Towards in vivo imaging applications and biomarker discovery. Trends Biochem. Sci. 2018, 43, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.; Stoka, V. Protease signalling in cell death: Caspases versus cysteine cathepsins. FEBS Lett. 2007, 581, 2761–2767. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.; Turk, D.; Turk, V. Protease signalling: The cutting edge. EMBO J. 2012, 31, 1630–1643. [Google Scholar] [CrossRef] [PubMed]
- Stoka, V.; Turk, B.; Schendel, S.L.; Kim, T.H.; Cirman, T.; Snipas, S.J.; Ellerby, L.M.; Bredesen, D.; Freeze, H.; Abrahamson, M.; et al. Lysosomal protease pathways to apoptosis. Cleavage of bid, not pro-caspases, is the most likely route. J. Biol. Chem. 2001, 276, 3149–3157. [Google Scholar] [CrossRef] [PubMed]
- Stoka, V.; Turk, B.; Turk, V. Lysosomal cysteine proteases: Structural features and their role in apoptosis. IUBMB Life 2005, 57, 347–353. [Google Scholar] [CrossRef]
- Droga-Mazovec, G.; Bojic, L.; Petelin, A.; Ivanova, S.; Romih, R.; Repnik, U.; Salvesen, G.S.; Stoka, V.; Turk, V.; Turk, B. Cysteine cathepsins trigger caspase-dependent cell death through cleavage of bid and antiapoptotic Bcl-2 homologues. J. Biol. Chem. 2008, 283, 19140–19150. [Google Scholar] [CrossRef]
- Wang, F.; Gómez-Sintes, R.; Boya, P. Lysosomal membrane permeabilization and cell death. Traffic 2018, 19, 918–931. [Google Scholar] [CrossRef]
- Mohamed, M.M.; Sloane, B.F. Cysteine cathepsins: Multifunctional enzymes in cancer. Nat. Rev. Cancer 2006, 6, 764–775. [Google Scholar] [CrossRef]
- Olson, O.C.; Joyce, J.A. Cysteine cathepsin proteases: Regulators of cancer progression and therapeutic response. Nat. Rev. Cancer 2015, 15, 712–729. [Google Scholar] [CrossRef] [PubMed]
- Vasiljeva, O.; Sevenich, L.; Reinheckel, T. Analyzing the role of proteases in breast cancer progression and metastasis using primary cells from transgenic oncomice. Methods Mol. Biol. 2021, 2294, 275–293. [Google Scholar] [CrossRef] [PubMed]
- Kos, J.; Mitrović, A.; Perišić Nanut, M.; Pišlar, A. Lysosomal peptidases-intriguing roles in cancer progression and neurodegeneration. FEBS Open Bio 2022, 12, 708–738. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.; Guo, J.; Zhang, X.; Sukhova, G.K.; Libby, P.; Shi, G.P. Cysteine protease cathepsins in cardiovascular disease: From basic research to clinical trials. Nat. Rev. Cardiol. 2018, 15, 351–370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luo, S.; Wang, M.; Shi, G.P. Cysteinyl cathepsins in cardiovascular diseases. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140360. [Google Scholar] [CrossRef]
- Stoka, V.; Turk, V.; Turk, B. Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Res. Rev. 2016, 32, 22–37. [Google Scholar] [CrossRef]
- Nixon, R.A. The aging lysosome: An essential catalyst for late-onset neurodegenerative diseases. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140443. [Google Scholar] [CrossRef]
- Vasiljeva, O.; Reinheckel, T.; Peters, C.; Turk, D.; Turk, V.; Turk, B. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr. Pharm. Des. 2007, 13, 387–403. [Google Scholar] [CrossRef]
- Hamon, Y.; Legowska, M.; Hervé, V.; Dallet-Choisy, S.; Marchand-Adam, S.; Vanderlynden, L.; Demonte, M.; Williams, R.; Scott, C.J.; Si-Tahar, M.; et al. Neutrophilic cathepsin C is maturated by a multistep proteolytic process and secreted by activated cells during inflammatory lung diseases. J. Biol. Chem. 2016, 291, 8486–8499. [Google Scholar] [CrossRef]
- Vizovišek, M.; Vidak, E.; Javoršek, U.; Mikhaylov, G.; Bratovš, A.; Turk, B. Cysteine cathepsins as therapeutic targets in inflammatory diseases. Expert Opin. Ther. Targets 2020, 24, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.M.; Yang, W.L.; Yang, F.Y.; Zhang, L.; Huang, W.J.; Hou, W.; Fan, C.F.; Jin, R.H.; Feng, Y.M.; Wang, Y.C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct Target Ther. 2021, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Nishiga, M.; Wang, D.W.; Han, Y.; Lewis, D.B.; Wu, J.C. COVID-19 and cardiovascular disease: From basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 2020, 17, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Ketterer, S.; Gomez-Auli, A.; Hillebrand, L.E.; Petrera, A.; Ketscher, A.; Reinheckel, T. Inherited diseases caused by mutations in cathepsin protease genes. FEBS J. 2017, 284, 1437–1454. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional enzymes in life and disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef] [PubMed]
- Lalmanach, G.; Saidi, A.; Bigot, P.; Chazeirat, T.; Lecaille, F.; Wartenberg, M. Regulation of the proteolytic activity of cysteine cathepsins by oxidants. Int. J. Mol. Sci. 2020, 21, 1944. [Google Scholar] [CrossRef]
- Turk, B.; Turk, D.; Salvesen, G.S. Regulating cysteine protease activity: Essential role of protease inhibitors as guardians and regulators. Curr. Pharm. Des. 2002, 8, 1623–1637. [Google Scholar] [CrossRef] [PubMed]
- Tušar, L.; Usenik, A.; Turk, B.; Turk, D. Mechanisms applied by protein inhibitors to inhibit cysteine proteases. Int. J. Mol. Sci. 2021, 22, 997. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Turk, V.; Bode, W. The cystatins: Protein inhibitors of cysteine proteinases. FEBS Lett. 1991, 285, 213–219. [Google Scholar] [CrossRef]
- Turk, V.; Stoka, V.; Turk, D. Cystatins: Biochemical and structural properties, and medical relevance. Front. Biosci. 2008, 13, 5406–5420. [Google Scholar] [CrossRef] [PubMed]
- Kordis, D.; Turk, V. Phylogenomic analysis of the cystatin superfamily in eukaryotes and prokaryotes. BMC Evol. Biol. 2009, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Bode, W.; Engh, R.; Musil, D.; Thiele, U.; Huber, R.; Karshikov, A.; Brzin, J.; Kos, J.; Turk, V. The 2.0 A X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J. 1988, 7, 2593–2599. [Google Scholar] [CrossRef] [PubMed]
- Stubbs, M.T.; Laber, B.; Bode, W.; Huber, R.; Jerala, R.; Lenarcic, B.; Turk, V. The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: A novel type of proteinase inhibitor interaction. EMBO J. 1990, 9, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
- Engh, R.A.; Dieckmann, T.; Bode, W.; Auerswald, E.A.; Turk, V.; Huber, R.; Oschkinat, H. Conformational variability of chicken cystatin. Comparison of structures determined by X-ray diffraction and NMR spectroscopy. J. Mol. Biol. 1993, 234, 1060–1069. [Google Scholar] [CrossRef]
- Unanue, E.R.; Turk, V.; Neefjes, J. Variations in MHC class II antigen processing and presentation in health and disease. Annu. Rev. Immunol. 2016, 34, 265–297. [Google Scholar] [CrossRef] [PubMed]
- Mihelic, M.; Turk, D. Two decades of thyroglobulin type-1 domain research. Biol. Chem. 2007, 388, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Katunuma, N. Structure-based development of specific inhibitors for individual cathepsins and their medical applications. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 29–39. [Google Scholar] [CrossRef]
- Turk, D.; Podobnik, M.; Popovic, T.; Katunuma, N.; Bode, W.; Huber, R.; Turk, V. Crystal structure of cathepsin B inhibited with CA030 at 2.0-A resolution: A basis for the design of specific epoxysuccinyl inhibitors. Biochemistry 1995, 34, 4791–4797. [Google Scholar] [CrossRef]
- Nägler, D.K.; Ménard, R. Human cathepsin X: A novel cysteine protease of the papain family with a very short proregion and unique insertions. FEBS Lett. 1998, 434, 135–139. [Google Scholar] [CrossRef]
- Santamaría, I.; Velasco, G.; Pendás, A.M.; Fueyo, A.; López-Otín, C. Cathepsin Z, a novel human cysteine proteinase with a short propeptide domain and a unique chromosomal location. J. Biol. Chem. 1998, 273, 16816–16823. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef] [PubMed]
- Musil, D.; Zucic, D.; Turk, D.; Engh, R.A.; Mayr, I.; Huber, R.; Popovic, T.; Turk, V.; Towatari, T.; Katunuma, N.; et al. The refined 2.15 A X-ray crystal structure of human liver cathepsin B: The structural basis for its specificity. EMBO J. 1991, 10, 2321–2330. [Google Scholar] [CrossRef] [PubMed]
- Nägler, D.K.; Storer, A.C.; Portaro, F.C.; Carmona, E.; Juliano, L.; Ménard, R. Major increase in endopeptidase activity of human cathepsin B upon removal of occluding loop contacts. Biochemistry 1997, 36, 12608–12615. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.; Dolenc, I.; Zerovnik, E.; Turk, D.; Gubensek, F.; Turk, V. Human cathepsin B is a metastable enzyme stabilized by specific ionic interactions associated with the active site. Biochemistry 1994, 33, 14800–14806. [Google Scholar] [CrossRef] [PubMed]
- Illy, C.; Quraishi, O.; Wang, J.; Purisima, E.; Vernet, T.; Mort, J.S. Role of the occluding loop in cathepsin B activity. J. Biol. Chem. 1997, 272, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Quraishi, O.; Nägler, D.K.; Fox, T.; Sivaraman, J.; Cygler, M.; Mort, J.S.; Storer, A.C. The occluding loop in cathepsin B defines the pH dependence of inhibition by its propeptide. Biochemistry 1999, 38, 5017–5023. [Google Scholar] [CrossRef] [PubMed]
- Renko, M.; Požgan, U.; Majera, D.; Turk, D. Stefin A displaces the occluding loop of cathepsin B only by as much as required to bind to the active site cleft. FEBS J. 2010, 277, 4338–4345. [Google Scholar] [CrossRef]
- Redzynia, I.; Ljunggren, A.; Abrahamson, M.; Mort, J.S.; Krupa, J.C.; Jaskolski, M.; Bujacz, G. Displacement of the occluding loop by the parasite protein, chagasin, results in efficient inhibition of human cathepsin B. J. Biol. Chem. 2008, 283, 22815–22825. [Google Scholar] [CrossRef]
- Hao, Y.; Purtha, W.; Cortesio, C.; Rui, H.; Gu, Y.; Chen, H.; Sickmier, E.A.; Manzanillo, P.; Huang, X. Crystal structures of human procathepsin H. PLoS ONE 2018, 13, e0200374. [Google Scholar] [CrossRef]
- Guncar, G.; Podobnik, M.; Pungercar, J.; Strukelj, B.; Turk, V.; Turk, D. Crystal structure of porcine cathepsin H determined at 2.1 A resolution: Location of the mini-chain C-terminal carboxyl group defines cathepsin H aminopeptidase function. Structure 1998, 6, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Jenko, S.; Dolenc, I.; Guncar, G.; Dobersek, A.; Podobnik, M.; Turk, D. Crystal structure of Stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases. J. Mol. Biol. 2003, 326, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Vasiljeva, O.; Dolinar, M.; Turk, V.; Turk, B. Recombinant human cathepsin H lacking the mini chain is an endopeptidase. Biochemistry 2003, 42, 13522–13528. [Google Scholar] [CrossRef]
- Rossi, A.; Deveraux, Q.; Turk, B.; Sali, A. Comprehensive search for cysteine cathepsins in the human genome. Biol. Chem. 2004, 385, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta 2012, 1824, 68–88. [Google Scholar] [CrossRef] [PubMed]
- Kirschke, H.; Wiederanders, B.; Brömme, D.; Rinne, A. Cathepsin S from bovine spleen. Purification, distribution, intracellular localization and action on proteins. Biochem. J. 1989, 264, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Almeida, P.C.; Nantes, I.L.; Chagas, J.R.; Rizzi, C.C.; Faljoni-Alario, A.; Carmona, E.; Juliano, L.; Nader, H.B.; Tersariol, I.L. Cathepsin B activity regulation. Heparin-like glycosaminogylcans protect human cathepsin B from alkaline pH-induced inactivation. J. Biol. Chem. 2001, 276, 944–951. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.C.; Hook, V.; O’Donoghue, A.J. Cathepsin B dipeptidyl carboxypeptidase and endopeptidase activities demonstrated across a broad pH range. Biochemistry 2022, 61, 1904–1914. [Google Scholar] [CrossRef]
- Yoon, M.C.; Phan, V.; Podvin, S.; Mosier, C.; O’Donoghue, A.J.; Hook, V. Distinct cleavage properties of cathepsin B compared to cysteine cathepsins enable the design and validation of a specific substrate for cathepsin B over a broad pH range. Biochemistry 2023, 62, 2289–2300. [Google Scholar] [CrossRef]
- Rozman, J.; Stojan, J.; Kuhelj, R.; Turk, V.; Turk, B. Autocatalytic processing of recombinant human procathepsin B is a bimolecular process. FEBS Lett. 1999, 459, 358–362. [Google Scholar] [CrossRef]
- Pungercar, J.R.; Caglic, D.; Sajid, M.; Dolinar, M.; Vasiljeva, O.; Pozgan, U.; Turk, D.; Bogyo, M.; Turk, V.; Turk, B. Autocatalytic processing of procathepsin B is triggered by proenzyme activity. FEBS J. 2009, 276, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Vasiljeva, O.; Dolinar, M.; Pungercar, J.R.; Turk, V.; Turk, B. Recombinant human procathepsin S is capable of autocatalytic processing at neutral pH in the presence of glycosaminoglycans. FEBS Lett. 2005, 579, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Caglic, D.; Pungercar, J.R.; Pejler, G.; Turk, V.; Turk, B. Glycosaminoglycans facilitate procathepsin B activation through disruption of propeptide-mature enzyme interactions. J. Biol. Chem. 2007, 282, 33076–33085. [Google Scholar] [CrossRef] [PubMed]
- Dahl, S.W.; Halkier, T.; Lauritzen, C.; Dolenc, I.; Pedersen, J.; Turk, V.; Turk, B. Human recombinant pro-dipeptidyl peptidase I (cathepsin C) can be activated by cathepsins L and S but not by autocatalytic processing. Biochemistry 2001, 40, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, J.; Nägler, D.K.; Zhang, R.; Ménard, R.; Cygler, M. Crystal structure of human procathepsin X: A cysteine protease with the proregion covalently linked to the active site cysteine. J. Mol. Biol. 2000, 295, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.J.; San Segundo, B.; McCormick, M.B.; Steiner, D.F. Nucleotide and predicted amino acid sequences of cloned human and mouse preprocathepsin B cDNAs. Proc. Natl. Acad. Sci. USA 1986, 83, 7721–7725. [Google Scholar] [CrossRef] [PubMed]
- Podobnik, M.; Kuhelj, R.; Turk, V.; Turk, D. Crystal structure of the wild-type human procathepsin B at 2.5 A resolution reveals the native active site of a papain-like cysteine protease zymogen. J. Mol. Biol. 1997, 271, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- Fuchs, R.; Gassen, H.G. Nucleotide sequence of human preprocathepsin H, a lysosomal cysteine proteinase. Nucleic Acids Res. 1989, 17, 9471. [Google Scholar] [CrossRef]
- Paris, A.; Strukelj, B.; Pungercar, J.; Renko, M.; Dolenc, I.; Turk, V. Molecular cloning and sequence analysis of human preprocathepsin C. FEBS Lett. 1995, 369, 326–330. [Google Scholar] [CrossRef]
- Lainé, D.; Palovich, M.; McCleland, B.; Petitjean, E.; Delhom, I.; Xie, H.; Deng, J.; Lin, G.; Davis, R.; Jolit, A.; et al. Discovery of novel cyanamide-based inhibitors of cathepsin C. ACS Med. Chem. Lett. 2011, 2, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Turk, D.; Podobnik, M.; Kuhelj, R.; Dolinar, M.; Turk, V. Crystal structures of human procathepsin B at 3.2 and 3.3 Angstroms resolution reveal an interaction motif between a papain-like cysteine protease and its propeptide. FEBS Lett. 1996, 384, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Coulombe, R.; Grochulski, P.; Sivaraman, J.; Ménard, R.; Mort, J.S.; Cygler, M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996, 15, 5492–5503. [Google Scholar] [CrossRef] [PubMed]
- Nägler, D.K.; Sulea, T.; Ménard, R. Full-length cDNA of human cathepsin F predicts the presence of a cystatin domain at the N-terminus of the cysteine protease zymogen. Biochem. Biophys. Res. Commun. 1999, 257, 313–318. [Google Scholar] [CrossRef]
- Fox, T.; de Miguel, E.; Mort, J.S.; Storer, A.C. Potent slow-binding inhibition of cathepsin B by its propeptide. Biochemistry 1992, 31, 12571–12576. [Google Scholar] [CrossRef]
- Jerala, R.; Zerovnik, E.; Kidric, J.; Turk, V. pH-induced conformational transitions of the propeptide of human cathepsin L. A role for a molten globule state in zymogen activation. J. Biol. Chem. 1998, 273, 11498–11504. [Google Scholar] [CrossRef]
- Dolenc, I.; Turk, B.; Pungercic, G.; Ritonja, A.; Turk, V. Oligomeric structure and substrate induced inhibition of human cathepsin C. J. Biol. Chem. 1995, 270, 21626–21631. [Google Scholar] [CrossRef]
- Turk, D.; Janjić, V.; Stern, I.; Podobnik, M.; Lamba, D.; Dahl, S.W.; Lauritzen, C.; Pedersen, J.; Turk, V.; Turk, B. Structure of human dipeptidyl peptidase I (cathepsin C): Exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 2001, 20, 6570–6582. [Google Scholar] [CrossRef]
- Rebernik, M.; Lenarčič, B.; Novinec, M. The catalytic domain of cathepsin C (dipeptidyl-peptidase I) alone is a fully functional endoprotease. Protein Expr. Purif. 2019, 157, 21–27. [Google Scholar] [CrossRef]
- Turk, B.; Turk, D.; Turk, V. Lysosomal cysteine proteases: More than scavengers. Biochim. Biophys. Acta 2000, 1477, 98–111. [Google Scholar] [CrossRef]
- Dolenc, I.; Štefe, I.; Turk, D.; Taler-Verčič, A.; Turk, B.; Turk, V.; Stoka, V. Human cathepsin X/Z is a biologically active homodimer. Biochim. Biophys. Acta Proteins Proteom. 2021, 1869, 140567. [Google Scholar] [CrossRef] [PubMed]
- Guncar, G.; Klemencic, I.; Turk, B.; Turk, V.; Karaoglanovic-Carmona, A.; Juliano, L.; Turk, D. Crystal structure of cathepsin X: A flip-flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease. Structure 2000, 8, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Puzer, L.; Cotrin, S.S.; Cezari, M.H.; Hirata, I.Y.; Juliano, M.A.; Stefe, I.; Turk, D.; Turk, B.; Juliano, L.; Carmona, A.K. Recombinant human cathepsin X is a carboxymonopeptidase only: A comparison with cathepsins B and L. Biol. Chem. 2005, 386, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Guncar, G.; Pungercic, G.; Klemencic, I.; Turk, V.; Turk, D. Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J. 1999, 18, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Turk, D. MAIN software for density averaging, model building, structure refinement and validation. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1342–1357. [Google Scholar] [CrossRef] [PubMed]
- Hook, V.; Yoon, M.; Mosier, C.; Ito, G.; Podvin, S.; Head, B.P.; Rissman, R.; O’Donoghue, A.J.; Hook, G. Cathepsin B in neurodegeneration of Alzheimer’s disease, traumatic brain injury, and related brain disorders. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140428. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.F.; Bahr, B.A.; Kinsey, S.T. Endosomal-lysosomal dysfunction in metabolic diseases and Alzheimer’s disease. Int. Rev. Neurobiol. 2020, 154, 303–324. [Google Scholar] [CrossRef]
- Drobny, A.; Prieto Huarcaya, S.; Dobert, J.; Kluge, A.; Bunk, J.; Schlothauer, T.; Zunke, F. The role of lysosomal cathepsins in neurodegeneration: Mechanistic insights, diagnostic potential and therapeutic approaches. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119243. [Google Scholar] [CrossRef]
- Sharma, A.; Swetha, R.; Bajad, N.G.; Ganeshpurkar, A.; Singh, R.; Kumar, A.; Singh, S.K. Cathepsin B-A neuronal death mediator in Alzheimer’s disease leading to neurodegeneration. Mini Rev. Med. Chem. 2022, 22, 2012–2023. [Google Scholar] [CrossRef]
- Boland, B.; Yu, W.H.; Corti, O.; Mollereau, B.; Henriques, A.; Bezard, E.; Pastores, G.M.; Rubinsztein, D.C.; Nixon, R.A.; Duchen, M.R.; et al. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2018, 17, 660–688. [Google Scholar] [CrossRef]
- Schattling, B.; Engler, J.B.; Volkmann, C.; Rothammer, N.; Woo, M.S.; Petersen, M.; Winkler, I.; Kaufmann, M.; Rosenkranz, S.C.; Fejtova, A.; et al. Bassoon proteinopathy drives neurodegeneration in multiple sclerosis. Nat. Neurosci. 2019, 22, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Telpoukhovskaia, M.A.; Bahr, B.A.; Chen, X.; Gan, L. Endo-lysosomal dysfunction: A converging mechanism in neurodegenerative diseases. Curr. Opin. Neurobiol. 2018, 48, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Ii, K.; Ito, H.; Kominami, E.; Hirano, A. Abnormal distribution of cathepsin proteinases and endogenous inhibitors (cystatins) in the hippocampus of patients with Alzheimer’s disease, parkinsonism-dementia complex on Guam, and senile dementia and in the aged. Virchows Arch. A Pathol. Anat. Histopathol. 1993, 423, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Mantle, D.; Falkous, G.; Ishiura, S.; Perry, R.H.; Perry, E.K. Comparison of cathepsin protease activities in brain tissue from normal cases and cases with Alzheimer’s disease, Lewy body dementia, Parkinson’s disease and Huntington’s disease. J. Neurol. Sci. 1995, 131, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Nixon, R.A. A “protease activation cascade” in the pathogenesis of Alzheimer’s disease. Ann. N. Y. Acad. Sci. 2000, 924, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, H.; Yamada, T.; Furuya, H.; Doh-ura, K.; Ohyagi, Y.; Iwaki, T.; Kira, J. Involvement of cathepsin B in the motor neuron degeneration of amyotrophic lateral sclerosis. Acta Neuropathol. 2003, 105, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Ratovitski, T.; Chighladze, E.; Waldron, E.; Hirschhorn, R.R.; Ross, C.A. Cysteine proteases bleomycin hydrolase and cathepsin Z mediate N-terminal proteolysis and toxicity of mutant huntingtin. J. Biol. Chem. 2011, 286, 12578–12589. [Google Scholar] [CrossRef]
- Pišlar, A.; Tratnjek, L.; Glavan, G.; Živin, M.; Kos, J. Upregulation of cysteine protease cathepsin X in the 6-hydroxydopamine model of Parkinson’s disease. Front. Mol. Neurosci. 2018, 11, 412. [Google Scholar] [CrossRef]
- Pišlar, A.H.; Zidar, N.; Kikelj, D.; Kos, J. Cathepsin X promotes 6-hydroxydopamine-induced apoptosis of PC12 and SH-SY5Y cells. Neuropharmacology 2014, 82, 121–131. [Google Scholar] [CrossRef]
- Fan, K.; Li, D.; Zhang, Y.; Han, C.; Liang, J.; Hou, C.; Xiao, H.; Ikenaka, K.; Ma, J. The induction of neuronal death by up-regulated microglial cathepsin H in LPS-induced neuroinflammation. J. Neuroinflammation 2015, 12, 54. [Google Scholar] [CrossRef]
- Fan, K.; Wu, X.; Fan, B.; Li, N.; Lin, Y.; Yao, Y.; Ma, J. Up-regulation of microglial cathepsin C expression and activity in lipopolysaccharide-induced neuroinflammation. J. Neuroinflammation 2012, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- von Bernhardi, R.; Eugenín-von Bernhardi, L.; Eugenín, J. Microglial cell dysregulation in brain aging and neurodegeneration. Front. Aging Neurosci. 2015, 7, 124. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, N.; Zhang, Y.; Hou, C.; Yang, X.; Shimizu, T.; Wang, X.; Ikenaka, K.; Fan, K.; Ma, J. Disinhibition of cathepsin C caused by cystatin F deficiency aggravates the demyelination in a cuprizone model. Front. Mol. Neurosci. 2016, 9, 152. [Google Scholar] [CrossRef]
- Allan, E.R.O.; Campden, R.I.; Ewanchuk, B.W.; Tailor, P.; Balce, D.R.; McKenna, N.T.; Greene, C.J.; Warren, A.L.; Reinheckel, T.; Yates, R.M. A role for cathepsin Z in neuroinflammation provides mechanistic support for an epigenetic risk factor in multiple sclerosis. J. Neuroinflammation 2017, 14, 103. [Google Scholar] [CrossRef] [PubMed]
- Pišlar, A.; Božić, B.; Zidar, N.; Kos, J. Inhibition of cathepsin X reduces the strength of microglial-mediated neuroinflammation. Neuropharmacology 2017, 114, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Pišlar, A.; Tratnjek, L.; Glavan, G.; Zidar, N.; Živin, M.; Kos, J. Neuroinflammation-induced upregulation of glial cathepsin X expression and activity in vivo. Front. Mol. Neurosci. 2020, 13, 575453. [Google Scholar] [CrossRef] [PubMed]
- Pišlar, A.; Bolčina, L.; Kos, J. New insights into the role of cysteine cathepsins in neuroinflammation. Biomolecules 2021, 11, 1796. [Google Scholar] [CrossRef] [PubMed]
- Campden, R.I.; Zhang, Y. The role of lysosomal cysteine cathepsins in NLRP3 inflammasome activation. Arch. Biochem. Biophys. 2019, 670, 32–42. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, Y.; Liu, S.; Liu, Y.; Yang, X.; Liu, G.; Shimizu, T.; Ikenaka, K.; Fan, K.; Ma, J. Cathepsin C promotes microglia M1 polarization and aggravates neuroinflammation via activation of Ca(2+)-dependent PKC/p38MAPK/NF-κB pathway. J. Neuroinflammation 2019, 16, 10. [Google Scholar] [CrossRef]
- Nakanishi, H. Cathepsin regulation on microglial function. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140465. [Google Scholar] [CrossRef]
- Colonna, M.; Butovsky, O. Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef] [PubMed]
- Bohlen, C.J.; Friedman, B.A.; Dejanovic, B.; Sheng, M. Microglia in brain development, homeostasis, and neurodegeneration. Annu. Rev. Genet. 2019, 53, 263–288. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jin, M.Z.; Yang, Z.Y.; Jin, W.L. Microglia in neurodegenerative diseases. Neural Regen Res. 2021, 16, 270–280. [Google Scholar] [CrossRef]
- Troubat, R.; Barone, P.; Leman, S.; Desmidt, T.; Cressant, A.; Atanasova, B.; Brizard, B.; El Hage, W.; Surget, A.; Belzung, C.; et al. Neuroinflammation and depression: A review. Eur. J. Neurosci. 2021, 53, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Réus, G.Z.; Fries, G.R.; Stertz, L.; Badawy, M.; Passos, I.C.; Barichello, T.; Kapczinski, F.; Quevedo, J. The role of inflammation and microglial activation in the pathophysiology of psychiatric disorders. Neuroscience 2015, 300, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, C.; Matosin, N.; Kaul, D.; Philipsen, A.; Gassen, N.C. The role of cathepsins in memory functions and the pathophysiology of psychiatric disorders. Front. Psychiatry 2020, 11, 718. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.C.; Vilalta, A. How microglia kill neurons. Brain Res. 2015, 1628, 288–297. [Google Scholar] [CrossRef]
- Kingham, P.J.; Pocock, J.M. Microglial secreted cathepsin B induces neuronal apoptosis. J. Neurochem. 2001, 76, 1475–1484. [Google Scholar] [CrossRef]
- Nakanishi, H. Neuronal and microglial cathepsins in aging and age-related diseases. Ageing Res. Rev. 2003, 2, 367–381. [Google Scholar] [CrossRef]
- Nakanishi, H. Microglial cathepsin B as a key driver of inflammatory brain diseases and brain aging. Neural Regen Res. 2020, 15, 25–29. [Google Scholar] [CrossRef]
- Hook, V.; Funkelstein, L.; Wegrzyn, J.; Bark, S.; Kindy, M.; Hook, G. Cysteine cathepsins in the secretory vesicle produce active peptides: Cathepsin L generates peptide neurotransmitters and cathepsin B produces beta-amyloid of Alzheimer’s disease. Biochim. Biophys. Acta 2012, 1824, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Murrell, J.R.; Goedert, M.; Farlow, M.R.; Klug, A.; Ghetti, B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl. Acad. Sci. USA 1998, 95, 7737–7741. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Ho, D.H.; Suk, J.E.; You, S.; Michael, S.; Kang, J.; Joong Lee, S.; Masliah, E.; Hwang, D.; Lee, H.J.; et al. Neuron-released oligomeric α-synuclein is an endogenous agonist of TLR2 for paracrine activation of microglia. Nat. Commun. 2013, 4, 1562. [Google Scholar] [CrossRef] [PubMed]
- Roodveldt, C.; Christodoulou, J.; Dobson, C.M. Immunological features of alpha-synuclein in Parkinson’s disease. J. Cell Mol. Med. 2008, 12, 1820–1829. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Guajardo, V.; Tentillier, N.; Romero-Ramos, M. The relation between α-synuclein and microglia in Parkinson’s disease: Recent developments. Neuroscience 2015, 302, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Gallegos, S.; Pacheco, C.; Peters, C.; Opazo, C.M.; Aguayo, L.G. Features of alpha-synuclein that could explain the progression and irreversibility of Parkinson’s disease. Front. Neurosci. 2015, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Ghio, S.; Kamp, F.; Cauchi, R.; Giese, A.; Vassallo, N. Interaction of α-synuclein with biomembranes in Parkinson’s disease--role of cardiolipin. Prog. Lipid Res. 2016, 61, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J.; Lansbury, P.J. Alzheimer’s disease is the most common neurodegenerative disorder. In Basic Neurochemistry: Molecular, Cellular and Medical Aspects, 6th ed.; Siegel, J.G., Agranoff, B.W., Albers, R.W., Fisher, S.K., Uhler, M., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 1999; pp. 101–102. [Google Scholar]
- DeTure, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef]
- Mohandas, E.; Rajmohan, V.; Raghunath, B. Neurobiology of Alzheimer’s disease. Indian J. Psychiatry 2009, 51, 55–61. [Google Scholar] [CrossRef]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef]
- Musiek, E.S.; Holtzman, D.M. Three dimensions of the amyloid hypothesis: Time, space and ‘wingmen’. Nat. Neurosci. 2015, 18, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, A.M.; Nixon, R.A. Enzymatically active lysosomal proteases are associated with amyloid deposits in Alzheimer brain. Proc. Natl. Acad. Sci. USA 1990, 87, 3861–3865. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, T.R.; Julian, R.R. Proteolysis of amyloid β by lysosomal enzymes as a function of fibril morphology. ACS Omega 2021, 6, 31520–31527. [Google Scholar] [CrossRef] [PubMed]
- McGeer, P.L.; McGeer, E.G. The amyloid cascade-inflammatory hypothesis of Alzheimer disease: Implications for therapy. Acta Neuropathol. 2013, 126, 479–497. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Takeda, M.; Suzuki, H.; Hattori, H.; Tada, K.; Hariguchi, S.; Hashimoto, S.; Nishimura, T. Abnormal distribution of cathepsins in the brain of patients with Alzheimer’s disease. Neurosci. Lett. 1991, 130, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Wendt, W.; Zhu, X.R.; Lübbert, H.; Stichel, C.C. Differential expression of cathepsin X in aging and pathological central nervous system of mice. Exp. Neurol. 2007, 204, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Hafner, A.; Glavan, G.; Obermajer, N.; Živin, M.; Schliebs, R.; Kos, J. Neuroprotective role of γ-enolase in microglia in a mouse model of Alzheimer’s disease is regulated by cathepsin X. Aging Cell 2013, 12, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhou, Y.; Halabisky, B.; Lo, I.; Cho, S.H.; Mueller-Steiner, S.; Devidze, N.; Wang, X.; Grubb, A.; Gan, L. Cystatin C-cathepsin B axis regulates amyloid beta levels and associated neuronal deficits in an animal model of Alzheimer’s disease. Neuron 2008, 60, 247–257. [Google Scholar] [CrossRef]
- Bernstein, H.G.; Keilhoff, G. Putative roles of cathepsin B in Alzheimer’s disease pathology: The good, the bad, and the ugly in one? Neural Regen Res. 2018, 13, 2100–2101. [Google Scholar] [CrossRef]
- Nixon, R.A. Amyloid precursor protein and endosomal-lysosomal dysfunction in Alzheimer’s disease: Inseparable partners in a multifactorial disease. FASEB J. 2017, 31, 2729–2743. [Google Scholar] [CrossRef]
- Bai, H.; Yang, B.; Yu, W.; Xiao, Y.; Yu, D.; Zhang, Q. Cathepsin B links oxidative stress to the activation of NLRP3 inflammasome. Exp. Cell Res. 2018, 362, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Palombella, V.J.; Rando, O.J.; Goldberg, A.L.; Maniatis, T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell 1994, 78, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Colleran, A.; Ryan, A.; O’Gorman, A.; Mureau, C.; Liptrot, C.; Dockery, P.; Fearnhead, H.; Egan, L.J. Autophagosomal IkappaB alpha degradation plays a role in the long term control of tumor necrosis factor-alpha-induced nuclear factor-kappaB (NF-kappaB) activity. J. Biol. Chem. 2011, 286, 22886–22893. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wu, Z. Inflammation spreading: Negative spiral linking systemic inflammatory disorders and Alzheimer’s disease. Front. Cell Neurosci. 2021, 15, 638686. [Google Scholar] [CrossRef] [PubMed]
- Viejo, L.; Noori, A.; Merrill, E.; Das, S.; Hyman, B.T.; Serrano-Pozo, A. Systematic review of human post-mortem immunohistochemical studies and bioinformatics analyses unveil the complexity of astrocyte reaction in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2022, 48, e12753. [Google Scholar] [CrossRef]
- Thygesen, C.; Ilkjær, L.; Kempf, S.J.; Hemdrup, A.L.; von Linstow, C.U.; Babcock, A.A.; Darvesh, S.; Larsen, M.R.; Finsen, B. Diverse protein profiles in CNS myeloid cells and CNS tissue from lipopolysaccharide- and vehicle-injected APP(SWE)/PS1(ΔE9) transgenic mice implicate cathepsin Z in Alzheimer’s disease. Front. Cell Neurosci. 2018, 12, 397. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Estick, C.M.; Ikonne, U.S.; Butler, D.; Pait, M.C.; Elliott, L.H.; Ruiz, S.; Smith, K.; Rentschler, K.M.; Mundell, C.; et al. The role of lysosomes in a broad disease-modifying approach evaluated across transgenic mouse models of Alzheimer’s disease and Parkinson’s disease and models of mild cognitive impairment. Int. J. Mol. Sci. 2019, 20, 4432. [Google Scholar] [CrossRef]
- Hook, G.; Reinheckel, T.; Ni, J.; Wu, Z.; Kindy, M.; Peters, C.; Hook, V. Cathepsin B gene knockout improves behavioral deficits and reduces pathology in models of neurologic disorders. Pharmacol. Rev. 2022, 74, 600–629. [Google Scholar] [CrossRef]
- Hook, G.; Kindy, M.; Hook, V. Cathepsin B deficiency improves memory deficits and reduces amyloid-β in hAβPP mouse models representing the major sporadic Alzheimer’s disease condition. J. Alzheimer’s Dis. 2023, 93, 33–46. [Google Scholar] [CrossRef]
- Hook, G.; Hook, V.; Kindy, M. The cysteine protease inhibitor, E64d, reduces brain amyloid-β and improves memory deficits in Alzheimer’s disease animal models by inhibiting cathepsin B, but not BACE1, β-secretase activity. J. Alzheimer’s Dis. 2011, 26, 387–408. [Google Scholar] [CrossRef]
- Cecarini, V.; Cuccioloni, M.; Zheng, Y.; Bonfili, L.; Gong, C.; Angeletti, M.; Mena, P.; Del Rio, D.; Eleuteri, A.M. Flavan-3-ol microbial metabolites modulate proteolysis in neuronal cells reducing amyloid-beta (1-42) levels. Mol. Nutr. Food Res. 2021, 65, e2100380. [Google Scholar] [CrossRef] [PubMed]
- Chitranshi, N.; Kumar, A.; Sheriff, S.; Gupta, V.; Godinez, A.; Saks, D.; Sarkar, S.; Shen, T.; Mirzaei, M.; Basavarajappa, D.; et al. Identification of novel cathepsin B inhibitors with implications in Alzheimer’s disease: Computational refining and biochemical evaluation. Cells 2021, 10, 1946. [Google Scholar] [CrossRef]
- Dunlop, R.A.; Carney, J.M. Mechanisms of L-serine-mediated neuroprotection include selective activation of lysosomal cathepsins B and L. Neurotox. Res. 2021, 39, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Saroha, B.; Kumar, G.; Kumari, M.; Kaur, R.; Raghav, N.; Sharma, P.K.; Kumar, N.; Kumar, S. A decennary update on diverse heterocycles and their intermediates as privileged scaffolds for cathepsin B inhibition. Int. J. Biol. Macromol. 2022, 222, 2270–2308. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Xie, A.; Yan, Z.; Zhu, X.; Song, Y.; Chen, T.J.B.X. Nanomedicines for Alzheimer’s disease: Therapies based on pathological mechanisms. Brain-X 2023, 1, e27. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Y.; Zhou, J. Neuroinflammation in Parkinson’s disease and its potential as therapeutic target. Transl. Neurodegener 2015, 4, 19. [Google Scholar] [CrossRef]
- Qiao, C.; Yin, N.; Gu, H.Y.; Zhu, J.L.; Ding, J.H.; Lu, M.; Hu, G. Atp13a2 Deficiency Aggravates Astrocyte-Mediated Neuroinflammation via NLRP3 Inflammasome Activation. CNS Neurosci. Ther. 2016, 22, 451–460. [Google Scholar] [CrossRef]
- Codolo, G.; Plotegher, N.; Pozzobon, T.; Brucale, M.; Tessari, I.; Bubacco, L.; de Bernard, M. Triggering of inflammasome by aggregated α-synuclein, an inflammatory response in synucleinopathies. PLoS ONE 2013, 8, e55375. [Google Scholar] [CrossRef]
- Freeman, D.; Cedillos, R.; Choyke, S.; Lukic, Z.; McGuire, K.; Marvin, S.; Burrage, A.M.; Sudholt, S.; Rana, A.; O’Connor, C.; et al. Alpha-synuclein induces lysosomal rupture and cathepsin dependent reactive oxygen species following endocytosis. PLoS ONE 2013, 8, e62143. [Google Scholar] [CrossRef]
- Zhou, Y.; Lu, M.; Du, R.H.; Qiao, C.; Jiang, C.Y.; Zhang, K.Z.; Ding, J.H.; Hu, G. MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol. Neurodegener. 2016, 11, 28. [Google Scholar] [CrossRef]
- McGlinchey, R.P.; Lee, J.C. Cysteine cathepsins are essential in lysosomal degradation of α-synuclein. Proc. Natl. Acad. Sci. USA 2015, 112, 9322–9327. [Google Scholar] [CrossRef] [PubMed]
- McGlinchey, R.P.; Lacy, S.M.; Huffer, K.E.; Tayebi, N.; Sidransky, E.; Lee, J.C. C-terminal α-synuclein truncations are linked to cysteine cathepsin activity in Parkinson’s disease. J. Biol. Chem. 2019, 294, 9973–9984. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Niu, J.Y.; Xiong, J.; Nie, S.K.; Zeng, F.; Zhang, Z.H. LRRK2 G2019S mutation inhibits degradation of α-Synuclein in an in vitro model of Parkinson’s disease. Curr. Med. Sci. 2018, 38, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Tsujimura, A.; Taguchi, K.; Watanabe, Y.; Tatebe, H.; Tokuda, T.; Mizuno, T.; Tanaka, M. Lysosomal enzyme cathepsin B enhances the aggregate forming activity of exogenous α-synuclein fibrils. Neurobiol. Dis. 2015, 73, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Blauwendraat, C.; Reed, X.; Krohn, L.; Heilbron, K.; Bandres-Ciga, S.; Tan, M.; Gibbs, J.R.; Hernandez, D.G.; Kumaran, R.; Langston, R.; et al. Genetic modifiers of risk and age at onset in GBA associated Parkinson’s disease and Lewy body dementia. Brain 2020, 143, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.P.; Boutin, M.; Tse, T.E.; Lu, H.; Haley, E.D.; Ouyang, X.; Zhang, J.; Auray-Blais, C.; Shacka, J.J. The lysosomal enzyme alpha-Galactosidase A is deficient in Parkinson’s disease brain in association with the pathologic accumulation of alpha-synuclein. Neurobiol. Dis. 2018, 110, 68–81. [Google Scholar] [CrossRef]
- Kim, M.J.; Jeong, H.; Krainc, D. Lysosomal ceramides regulate cathepsin B-mediated processing of saposin C and glucocerebrosidase activity. Hum. Mol. Genet. 2022, 31, 2424–2437. [Google Scholar] [CrossRef] [PubMed]
- Senkevich, K.; Rudakou, U.; Gan-Or, Z. Genetic mechanism vs. genetic subtypes: The example of GBA. Handb. Clin. Neurol. 2023, 193, 155–170. [Google Scholar] [CrossRef]
- Lee, D.C.; Close, F.T.; Goodman, C.B.; Jackson, I.M.; Wight-Mason, C.; Wells, L.M.; Womble, T.A.; Palm, D.E. Enhanced cystatin C and lysosomal protease expression following 6-hydroxydopamine exposure. Neurotoxicology 2006, 27, 260–276. [Google Scholar] [CrossRef]
- Wu, Z.; Nakanishi, H. Lessons from microglia aging for the link between inflammatory bone disorders and Alzheimer’s disease. J. Immunol. Res. 2015, 2015, 471342. [Google Scholar] [CrossRef]
- Milanowski, L.M.; Hou, X.; Bredenberg, J.M.; Fiesel, F.C.; Cocker, L.T.; Soto-Beasley, A.I.; Walton, R.L.; Strongosky, A.J.; Faroqi, A.H.; Barcikowska, M.; et al. Cathepsin B p.Gly284Val variant in Parkinson’s disease pathogenesis. Int. J. Mol. Sci. 2022, 23, 7086. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.; Lim, K.L. Genetic insights into sporadic Parkinson’s disease pathogenesis. Curr. Genomics 2013, 14, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Kalinderi, K.; Bostantjopoulou, S.; Fidani, L. The genetic background of Parkinson’s disease: Current progress and future prospects. Acta Neurol. Scand. 2016, 134, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, G.; Zhou, Q.; Xie, Y.; Wang, Z.; Fang, Z.; Lu, B.; Qin, L.; Zhao, Y.; Zhang, R.; et al. Gene4PD: A comprehensive genetic database of Parkinson’s disease. Front. Neurosci. 2021, 15, 679568. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S. Huntington’s Disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a007476. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Tabrizi, S.J. Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 2011, 10, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Sanchez, M.; Licitra, F.; Underwood, B.R.; Rubinsztein, D.C. Huntington’s Disease: Mechanisms of pathogenesis and therapeutic strategies. Cold Spring Harb. Perspect. Med. 2017, 7, a024240. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.; Rajamma, U. Huntington’s disease: The coming of age. J. Genet 2018, 97, 649–664. [Google Scholar] [CrossRef]
- Nagata, E.; Sawa, A.; Ross, C.A.; Snyder, S.H. Autophagosome-like vacuole formation in Huntington’s disease lymphoblasts. Neuroreport 2004, 15, 1325–1328. [Google Scholar] [CrossRef]
- Zhang, X.D.; Wang, Y.; Wang, Y.; Zhang, X.; Han, R.; Wu, J.C.; Liang, Z.Q.; Gu, Z.L.; Han, F.; Fukunaga, K.; et al. p53 mediates mitochondria dysfunction-triggered autophagy activation and cell death in rat striatum. Autophagy 2009, 5, 339–350. [Google Scholar] [CrossRef]
- Kegel, K.B.; Kim, M.; Sapp, E.; McIntyre, C.; Castaño, J.G.; Aronin, N.; DiFiglia, M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J. Neurosci. 2000, 20, 7268–7278. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.H.; Gu, Z.L. Huntingtin processing in pathogenesis of Huntington disease. Acta Pharmacol. Sin. 2004, 25, 1243–1249. [Google Scholar] [PubMed]
- Kim, Y.J.; Sapp, E.; Cuiffo, B.G.; Sobin, L.; Yoder, J.; Kegel, K.B.; Qin, Z.H.; Detloff, P.; Aronin, N.; DiFiglia, M. Lysosomal proteases are involved in generation of N-terminal huntingtin fragments. Neurobiol. Dis. 2006, 22, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.P.; Holcomb, J.; Al-Ramahi, I.; de Haro, M.; Gafni, J.; Zhang, N.; Kim, E.; Sanhueza, M.; Torcassi, C.; Kwak, S.; et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington’s disease. Neuron 2010, 67, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Ouyang, X.; Schneider, L.; Zhang, J. Reduction of mutant huntingtin accumulation and toxicity by lysosomal cathepsins D and B in neurons. Mol. Neurodegener. 2011, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Bhutani, N.; Piccirillo, R.; Hourez, R.; Venkatraman, P.; Goldberg, A.L. Cathepsins L and Z are critical in degrading polyglutamine-containing proteins within lysosomes. J. Biol. Chem. 2012, 287, 17471–17482. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.Y.; Lan, C.P.; Hasan, S.; Brown, M.E.; McLaurin, J. scyllo-Inositol promotes robust mutant Huntingtin protein degradation. J. Biol. Chem. 2014, 289, 3666–3676. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.K.H.; Thombre, R.; Wang, J. Autophagy as a common pathway in amyotrophic lateral sclerosis. Neurosci. Lett. 2019, 697, 34–48. [Google Scholar] [CrossRef]
- Mori, F.; Miki, Y.; Kon, T.; Tanji, K.; Wakabayashi, K. Autophagy is a common degradation pathway for Bunina bodies and TDP-43 inclusions in amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2019, 78, 910–921. [Google Scholar] [CrossRef]
- Lee, J.P.; Gerin, C.; Bindokas, V.P.; Miller, R.; Ghadge, G.; Roos, R.P. No correlation between aggregates of Cu/Zn superoxide dismutase and cell death in familial amyotrophic lateral sclerosis. J. Neurochem. 2002, 82, 1229–1238. [Google Scholar] [CrossRef]
- Offen, D.; Barhum, Y.; Melamed, E.; Embacher, N.; Schindler, C.; Ransmayr, G. Spinal cord mRNA profile in patients with ALS: Comparison with transgenic mice expressing the human SOD-1 mutant. J. Mol. Neurosci. 2009, 38, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Boutahar, N.; Wierinckx, A.; Camdessanche, J.P.; Antoine, J.C.; Reynaud, E.; Lassabliere, F.; Lachuer, J.; Borg, J. Differential effect of oxidative or excitotoxic stress on the transcriptional profile of amyotrophic lateral sclerosis-linked mutant SOD1 cultured neurons. J. Neurosci. Res. 2011, 89, 1439–1450. [Google Scholar] [CrossRef] [PubMed]
- Saris, C.G.; Groen, E.J.; Koekkoek, J.A.; Veldink, J.H.; van den Berg, L.H. Meta-analysis of gene expression profiling in amyotrophic lateral sclerosis: A comparison between transgenic mouse models and human patients. Amyotroph Lateral Scler Front. Degener 2013, 14, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Fukada, Y.; Yasui, K.; Kitayama, M.; Doi, K.; Nakano, T.; Watanabe, Y.; Nakashima, K. Gene expression analysis of the murine model of amyotrophic lateral sclerosis: Studies of the Leu126delTT mutation in SOD1. Brain Res. 2007, 1160, 1–10. [Google Scholar] [CrossRef]
- Gonzalez de Aguilar, J.L.; Niederhauser-Wiederkehr, C.; Halter, B.; De Tapia, M.; Di Scala, F.; Demougin, P.; Dupuis, L.; Primig, M.; Meininger, V.; Loeffler, J.P. Gene profiling of skeletal muscle in an amyotrophic lateral sclerosis mouse model. Physiol. Genomics 2008, 32, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Chiu, I.M.; Morimoto, E.T.; Goodarzi, H.; Liao, J.T.; O’Keeffe, S.; Phatnani, H.P.; Muratet, M.; Carroll, M.C.; Levy, S.; Tavazoie, S.; et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013, 4, 385–401. [Google Scholar] [CrossRef] [PubMed]
- Ulbrich, L.; Cozzolino, M.; Marini, E.S.; Amori, I.; De Jaco, A.; Carrì, M.T.; Augusti-Tocco, G. Cystatin B and SOD1: Protein–protein interaction and possible relation to neurodegeneration. Cell. Mol. Neurobiol. 2014, 34, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Hayakawa, T.; Wakasugi, K.; Yamanaka, K. Cystatin C protects neuronal cells against mutant copper-zinc superoxide dismutase-mediated toxicity. Cell Death Dis. 2014, 5, e1497. [Google Scholar] [CrossRef]
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Stadelmann, C.; Wegner, C.; Brück, W. Inflammation, demyelination, and degeneration—Recent insights from MS pathology. Biochim. Biophys. Acta 2011, 1812, 275–282. [Google Scholar] [CrossRef]
- Bever, C.T., Jr.; Panitch, H.S.; Johnson, K.P. Increased cathepsin B activity in peripheral blood mononuclear cells of multiple sclerosis patients. Neurology 1994, 44, 745–748. [Google Scholar] [CrossRef] [PubMed]
- Bever, C.T., Jr.; Garver, D.W. Increased cathepsin B activity in multiple sclerosis brain. J. Neurol. Sci. 1995, 131, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Bizzozero, O.A. Decreased activity of the 20S proteasome in the brain white matter and gray matter of patients with multiple sclerosis. J. Neurochem. 2011, 117, 143–153. [Google Scholar] [CrossRef]
- Ma, J.; Tanaka, K.F.; Yamada, G.; Ikenaka, K. Induced expression of cathepsins and cystatin C in a murine model of demyelination. Neurochem. Res. 2007, 32, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Allan, E.R.; Yates, R.M. Redundancy between cysteine cathepsins in murine experimental autoimmune encephalomyelitis. PLoS ONE 2015, 10, e0128945. [Google Scholar] [CrossRef] [PubMed]
- Okada, R.; Zhang, X.; Harada, Y.; Wu, Z.; Nakanishi, H. Cathepsin H deficiency in mice induces excess Th1 cell activation and early-onset of EAE though impairment of toll-like receptor 3 cascade. Inflamm. Res. 2018, 67, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Wisessmith, W.; Li, J.; Abe, M.; Sakimura, K.; Chetsawang, B.; Sahara, Y.; Tohyama, K.; Tanaka, K.F.; Ikenaka, K. The balance between cathepsin C and cystatin F controls remyelination in the brain of Plp1-overexpressing mouse, a chronic demyelinating disease model. Glia 2017, 65, 917–930. [Google Scholar] [CrossRef]
- Durose, W.W.; Shimizu, T.; Li, J.; Abe, M.; Sakimura, K.; Chetsawang, B.; Tanaka, K.F.; Suzumura, A.; Tohyama, K.; Ikenaka, K. Cathepsin C modulates myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis. J. Neurochem. 2019, 148, 413–425. [Google Scholar] [CrossRef]
- Haves-Zburof, D.; Paperna, T.; Gour-Lavie, A.; Mandel, I.; Glass-Marmor, L.; Miller, A. Cathepsins and their endogenous inhibitors cystatins: Expression and modulation in multiple sclerosis. J. Cell Mol. Med. 2011, 15, 2421–2429. [Google Scholar] [CrossRef]
- Czibere, L.; Baur, L.A.; Wittmann, A.; Gemmeke, K.; Steiner, A.; Weber, P.; Pütz, B.; Ahmad, N.; Bunck, M.; Graf, C.; et al. Profiling trait anxiety: Transcriptome analysis reveals cathepsin B (Ctsb) as a novel candidate gene for emotionality in mice. PLoS ONE 2011, 6, e23604. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, K.; Liu, Y.; Liu, G.; Yang, X.; Ma, J. Cathepsin C aggravates neuroinflammation involved in disturbances of behaviour and neurochemistry in acute and chronic stress-induced murine model of depression. Neurochem. Res. 2018, 43, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Höger, N.; Feige, B.; Blechert, J.; Normann, C.; Nissen, C. Fear extinction as a model for synaptic plasticity in major depressive disorder. PLoS ONE 2014, 9, e115280. [Google Scholar] [CrossRef] [PubMed]
- Padamsey, Z.; McGuinness, L.; Bardo, S.J.; Reinhart, M.; Tong, R.; Hedegaard, A.; Hart, M.L.; Emptage, N.J. Activity-dependent exocytosis of lysosomes regulates the structural plasticity of dendritic spines. Neuron 2017, 93, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.P.; Silver, J. Cathepsins in neuronal plasticity. Neural Regen Res. 2021, 16, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Khaket, T.P.; Kwon, T.K.; Kang, S.C. Cathepsins: Potent regulators in carcinogenesis. Pharmacol. Ther. 2019, 198, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Talieri, M.; Papadopoulou, S.; Scorilas, A.; Xynopoulos, D.; Arnogianaki, N.; Plataniotis, G.; Yotis, J.; Agnanti, N. Cathepsin B and cathepsin D expression in the progression of colorectal adenoma to carcinoma. Cancer Lett. 2004, 205, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.P.; Krüger, S.; Fogeron, M.L.; Lamer, S.; Chen, J.; Pross, M.; Schulz, H.U.; Lage, H.; Heim, S.; Roessner, A.; et al. Overexpression of cathepsin B in gastric cancer identified by proteome analysis. Proteomics 2005, 5, 1693–1704. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.H.; Lee, S.H.; Lee, K.H.; Lee, K.Y.; Kim, H.; Ryu, J.K.; Yoon, Y.B.; Kim, Y.T. Cathepsin B is a target of Hedgehog signaling in pancreatic cancer. Cancer Lett. 2009, 273, 266–272. [Google Scholar] [CrossRef]
- Nouh, M.A.; Mohamed, M.M.; El-Shinawi, M.; Shaalan, M.A.; Cavallo-Medved, D.; Khaled, H.M.; Sloane, B.F. Cathepsin B: A potential prognostic marker for inflammatory breast cancer. J. Transl. Med. 2011, 9, 1. [Google Scholar] [CrossRef]
- Gong, F.; Peng, X.; Luo, C.; Shen, G.; Zhao, C.; Zou, L.; Li, L.; Sang, Y.; Zhao, Y.; Zhao, X. Cathepsin B as a potential prognostic and therapeutic marker for human lung squamous cell carcinoma. Mol. Cancer 2013, 12, 125. [Google Scholar] [CrossRef]
- Abdulla, M.H.; Valli-Mohammed, M.A.; Al-Khayal, K.; Al Shkieh, A.; Zubaidi, A.; Ahmad, R.; Al-Saleh, K.; Al-Obeed, O.; McKerrow, J. Cathepsin B expression in colorectal cancer in a Middle East population: Potential value as a tumor biomarker for late disease stages. Oncol. Rep. 2017, 37, 3175–3180. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Jiang, D.; Zhang, L.; Su, Q.; Mao, W.; Jiang, C. Expression profile of cathepsins indicates the potential of cathepsins B and D as prognostic factors in breast cancer patients. Oncol. Lett. 2016, 11, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.E.; Ho, C.C.; Yang, S.F.; Lin, S.H.; Yeh, K.T.; Lin, C.W.; Chen, M.K. Cathepsin B expression and the correlation with clinical aspects of oral squamous cell carcinoma. PLoS ONE 2016, 11, e0152165. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wang, H.; Li, Z.; Wang, L.; Zheng, F.; Jiang, J.; Gao, Y.; Zhong, H.; Huang, Y.; Suo, Z. Cathepsin B may be a potential biomarker in cervical cancer. Histol. Histopathol. 2012, 27, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Devetzi, M.; Scorilas, A.; Tsiambas, E.; Sameni, M.; Fotiou, S.; Sloane, B.F.; Talieri, M. Cathepsin B protein levels in endometrial cancer: Potential value as a tumour biomarker. Gynecol. Oncol. 2009, 112, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Jechorek, D.; Votapek, J.; Meyer, F.; Kandulski, A.; Roessner, A.; Franke, S. Characterization of cathepsin X in colorectal cancer development and progression. Pathol. Res. Pract. 2014, 210, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Zhang, D.; Hu, T.; Zhao, H.; Zhao, X.; Lou, Z.; He, Y.; Qin, W.; Xia, J.; Zhang, X.; et al. KMT2A histone methyltransferase contributes to colorectal cancer development by promoting cathepsin Z transcriptional activation. Cancer Med 2019, 8, 3544–3552. [Google Scholar] [CrossRef]
- Krueger, S.; Kalinski, T.; Hundertmark, T.; Wex, T.; Küster, D.; Peitz, U.; Ebert, M.; Nägler, D.K.; Kellner, U.; Malfertheiner, P.; et al. Up-regulation of cathepsin X in Helicobacter pylori gastritis and gastric cancer. J. Pathol. 2005, 207, 32–42. [Google Scholar] [CrossRef]
- Wang, J.; Chen, L.; Li, Y.; Guan, X.Y. Overexpression of cathepsin Z contributes to tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. PLoS ONE 2011, 6, e24967. [Google Scholar] [CrossRef]
- Lines, K.E.; Chelala, C.; Dmitrovic, B.; Wijesuriya, N.; Kocher, H.M.; Marshall, J.F.; Crnogorac-Jurcevic, T. S100P-binding protein, S100PBP, mediates adhesion through regulation of cathepsin Z in pancreatic cancer cells. Am. J. Pathol. 2012, 180, 1485–1494. [Google Scholar] [CrossRef]
- del Re, E.C.; Shuja, S.; Cai, J.; Murnane, M.J. Alterations in cathepsin H activity and protein patterns in human colorectal carcinomas. Br. J. Cancer 2000, 82, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Staack, A.; Tolic, D.; Kristiansen, G.; Schnorr, D.; Loening, S.A.; Jung, K. Expression of cathepsins B, H, and L and their inhibitors as markers of transitional cell carcinoma of the bladder. Urology 2004, 63, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.M.; Huang, Y.H.; Yeh, C.T.; Tsai, M.M.; Liao, C.H.; Cheng, W.L.; Chen, W.J.; Lin, K.H. Cathepsin H regulated by the thyroid hormone receptors associate with tumor invasion in human hepatoma cells. Oncogene 2011, 30, 2057–2069. [Google Scholar] [CrossRef] [PubMed]
- Khaket, T.P.; Singh, M.P.; Khan, I.; Bhardwaj, M.; Kang, S.C. Targeting of cathepsin C induces autophagic dysregulation that directs ER stress mediated cellular cytotoxicity in colorectal cancer cells. Cell Signal. 2018, 46, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Jevnikar, Z.; Rojnik, M.; Jamnik, P.; Doljak, B.; Fonovic, U.P.; Kos, J. Cathepsin H mediates the processing of talin and regulates migration of prostate cancer cells. J. Biol. Chem. 2013, 288, 2201–2209. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, S.S.; Goldstein, L.J.; Gottesman, M.M. Expression of cathepsin L in human tumors. Cancer Res. 1991, 51, 1478–1481. [Google Scholar] [PubMed]
- Sudhan, D.R.; Siemann, D.W. Cathepsin L targeting in cancer treatment. Pharmacol. Ther. 2015, 155, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xiang, Z.; Zhu, T.; Chen, J.; Zhong, M.Z.; Huang, J.; Wang, K.S.; Li, L.; Sun, L.Q.; Zhou, W.B. Cathepsin L interacts with CDK2-AP1 as a potential predictor of prognosis in patients with breast cancer. Oncol. Lett. 2020, 19, 167–176. [Google Scholar] [CrossRef]
- Keerthivasan, S.; Keerthivasan, G.; Mittal, S.; Chauhan, S.S. Transcriptional upregulation of human cathepsin L by VEGF in glioblastoma cells. Gene 2007, 399, 129–136. [Google Scholar] [CrossRef]
- Gao, L.; Fang, Y.Q.; Zhang, T.Y.; Ge, B.; Tang, R.J.; Huang, J.F.; Jiang, L.M.; Tan, N. Acidic extracellular microenvironment promotes the invasion and cathepsin B secretion of PC-3 cells. Int. J. Clin. Exp. Med. 2015, 8, 7367–7373. [Google Scholar]
- Jakoš, T.; Pišlar, A.; Jewett, A.; Kos, J. Cysteine cathepsins in tumor-associated immune cells. Front. Immunol. 2019, 10, 2037. [Google Scholar] [CrossRef] [PubMed]
- Jakoš, T.; Pišlar, A.; Pečar Fonović, U.; Kos, J. Lysosomal peptidases in innate immune cells: Implications for cancer immunity. Cancer Immunol. Immunother. 2020, 69, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Vasiljeva, O.; Papazoglou, A.; Krüger, A.; Brodoefel, H.; Korovin, M.; Deussing, J.; Augustin, N.; Nielsen, B.S.; Almholt, K.; Bogyo, M.; et al. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006, 66, 5242–5250. [Google Scholar] [CrossRef] [PubMed]
- Gocheva, V.; Wang, H.W.; Gadea, B.B.; Shree, T.; Hunter, K.E.; Garfall, A.L.; Berman, T.; Joyce, J.A. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010, 24, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Akkari, L.; Gocheva, V.; Kester, J.C.; Hunter, K.E.; Quick, M.L.; Sevenich, L.; Wang, H.W.; Peters, C.; Tang, L.H.; Klimstra, D.S.; et al. Distinct functions of macrophage-derived and cancer cell-derived cathepsin Z combine to promote tumor malignancy via interactions with the extracellular matrix. Genes Dev. 2014, 28, 2134–2150. [Google Scholar] [CrossRef] [PubMed]
- Vasiljeva, O.; Korovin, M.; Gajda, M.; Brodoefel, H.; Bojic, L.; Krüger, A.; Schurigt, U.; Sevenich, L.; Turk, B.; Peters, C.; et al. Reduced tumour cell proliferation and delayed development of high-grade mammary carcinomas in cathepsin B-deficient mice. Oncogene 2008, 27, 4191–4199. [Google Scholar] [CrossRef] [PubMed]
- Edgington-Mitchell, L.E.; Rautela, J.; Duivenvoorden, H.M.; Jayatilleke, K.M.; van der Linden, W.A.; Verdoes, M.; Bogyo, M.; Parker, B.S. Cysteine cathepsin activity suppresses osteoclastogenesis of myeloid-derived suppressor cells in breast cancer. Oncotarget 2015, 6, 27008–27022. [Google Scholar] [CrossRef] [PubMed]
- Gocheva, V.; Zeng, W.; Ke, D.; Klimstra, D.; Reinheckel, T.; Peters, C.; Hanahan, D.; Joyce, J.A. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006, 20, 543–556. [Google Scholar] [CrossRef]
- Victor, B.C.; Anbalagan, A.; Mohamed, M.M.; Sloane, B.F.; Cavallo-Medved, D. Inhibition of cathepsin B activity attenuates extracellular matrix degradation and inflammatory breast cancer invasion. Breast Cancer Res. 2011, 13, R115. [Google Scholar] [CrossRef]
- Mitrović, A.; Mirković, B.; Sosič, I.; Gobec, S.; Kos, J. Inhibition of endopeptidase and exopeptidase activity of cathepsin B impairs extracellular matrix degradation and tumour invasion. Biol. Chem. 2016, 397, 165–174. [Google Scholar] [CrossRef]
- Mai, J.; Sameni, M.; Mikkelsen, T.; Sloane, B.F. Degradation of extracellular matrix protein tenascin-C by cathepsin B: An interaction involved in the progression of gliomas. Biol. Chem. 2002, 383, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.P.; Yue, X.; Li, S.Q. Cathepsin C interacts with TNF-α/p38 MAPK signaling pathway to promote proliferation and metastasis in hepatocellular carcinoma. Cancer Res. Treat. 2020, 52, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Gogineni, V.R.; Gupta, R.; Nalla, A.K.; Velpula, K.K.; Rao, J.S. uPAR and cathepsin B shRNA impedes TGF-β1-driven proliferation and invasion of meningioma cells in a XIAP-dependent pathway. Cell Death Dis. 2012, 3, e439. [Google Scholar] [CrossRef]
- Tummalapalli, P.; Spomar, D.; Gondi, C.S.; Olivero, W.C.; Gujrati, M.; Dinh, D.H.; Rao, J.S. RNAi-mediated abrogation of cathepsin B and MMP-9 gene expression in a malignant meningioma cell line leads to decreased tumor growth, invasion and angiogenesis. Int. J. Oncol. 2007, 31, 1039–1050. [Google Scholar]
- Sevenich, L.; Schurigt, U.; Sachse, K.; Gajda, M.; Werner, F.; Müller, S.; Vasiljeva, O.; Schwinde, A.; Klemm, N.; Deussing, J.; et al. Synergistic antitumor effects of combined cathepsin B and cathepsin Z deficiencies on breast cancer progression and metastasis in mice. Proc. Natl. Acad. Sci. USA 2010, 107, 2497–2502. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Bhasin, S.; Khanna, P.; Joshi, M.; Joslin, P.M.; Saxena, R.; Amin, S.; Liu, S.; Sindhu, S.; Walker, S.R.; et al. Study of cathepsin B inhibition in VEGFR TKI treated human renal cell carcinoma xenografts. Oncogenesis 2019, 8, 15. [Google Scholar] [CrossRef]
- Gocheva, V.; Chen, X.; Peters, C.; Reinheckel, T.; Joyce, J.A. Deletion of cathepsin H perturbs angiogenic switching, vascularization and growth of tumors in a mouse model of pancreatic islet cell cancer. Biol. Chem. 2010, 391, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Ruffell, B.; Affara, N.I.; Cottone, L.; Junankar, S.; Johansson, M.; DeNardo, D.G.; Korets, L.; Reinheckel, T.; Sloane, B.F.; Bogyo, M.; et al. Cathepsin C is a tissue-specific regulator of squamous carcinogenesis. Genes Dev. 2013, 27, 2086–2098. [Google Scholar] [CrossRef]
- Overall, C.M.; Dean, R.A. Degradomics: Systems biology of the protease web. Pleiotropic roles of MMPs in cancer. Cancer Metastasis Rev. 2006, 25, 69–75. [Google Scholar] [CrossRef]
- Vasiljeva, O.; Turk, B. Dual contrasting roles of cysteine cathepsins in cancer progression: Apoptosis versus tumour invasion. Biochimie 2008, 90, 380–386. [Google Scholar] [CrossRef]
- Affara, N.I.; Andreu, P.; Coussens, L.M. Delineating protease functions during cancer development. Methods Mol. Biol. 2009, 539, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Skrzydlewska, E.; Sulkowska, M.; Koda, M.; Sulkowski, S. Proteolytic-antiproteolytic balance and its regulation in carcinogenesis. World J. Gastroenterol. 2005, 11, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Mason, S.D.; Joyce, J.A. Proteolytic networks in cancer. Trends Cell Biol. 2011, 21, 228–237. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).