Abstract
The gut microbiome is intimately intertwined with the host immune system, having effects on the systemic immune system. Dysbiosis of the gut microbiome has been linked not only to gastrointestinal disorders but also conditions of the skin, lungs, and brain. Commensal bacteria can affect the immune status of the host through a stimulation of the innate immune system, training of the adaptive immune system, and competitive exclusion of pathogens. Commensal bacteria improve immune response through the production of immunomodulating compounds such as microbe-associated molecular patterns (MAMPs), short-chain fatty acids (SCFAs), and secondary bile acids. The microbiome, especially when in dysbiosis, is plastic and can be manipulated through the introduction of beneficial bacteria or the adjustment of nutrients to stimulate the expansion of beneficial taxa. The complex nature of the gastrointestinal tract (GIT) ecosystem complicates the use of these methods, as similar treatments have various results in individuals with different residential microbiomes and differential health statuses. A more complete understanding of the interaction between commensal species, host genetics, and the host immune system is needed for effective microbiome interventions to be developed and implemented in a clinical setting.
1. Introduction
The human body is host to many diverse microbes, with the largest community inhabiting the gastrointestinal tract (GIT). There has recently been an increased interest in how these communities interact with the human host and affect its overall health. These microbial communities consist of viruses, archaea, eukaryotes (fungi, protists, arthropods, etc.), and bacteria. The bacterial members of this community are the most well studied and will be the focus of this review. Strains of commensal bacteria have often coevolved alongside a specific taxonomic group of hosts, sometimes being specific to a single species [1,2]. An extreme example are the “infant type” Bifidobacterium spp., which have co-evolved alongside humans and specialize in utilizing carbohydrates found in human breastmilk, allowing extensive colonization only during nursing [3,4]. The microbiome is also specific to each area of the host GIT with the community changing along its length [5,6]. Many studies to date have focused on the fecal microbiome because it is the most accessible for sampling. The bacterial community present in the feces is most like that in the colon; however, it is not a good representative of the bacterial communities in more proximal areas of the GIT [7]. It is generally accepted that the fetus is sterile within the womb and colonization of the GIT begins during birth [8,9], although some have argued that microbiome colonization begins in utero [10]. The GIT microbiome changes rapidly during early life and stabilizes as we age. The neonatal intestine is aerobic and initial colonization is by facultative anaerobes belonging to the phyla Actinomycetota and Bacillota (formerly Actinobacter and Firmicutes); as the intestine matures and becomes anaerobic, Pseudomonadota and Bacteroidota (formerly Proteobacteria and Bacteriodetes) become more prevalent [4,11]. This succession pattern to a healthy adult microbiome has long term implications for host health, and disruptions of the GIT community early in life can affect vaccine response and increase the chances of autoimmune, neurological, lung, and gastrointestinal disorders later in life [11,12,13,14,15,16,17].
The advent of “omic” studies has made identifying and studying the large number of unculturable bacteria within the human GIT possible. Metagenomic studies allow us to identify the bacteria within the community with more accuracy than the use of marker genes (16S rRNA being the most commonly used). With the improvements in sequencing and data analysis, it has become clear that marker genes often underestimate the diversity of the community [18]. The use of metagenomic sequencing also allows for the prediction of the metabolic pathways present, giving insight into the metabolic potential of the community [19]. Large scale collaborations have created datasets of human genome information paired with microbiome information. Differences in sampling, sample preparation, sequencing technology, and species identification are challenges that must be addressed when utilizing data from multiple studies [20]. Despite these challenges, researchers have been able to identify a link between Bifidobacterium species and specific genotypes related to the LCT gene [21,22,23]. A link between ABO blood groups and multiple bacteria taxa has also been identified [21,22]. Identifying genetic variation with links to the microbiome is challenging. There are usually confounding social, diet, and environmental factors, which also affect the microbiome. Further studies with consistent methods in diverse populations are needed to untangle these factors [20]. With this burst of available information, researchers have been able to identify specific changes in the GIT microbiome of people suffering from many diseases and disorders, as well as possible genetic links to disease susceptibility.
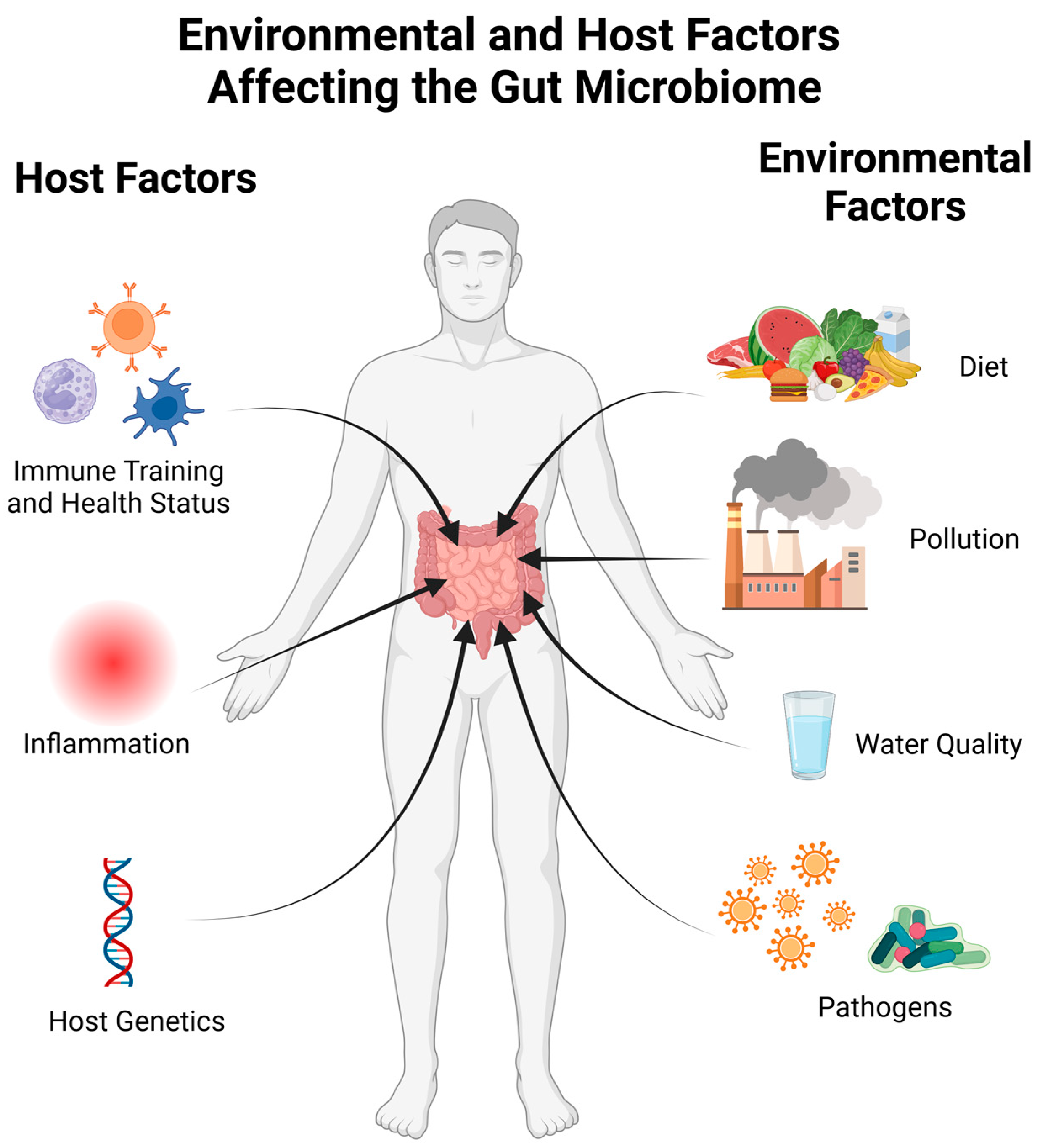
The gut microbiome is also susceptible to perturbation by a number of environmental factors. Exposure to environmental pollutants from air, water, or food contamination affects the gut microbiome [24,25,26]. Air pollution can cause damage and inflammation in the lungs. This activation of the innate immune system can have systemic affects and cause changes to the intestinal microbiome [25,27,28]. A study in Dutch individuals also showed a link between smoking—past and present, as well as secondhand smoke—and the gut microbiome [29]. The contamination of water with heavy metals and other oxidizers disrupts the redox balance within the intestinal environment. This can directly affect bacteria sensitive to oxidative stress, such as Faecalibacterium, or cause damage to intestinal tissues, resulting in a disruptive immune response [24,26]. Bisphenol A (BPA) is a common contaminant known to affect the human endocrine system. Exposure to this chemical has been linked to decreases in Bifidobacteria and Akkermansia levels [25]. The factors affecting the microbiome can be thought of as originating from the host or the host’s environment, as highlighted in Figure 1.
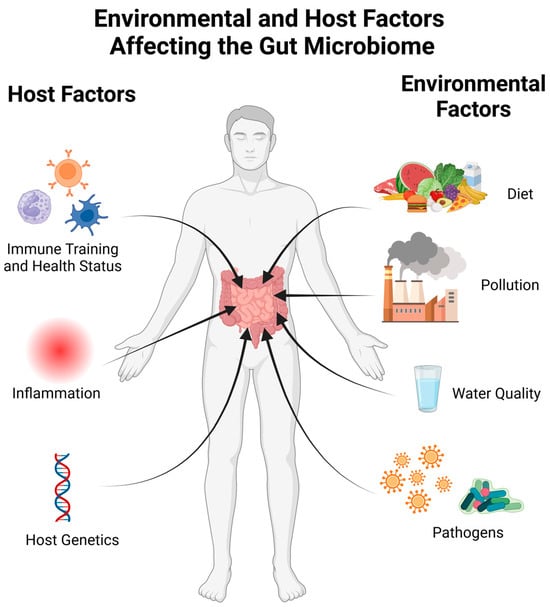
Figure 1.
A visualization of factors influencing the microbiome. The human host affects the microbiome through their current health status as well as immune training from previous infections or vaccinations. Both systemic and gut inflammation can perturb the microbiome, and a number of host genetic factors have been linked to the gut microbiome composition. Environmental factors such as diet, water quality, pollution, and exposure to pathogens also have consequences within the gut microbiome. Created with BioRender.com (accessed on 23 October 2023).
Healthy adults with an intact microbiome are unlikely to be colonized by a number of pathogens; however, the disruption of the microbiome allows for colonization by these pathogens. Clostridioides difficile pathogenesis often occurs after prolonged antibiotic treatment has disrupted the microbiome; however, it is present in most healthy adults with no adverse effects. The reconstitution of the microbiome has become an approved medical intervention for prolonged C. difficile cases [30,31]. Vibrio cholera is able to colonize infant mice with undeveloped immune systems, causing a disease state, but adult mice cannot be colonized without first disrupting the GIT microbiome [32,33]. Similarly, it has been proposed that cholera is only able to colonize and cause disease in humans when the microbiome is disrupted, often by malnutrition or diarrhea of an unrelated source [34,35]. Listeria monocytogens is an agent of food borne illnesses that is also better able to infect mice when the microbiome is disturbed; resistance has been linked to products of the microbiome such as SCFAs and antimicrobials [36]. The microbiome also provides protection against Salmonella colonization, recently reviewed in Rogers et al. [37]. A healthy microbiome provides protection against intestinal pathogens through direct competition as well as the priming and training of the host immune system. A more in-depth discussion of how the microbiome restricts pathogen growth, the bacterial products involved, which commensal bacteria produce them, and interventions to harness this protection are discussed below. With this review, we hope to present a comprehensive look at how the gut microbiome affects human health and present possible modes of action for potential therapeutic approaches.
2. Mechanisms by Which the Microbiome Provides Colonization Resistance
Studies in gnotobiotic models have shown that in the absence of commensal bacteria, the host immune system fails to develop properly, with little innate immune response and a poor response of the adaptive immune system when exposed to bacteria. Commensal organisms are needed to train the host immune system and improve their resistance to pathogens [16,17,38]. This immune training has implications throughout the host body and is not localized to the GIT. The microbiome improves pathogen resistance through nutrient and oxygen sequestering, the production of antimicrobial compounds, occupying attachment sites, the stimulation of mucus production, the tightening of tight junctions, the regulation of inflammation, and the training of innate and adaptive immunity [39,40,41,42,43,44]. Colonization resistance is often the result of immunoactive compounds produced by the bacterial community. These include microbe-associated molecular patterns (MAMPs), extracellular vesicles (EVs), microbe-derived anti-inflammatory compounds, the degradation of harmful compounds, and beneficial microbial metabolic products that stimulate the immune system, increasing its effectiveness against pathogens [38,43,45,46].
The microbiome provides colonization resistance through environmental engineering, the production of antibacterial compounds, and direct competition for electron acceptors, nutrients, and physical space within the intestine [37,47]. Some commensal bacteria produce antimicrobial compounds. These compounds reduce the competition from other species and allow the commensal to establish itself within the intestine [48,49]. These compounds are discussed in more detail in the Section 3. Production of organic acids lowers the pH of the intestinal lumen and changes the redox potential of the environment, which can interfere with virulence factors [37]. Of particular interest are short-chain fatty acids (SCFAs) [37,50]. Increased oxidative stress in the intestinal lumen has been linked to changes in the microbiome that lead to dysbiosis and proliferation of pathogens [51,52]. Conversely, oxygen sequestering by commensal bacteria can limit the growth of pathogens. This is the proposed mode of action for the colonization resistance provided by resident Enterobacterales against virulent Salmonella sp. [37]. The residential bacteria utilize the available nutrients, and an established microbiome is resistant to the introduction of new species [37,53,54]. This is true for pathogens but also the introduction of beneficial bacteria, and it is a challenge for microbiome interventions involving probiotics. Finally, commensal bacteria can occupy attachment sites that are required for pathogen virulence. Segmented filamentous bacteria (SFB) attach to intestinal epithelial cells in a similar manner to Salmonella, effectively blocking Salmonella from attaching to the intestinal epithelium and preventing disease [2,55,56]. Similarly, non-infectious E. coli can occupy attachment sites used by virulent E. coli strains [53]. Commensal bacteria that are closely related to pathogens are often able to occupy the ecological niche preferred by the pathogen and, once established, are effective in excluding them from the intestine [37,53,54]. The innate immune system in the GIT is constitutively active, as it is always stimulated by commensal bacteria. A healthy gut has effective barrier function and appropriate inflammation responses. An overly sensitive inflammation response can lead to a loss in barrier function, referred to as a “leaky gut”, allowing metabolites, proteins, and even bacteria to cross the epithelium, causing sepsis. This can lead to poor health outcomes throughout the body, and has been linked to intestinal, metabolic, lung, skin, and neurological disorders [57,58,59]. However, changes to the microbiome associated with these conditions are not consistent, and it is unclear if the microbiome changes that are observed are a cause or effect of the condition in humans, although fecal transplantation can transfer these disorders to recipient mice [59,60,61].
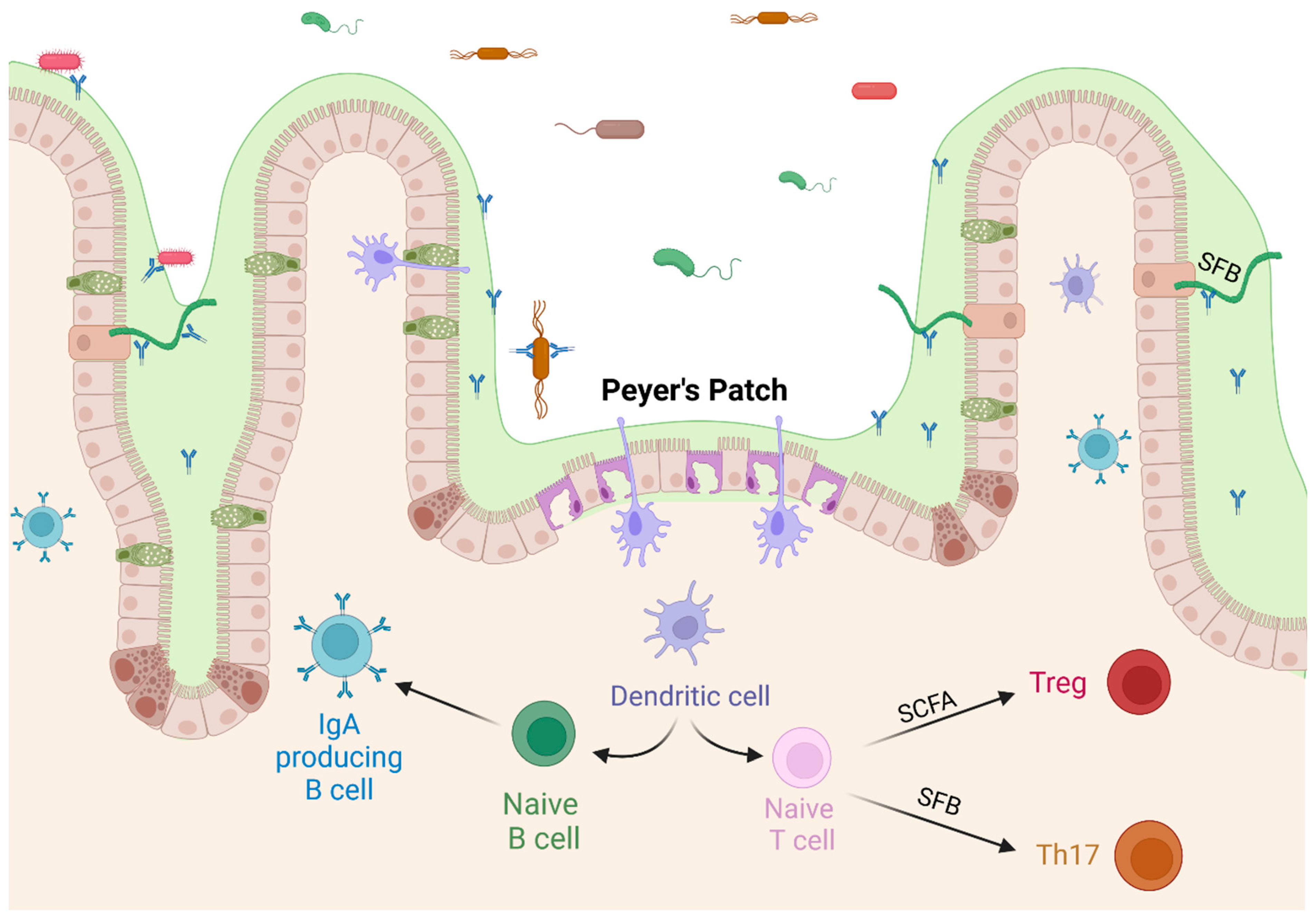
The adaptive immune system is also very active within the GIT. Antigens in the lumen are constantly sampled, especially in the specialized areas of the Peyer’s Patches, which are located along the small intestine, most densely in the ileum. The development of Peyer’s Patches is reduced in germ-free mice [62]. They are areas with a thinner mucus coating, no villi, and specialized M-cells that allow dendrites to extend into the lumen and sample antigens, including food, viruses, and bacteria. Located below the Peyer’s Patch is an area of lymph tissue where antigen-presenting cells can interact with naïve B-cells, triggering the subsequent production of sIgA in the lamina propria [63]. Sampling through the Peyer’s Patch is a source of undiversified IgA in the intestine [64], although luminal sampling near goblet cells has also been observed [65,66]. The differentiation of T-cells is also directly affected by the microbiome. CD4+ naïve T-cells differentiate into regulatory T-cells (Treg) or Th17 cells in the presence of TGF-β, with Th17 cells requiring the additional presence of IL-6. Beneficial commensal bacteria lower the production of IL-6, promoting the differentiation into Treg cells, which produce IL-10 and reduce inflammation levels [67,68]. Antigen sampling within the gut is likely to stimulate immunotolerance because of regulatory cytokine ratios present in the healthy intestine. This propensity for tolerance is extended to other areas of the body, as the immune cells trained in the GIT migrate to other parts of the body [69]. Peyer’s Patches are also the most prominent areas for T-cell dependent class switching, while T-cell independent class switching takes place within the laminal propria [17,63]. Immune training through luminal sampling in the GIT trains the adaptive immune system and affects the systemic immune response.
3. Beneficial Bacterial Products
Beneficial bacterial products are diverse in nature, and some are structural compounds of the microbial cell (LPS, peptidoglycan, unmethylated DNA, etc.), while others are products of microbial metabolism (microbial anti-inflammatory molecule, SCFA, bacteriocins, etc.). These bacterial products are involved in crosstalk with the host. In many experiments and clinical trials, the exact compounds responsible for the benefit to the host is not clear, and the mechanism of action is often expounded by in vitro experiments involving cell lines or in animal models.
The innate immune system is stimulated by MAMPs such as peptidoglycans, unmethylated CpG DNA, LPS, etc. These patterns are recognized by pattern recognition receptors (PRRs) that can be attached to the cell membrane within the periplasm or secreted outside the cell. Membrane-bound PRRs include Toll-like receptors (TLRs), C-type lectin receptors (CLRs), and Nod-like receptors (NLRs), which are found on a variety of cells within the GIT and throughout the body [44]. Peroxisome proliferator-activated receptors (PPARs) are nuclear receptors that regulate inflammatory and metabolic processes and are found in cells throughout the body [70]. The innate immune system, including the epithelium barrier, is the first line of defense against pathogens and, until recently, was believed to have little or no specificity and no immune memory, providing a rapid and consistent response to all potential pathogens [43,71,72,73,74]. Recent work has shown that the innate immune system does respond differently to repeated challenges. Vaccination with live attenuated Bacillus Calmette-Guerin provides non-specific protection against multiple pathogens [40,71], possibly from changes to glucose metabolism by immune cells [72] and the reprogramming of stem cells within the germinal centers of the bone [73]. By increasing innate immune cell production and cytokine production, the innate immune system may have more immune memory than previously thought. While MAMPs are non-specific, the presence of non-pathogenic commensals can provide some cross protection against similar pathogens.
Extracellular vesicles (EVs) are spherical membranous vesicles that can contain molecules such as proteins and nucleic acids [45,70,74] as well as the MAMPs associated with cell wall components. EVs are produced by both prokaryotic and eukaryotic cells and are utilized for intercellular communication [75]. EVs produced by Faecalibacterium prausnitzii [70,74], Akkermansia muciniphila [76], and Bacteroides thetaiotaomicron [45] have all been shown to interact with TLR and PPARs, affecting cytokine production. B. thetaiotaomicron were shown to interact with TLR1,2,3, and 7 to help immune cell survival, reducing inflammatory signals [45]. EVs from A. muciniphila induced the phosphorylation of AMPK, a tight junction protein activator. This was supported by reduced intestinal permeability in mice colonized by A. muciniphila [76]. The production of IL-10 was increased by F. prausnitzii and A. muciniphila EVs through reduction in the NF-kB inhibitor, NFKBIL1 [74,76]. Commensal EVs also reduce the expression of inflammatory cytokines such as IL-1, IL-2, IL-6, IL-12, and IFN- [70,77].
SCFAs are the product of the bacterial fermentation of indigestible fiber in the large intestine. The most common SCFAs are acetate, propionate, and butyrate [47,78,79], although less abundant SCFAs can also influence the immune system [80]. SCFAs interact with G protein-coupled receptors (GPR) 41 and 43; these receptors are found in intestinal epithelial cells but also peripheral organs and blood cells, affecting inflammation throughout the body [81,82]. Butyrate has been reported to reduce inflammation throughout the body, reducing the production of IL-1β, IL-2, IL-12, IFN-and TNF-α, while upregulating the production of IL-10 [47,83]. Butyrate also promotes the differentiation of Treg cells, which increases tolerogenic responses [67,84]. Butyrate can be directly used by colonocytes as an energy source, promoting a healthy intestinal epithelium. Healthy colonocytes promote barrier function by increasing mucus production, promoting healthy tight junctions and the increased production of antimicrobial proteins in Paneth cells [47,80,81,82]. SCFAs also lower the pH of the intestinal environment, inhibiting the growth of many pathogens [37,85]. Propionate was shown to limit the growth of Salmonella enterica serovar Typhimurium in vitro through changes to the intercellular pH [86]. Butyrate is able to reduce the growth of multiple C. difficile strains [87] and L. monocytogens [50] in culture, showing an inhibition of this pathogen unrelated to host immune response [78].
4. Beneficial Bacteria
The benefit of fermented foods such as yogurt, cheese, kefir, and natto to gut health has been known for centuries [88]. Many probiotics that are currently on the market are species that were historically used in food production, particularly lactic acid-producing bacteria (LAB). LAB include bacteria belonging to the genera Lactobacillus, Lactococcus, and Bifidobacterium and are commonly found in fermented milk products. LAB are well studied, and their probiotic benefits and mechanisms of action have been recently reviewed in detail by Tiwari and Tiwari (2022) [48]. LAB produce bacteriocins, bacteriocin-like molecules, hydrogen peroxide, and carbon dioxide, which restrict the growth of the major pathogens Listeria [49], Clostridium, and Salmonella [89]. LAB also interact with the immune system, influencing immunotolerance and generally promoting Treg differentiation as well as reducing inflammation responses within the intestine [48,78,89,90]. The use of LAB in food production has allowed for a large-scale production of probiotics for commercial sale with relatively little regulatory opposition.
A healthy GIT microbiome is hard to define because of the high variation seen across populations [91,92,93]. Generally, a high alpha diversity is indicative of a healthy microbial community. A loss of diversity and a low evenness score are commonly seen in the microbiomes of people suffering from dysbiosis and many medical conditions [16,92,94]. The most abundant bacterial phyla in the mammalian GIT are Bacillota and Bacteroidota; these two phyla constitute ~80% of a healthy microbiome. These abundances are often compared and referred to as the older nomenclature of Firmicutes:Bacteroidetes or the F:B ratio. A large F:B ratio is often an indication of a healthy gut microbiome, while a low F:B ratio often indicates dysbiosis [95,96]; however, an increased F:B ratio has been linked to obesity [96,97,98]. The age of the host is a major factor when using this ratio and must be taken into consideration [16,94,99,100]. Some Bacillota are oxygen tolerant and more abundant in the GIT of neonatal and young hosts, with the oxygen sensitive Bacteroidota being a secondary colonizer [6,14,101].
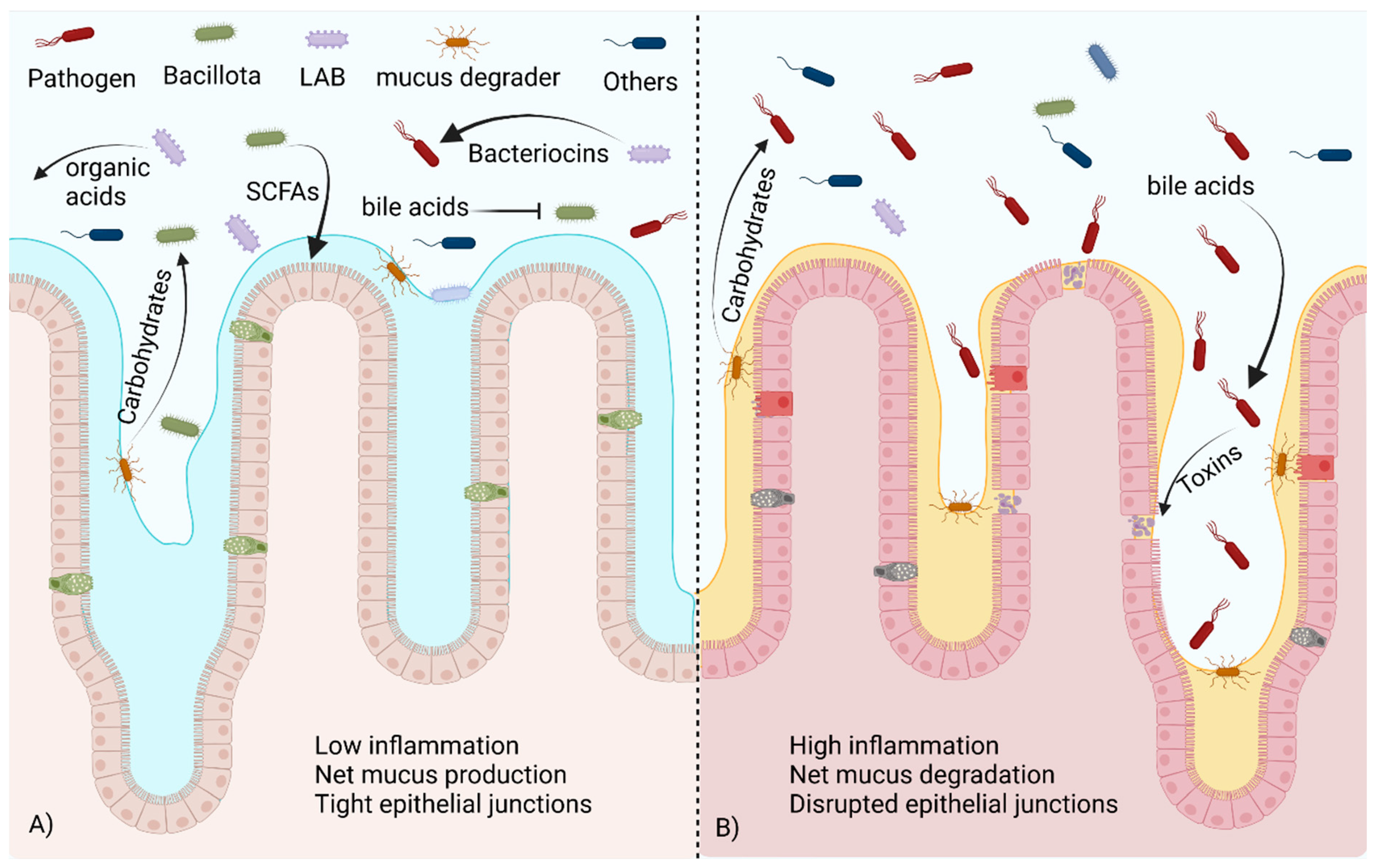
The majority of bacteria within the phylum Bacillota are gram-positive, spore-forming bacteria with various levels of oxygen tolerance. This phylum was identified as beneficial to gut health early in the study of the GIT microbiome. Bacillota are often involved with the fermentation of fiber in the colon. The main products of interest in this fermentation are SCFAs. These products of bacterial fermentation can be used as an energy source by colonocytes, and they can enter the bloodstream and react with cell receptors located in distant organs [75,83,85,102,103]. The diversity found within the phyla Bacillota likely plays a role in the inconsistent results when this high-level classification is considered. While total Bacillota present is often correlated to SCFA production/levels [96], this is not always the case. Strategies such as this rely on the accurate identification and prior knowledge of fermentation abilities, both of which may be lacking, resulting in inaccuracies. To complicate matters further, there are a number of pathogenic species within the Bacillota, the most common of which are Clostridiodes difficile and species belonging to the genus Staphylococcus. While high-level generalizations are appealing, it is an oversimplification of the complex nature of the gut ecosystem, and identification to a more specific classification. A visualization of many of these concepts can be seen in Figure 2. Below, we discuss specific groups that are thought to be beneficial to host health and immune function.
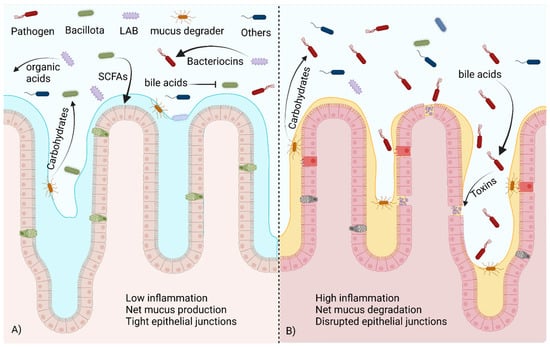
Figure 2.
A healthy commensal bacteria population versus one in dysbiosis. (A) In the healthy gut microbiome, beneficial Bacillota metabolize bile acids and utilize carbohydrates freed by mucus degraders to produce SCFAs, promoting mucus production and healthy tight junctions in the epithelium. LABs produce organic acids and bacteriocins that control pathogen populations. (B) In dysbiosis, the presence of unmodified bile acids stimulates toxin production via pathogens. Without competition from beneficial species, pathogens are able to utilize carbohydrates released by mucus degraders to expand their population, damaging the epithelium. Created with BioRender.com (accessed on 23 October 2023).
The genus Faecalibacterium contains commensal bacteria that are also known to ferment indigestible fiber into SCFAs [75]. Faecalibacterium are obligate anaerobes and are unable to colonize the human GIT early in life but become common after 2–3 years, becoming ~5% of the microbiome in adults [104]. F. prausnitzii produces a protein that reduces inflammatory signals in mammalian cells [105]. This has been shown to be beneficial in Crohn’s disease and IBS [105,106]. Increased oxidative stress is associated with gut inflammation, and Faecalibacterium has been suggested as a potential marker species because of its relatively high abundance and oxygen sensitivity [107,108]. An increase in oxidative stress during inflammation has been suggested as one cause of dysbiosis [109]. Many SCFA producers are sensitive to oxygenation stress, while opportunistic pathogens tend to have a higher oxidative stress tolerance. Members of the phylum Verrucomicrobiota, such as Akkermansia muciniphila, also produce SCFAs but have a higher oxygen tolerance [83,110]. This phylum is most abundant early in life but is present in most adult microbiomes [94,101]. Akkermansia spp. and other members of the Verrucomicrobiota digest the mucus lining the GIT as a nutrient source, releasing metabolites and freeing carbohydrates that can be further degraded by other members of the community [83]. A. muciniphila and Bifidobacterium sp. have been considered for use as a probiotic because of their ability to promote the growth of other beneficial bacteria [85,111], Figure 2A. It can be considered an environmental engineer as it can colonize intestines with oxidative stress, reduce the oxygen levels through reduced inflammation and metabolism, and produce nutrients for the more oxygen sensitive SCFA producing bacteria allowing them to colonize further improving gut health [76,83,112]. Species from the genus Blautia, particularly Blautia obeum, have been shown to provide protection from a Vibrio cholera infection [113]. The presence of bile acids induces the production of toxin-coregulated pilus (TCP) and type VI secretion system (T6SS) proteins in V. cholera. Blautia species reduce primary bile acids into secondary bile acids, downregulating these virulence factors [114,115], as shown in Figure 2B. However, bile acids are toxic to L. monocytogenes, and the degradation of these products may lead to a higher susceptibility to listeria infection [36]. Commensal bacteria can also mimic quorum-sensing signals essential for virulence in cholera colonization. The most well documented is the production of autoinducer 2 (AI2) by B. obeum, which blocks production of TCP, reducing virulence within the intestine [33,34].
Segmented filamentous bacteria (SFB) have both a unique structure and life cycle, discussed in some detail in Hedblom et al., 2018 [2]. These bacteria attach directly to the intestinal epithelium, most often in the ileum, causing changes to the host cell and resulting in a pedestal-like structure at the site of attachment and a loss of microvilli in the immediate area. The preference of attachment site varies by species and includes enterocytes, M-cells, goblet cells, and junctions between cells in the ileum. SFB are often host specific [56], possibly through the recognition of differences within the flagellar proteins [116]. The attachment method of SFB is similar to Salmonella sp., blocking attachment sites from the pathogen [2]. Despite this similarity in attachment methods, SFB do not incite an inflammatory response; however, they do elicit an immune response.
SFB have been shown to induce immunological changes in mice, and the abundance of SFB correlates with vaccine response in a number of studies. The relationship between SFB and immune response has been extensively investigated in mice. Ivanov and colleagues (2009) showed that the presence of SFB induces Th17 cell differentiation through Toll Like Receptor 5 (TLR5) signaling [117]. SFB also induce the production of serum amyloid A (SAA), IL-17, and IL-22, which stimulate TH17 cell differentiation [55,117,118]. This increased antimicrobial defense was sufficient to protect against Citrobacter rodentium [117]. TH17 cells can be subdivided into those that clear pathogens from mucosal surfaces and those that promote immunotolerance [119,120]; the relative abundance of these cells is dependent on cytokine concentrations with pathogenic TH17 cells being induced by IL-23 exposure [68]. The overproduction of TH17 cells has been implicated in autoimmune conditions, including those centered in the GIT such as Crohn’s disease, irritable bowel syndrome (IBS), and others [118,119]. However, the negative effects of TH17 upregulation can be mitigated by the upregulation of anti-inflammatory cytokines by butyrate production in the GIT [47], depicted in Figure 3.
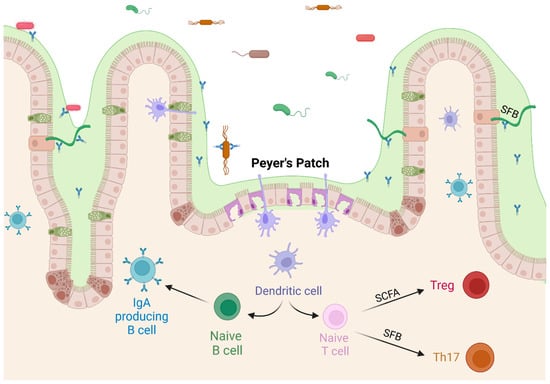
Figure 3.
Commensal bacteria affect T cell differentiation. SCFAs produced by commensal bacteria upregulate the differentiation of regulatory T cells, and SFB stimulate differentiation of Th17 cells, both pathogenic and regulatory. Created with BioRender.com (accessed on 23 October 2023).
5. Therapeutic Interventions to Improve the GIT Microbiome
As we discussed, commensal microbes and their effects of “colonization resistance” contribute to preventing host diseases. Therefore, commensal bacteria as a therapeutic intervention have garnered attention in the medical world with the rise of antibiotic resistant bacteria and the push to reduce antibiotic use. A better understanding of the mechanism of action of the microbiome in immune response and pathogen resistance does suggest that there is potential for both the treatment and prevention of microbiome-related disorders. However, the complex ecology of the GIT means that individuals will have unique reactions to microbiome intervention. Bacterial products will elicit different responses dependent on the bacterial community present, the preexisting inflammation levels, and the immune state and genetics of the host. Because of these factors, microbiome interventions may have to be personalized to each patient, requiring pretreatment investigations into the microbial community present to identify proper treatment options. However, there is less concern about this when the microbiome has been previously decimated by antibiotics, radiation, or chemotherapy.
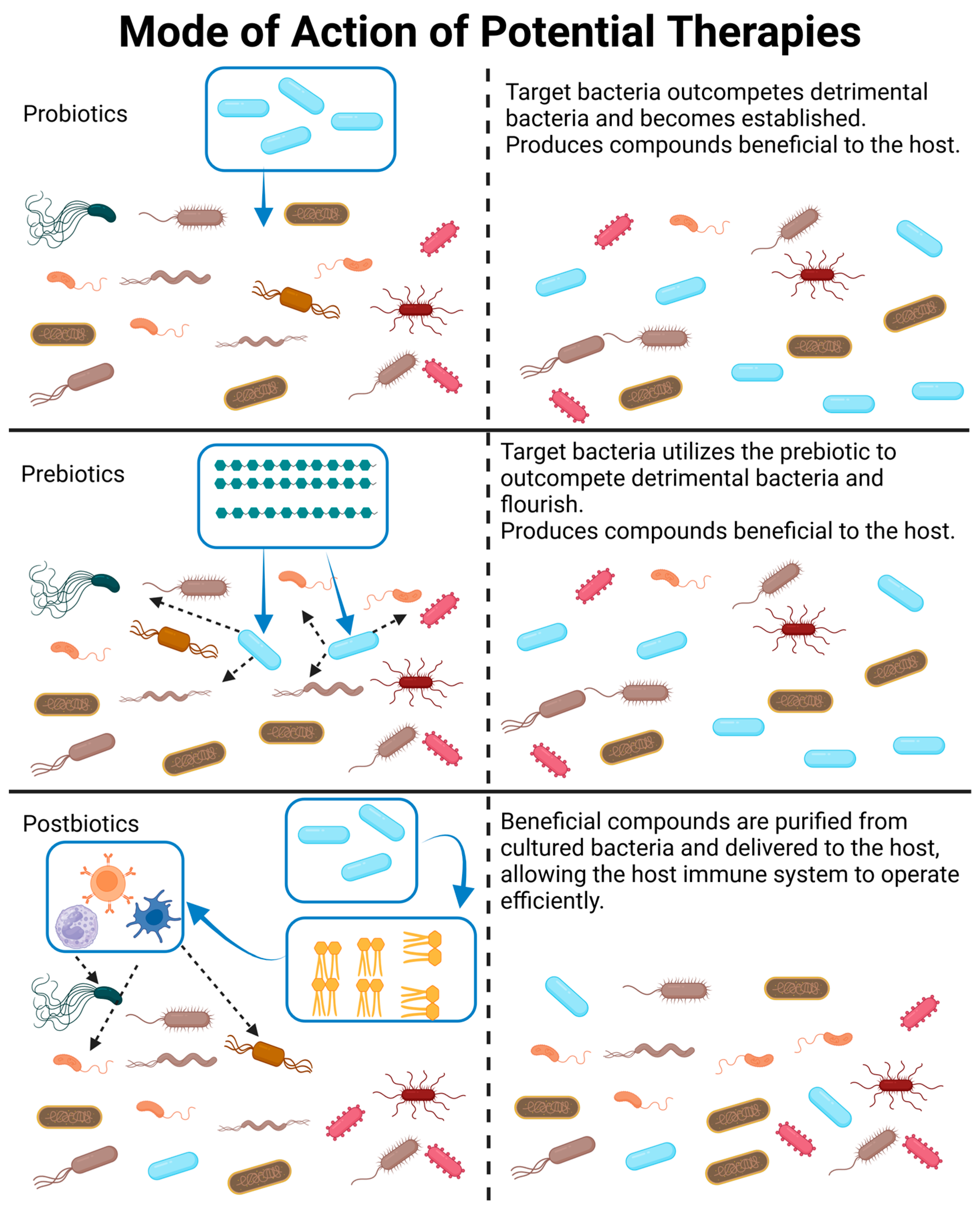
Therapeutic interventions can broadly fall into four categories: (1) the introduction of bacterial communities via fecal transplantation (FT) or the introduction of a predetermined bacterial consortia, (2) probiotics, (3) postbiotics, and (4) prebiotics. Categories 1 and 2 involve the introduction of live organisms, which can be a major concern. It is difficult to receive government approval for the development of new probiotics due to concerns regarding safety and quality control. This difficulty has led to the search for non-living alternatives, such as prebiotics, which stimulate the expansion of existing beneficial species, or postbiotics, which are non-living bacterial products. Potential uses and concerns with each of these categories will be discussed in the sections below and are visualized in Figure 4.
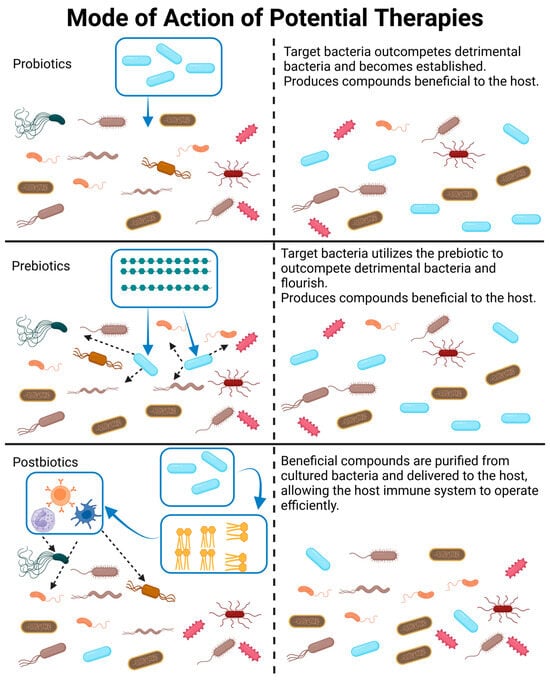
Figure 4.
A visual representation and description of how probiotics, prebiotics, and postbiotics modify the gut microbiome. Please note that probiotics may be a combination of bacterial species, which is not depicted in this figure. Likewise, postbiotics may be a combination of bacterial products or whole killed bacteria delivered to the GIT. Created with BioRender.com (accessed on 23 October 2023).
5.1. Fecal Transplantation and Bacterial Consortiums
FT has been effective in treating chronic Clostridioides difficile infections when antibiotics have been ineffective [30,31]. FT can be conducted through oral administration, enema, or placed directly into the small intestine via colonoscopy or transendoscopic enteral tubing [121]. When administering FT through the oral route, it is important to protect the bacterial community from the acidic environment of the stomach, while the enema route places the community within the colon and does not immediately seed bacteria into the small intestine. An advantage of FT is that a functional community with intact interspecies relationships is transplanted [121,122]. Identifying appropriate donors is challenging, and the microbiome changes throughout the host’s lifetime, meaning previously successful donors may no longer be suitable [123]. The largest downfall of FT is that opportunistic pathogens are present in all fecal samples, even from healthy donors. The current state of FT as a clinical treatment has been reviewed in Gupta et al., 2022 [31]; Biazzo and Deidda, 2022 [121]; and Mahmoudi and Hossainpour, 2022 [122]. The adult immune system is trained and primed by the host’s native GIT microbiome. Some studies have suggested that FT is most successful when the donor has a similar genetic background and diet to the recipient [124]. As host genetics and diet are two major factors affecting the microbiome, selecting donors based on these factors will increase the similarity between the donor microbiome and the recipient’s natural microbiome, increasing the likelihood of successful recolonization [124]. In the case of patients undergoing treatments for a condition unrelated to the GIT that is likely to disrupt the microbiome (i.e., prolonged antibiotic use, chemotherapy, etc.), it is possible to store fecal samples pretreatment, recolonizing the patient with their own native microflora [125]. There is also the possibility of the creation of autoprobiotics, where beneficial bacteria from fecal samples are cultured to probiotic levels and then reintroduced to the host to boost beneficial bacteria levels [126].
An alternative to FT is the production of bacterial consortia. This eliminates the need to find healthy donors, and potential pathogens can be excluded while still introducing beneficial bacteria with healthy interactions, which is a major concern in immunocompromised patients [126,127,128]. The development and maintenance of these consortia is a major hurdle in their production and has similar constraints to probiotics, as discussed below. Many gut bacteria are difficult to culture, as they require stable temperatures, anaerobic or microaerobic conditions, and molecular cross feeding to flourish.
5.2. Probiotics
The health benefits of fermented foods have been known for centuries, and probiotics are often delivered through fermented foods such as yogurt, cheese, natto, etc. [88,127,129]. The ideal probiotic is easily cultured, stored, and delivered to the patients. It must also be possible to deliver the living bacteria to the proper area of the intestine for it to colonize, which means surviving passage through the stomach into the intestines. The effectiveness of the probiotic is also dependent on its ability to colonize despite competition by resident bacteria. The genera Lactobacillus and Bifidobacter have many advantages as probiotics [126,130]. They are found in many fermented foods and known to be safe for human consumption. They are also acid resistant, and a small population can survive past the stomach to colonize the intestine. Finally, they are LAB and provide all the previously discussed benefits [89,131,132].
Species from the genus Bacillus, such as B. subtilus and B. ceres, are currently sold as probiotics for both livestock and humans [133,134,135,136]. These bacteria are capable of sporulation and have been shown to survive passage through the stomach in livestock [137,138]. In livestock, bacilli probiotics improve immune function, increase barrier function within the intestines, and reduce oxidation stress within the intestine [133,139,140]. The ability of Bacillus sp. to colonize the intestine long term is under debate, and the levels within the feces are significantly reduced once oral ingestion is stopped [141].
5.3. Postbiotics
Postbiotics can include purified bacterial products or whole killed bacteria. Postbiotics avoid the concerns of administering live organisms to patients, but the benefits are temporary and are lost when treatment is concluded [90,142,143]. Often inactivated bacteria are administered with the culture medium, providing any beneficial compounds produced alongside the MAMPs present in the killed bacterial cells, having similar effects on the host immune response [144]. Purified EVs have also been shown to have similar effects as live cell cultures on Caco-2 cells in a culture [45,70]. Bacteriocins have been purified from a number of species and used in food preservation and livestock studies [145,146,147]. However, some compounds can be altered via passage through the stomach or absorbed in the small intestine before reaching the colon, reducing effectiveness, and the compounds may need protective preprocessing. For example, SCFAs can be esterified to ensure delivery to the colon where their effects are most beneficial [47]. Further work is needed to identify purification methods and the effects of postbiotics on human health.
5.4. Prebiotics
Prebiotics are compounds that cannot be digested by the host but are accessible to the microbiome. Often the goal of prebiotics is to stimulate the production of beneficial products, but they may also be chosen to simply expand the population of beneficial bacteria. The most common prebiotics are indigestible fibers. These compounds are undigested by the host and reach the lower intestine intact, where they can be fermented by commensal bacteria, often producing SCFAs as a result [51,85,148]. Pascale et al., 2022 [148] recently reviewed the use of pectins as a prebiotic. They found that pectins promoted SCFA production, particularly acetate. The conversion of acetate to propionate and butyrate varied, likely influenced by the resident microbiome community [75]. Fructooligosaccharides are another common prebiotic. Costa et al., 2022, discuss the effects of this prebiotic on inflammation and gut immune response [51]. The increased intake of non-digestible starches increased the SCFA production of test subjects on a controlled diet [85]. As previously discussed, SCFAs are directly beneficial to the host; by promoting the growth of beneficial SCFA-producing bacteria, the growth of potential pathogens is also restricted through competitive exclusion and colonization resistance.
One challenge of prebiotics is that they can only promote the growth of bacteria that are present in the microbiome. This can be addressed by administering probiotics alongside prebiotics, often referred to as synbiotics. By providing the nutrients preferred by the probiotic, they are given an advantage over other bacteria within the GIT. The administration of a commercial synbiotic alongside Bifidobacterium longum significantly reduced mucosal inflammation compared to the probiotic alone [149]. Similar results were observed when a fiber synbiotic was combined with a five species Bacillus spp. bacterial consortium [150]. Identifying appropriate prebiotic and probiotic combinations could improve the efficacy of such treatments in a clinical setting.
In some cases, the restriction of bacterial nutrients is a more beneficial course of action. This requires dietary changes rather than the addition of a prebiotic. Diets low in fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAP) have been suggested to treat inflammatory bowel disorders. The microbial community is changed by removing the resources. It is the act of restricting the nutrient that causes the change [151]. However, the efficacy of this treatment is debated [152], and the suitability of this treatment may rely on the pre-existing microbial community [153].
6. Future Direction
While microbiome interventions are a promising area of study, there is still much to be learned in how they interact with the resident microbiome and the host. Introduction of a new species to a complex ecosystem can be challenging, as they are often unable to establish themselves within the preexisting community. It is also difficult to predict the effects this will have on the ecosystem and, consequentially, the host. A more complete knowledge of the interactions between species within the GIT, such as cross feeding and the production of antimicrobial compounds, will improve our ability to accurately predict the effects of microbiome interventions. Similarly, a better understanding of which species can utilize prebiotic compounds will help avoid the unintended proliferation of potential pathogens.
Microbial interventions have shown the most consistent results when the resident population has been disrupted, most often by medical interventions such as antibiotic use or chemotherapy. In these cases, the GIT is more easily colonized by the desired bacteria, and there are less unpredicted consequences due to interactions with the resident bacteria. But, even in disorder, the established microbiome is highly variable, and the circumstances of each individual patient must be taken into consideration when planning microbiome interventions. Ideally, each patient would have a fecal sample sequenced to identify the established microbiome and a customized plan created; however, this is unrealistic. Microbial interventions could be semi tailored through the identification of pre-existing conditions and a general understanding of the patient’s diet, allowing assumptions of the resident microbiome to be made.
7. Conclusions
The GIT microbiome influences the host immune system and can have serious effects on the systemic immune response and the overall health of the host. Recent work has highlighted the connection between the microbiome and intestinal, metabolic, cardiovascular, respiratory, and neurological conditions. While the GIT microbiome is a complex ecosystem, we can influence the composition of the community using fecal transplants, probiotics, prebiotics, and postbiotics. However, the same treatment may have opposing effects in different individuals because of the variable nature of the resident bacteria, the immune status of the host, and the nutrients available to the microbiome. Therefore, it is important that further research is conducted to improve our understanding of the interspecies relationships and the consequences of microbiome interventions under different conditions. The development of protocols for the creation of disorder-specific or personalized treatments will improve the efficacy of microbiome interventions.
Author Contributions
Conceptualization, Y.G. and B.M.; resources, Y.G.; writing—original draft preparation, B.M.; writing—review and editing, Y.G. and B.M.; visualization, B.M.; funding acquisition, Y.G. and B.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by a Grant-in-Aid for Challenging Research Exploratory (23K18285 to Y.G.) and JSPS Fellows from JSPS (23KF0133 to B.M., Y.G.); the Japan Agency for Medical Research and Development (AMED) Japan Initiative for World-leading Vaccine Research and Development Centers (JP223fa627003 to Y.G.); AMED-PRIME (JP19gm6010005 to Y.G.); JST FOREST Program (JPMJFR225D to Y.G.); Terumo Life Science Foundation (to Y.G.).
Data Availability Statement
No new data were created or analyzed in generating this review. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wong, C.B.; Odamaki, T.; Xiao, J.-Z. Insights into the reason of Human-Residential Bifidobacteria (HRB) being the natural inhabitants of the human gut and their potential health-promoting benefits. FEMS Microbiol. Rev. 2020, 44, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Hedblom, G.A.; Reiland, H.A.; Sylte, M.J.; Johnson, T.J.; Baumler, D.J. Segmented Filamentous Bacteria—Metabolism Meets Immunity. Front. Microbiol. 2018, 9, 1991. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Lin, Y.; Zhang, H.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Intestinal ‘Infant-Type’ Bifidobacteria Mediate Immune System Development in the First 1000 Days of Life. Nutrients 2022, 14, 1498. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Milani, C.; Duranti, S.; Ferrario, C.; Lugli, G.A.; Mancabelli, L.; van Sinderen, D.; Ventura, M. Bifidobacteria and the infant gut: An example of co-evolution and natural selection. Cell. Mol. Life Sci. 2018, 75, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zeng, X.; Zhang, G.; Hou, C.; Li, N.; Yu, H.; Shang, L.; Zhang, X.; Trevisi, P.; Yang, F.; et al. Maternal milk and fecal microbes guide the spatiotemporal development of mucosa-associated microbiota and barrier function in the porcine neonatal gut. BMC Biol. 2019, 17, 106. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zheng, Z.; Yu, L.; Wu, S.; Sun, L.; Wu, S.; Xu, Q.; Cai, S.; Qin, N.; Bao, W. Examination of the temporal and spatial dynamics of the gut microbiome in newborn piglets reveals distinct microbial communities in six intestinal segments. Sci. Rep. 2019, 9, 3453. [Google Scholar] [CrossRef] [PubMed]
- Leite, G.G.S.; Weitsman, S.; Parodi, G.; Celly, S.; Sedighi, R.; Sanchez, M.; Morales, W.; Villanueva-Millan, M.J.; Barlow, G.M.; Mathur, R.; et al. Mapping the Segmental Microbiomes in the Human Small Bowel in Comparison with Stool: A REIMAGINE Study. Dig. Dis. Sci. 2020, 65, 2595–2604. [Google Scholar] [CrossRef]
- Winters, A.D.; Romero, R.; Greenberg, J.M.; Galaz, J.; Shaffer, Z.D.; Garcia-Flores, V.; Kracht, D.J.; Gomez-Lopez, N.; Theis, K.R. Does the Amniotic Fluid of Mice Contain a Viable Microbiota? Front. Immunol. 2022, 13, 820366. [Google Scholar] [CrossRef]
- Blaser, M.J.; Devkota, S.; McCoy, K.D.; Relman, D.A.; Yassour, M.; Young, V.B. Lessons learned from the prenatal microbiome controversy. Microbiome 2021, 9, 8. [Google Scholar] [CrossRef]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef]
- Wiertsema, S.P.; van Bergenhenegouwen, J.; Garssen, J.; Knippels, L.M.J. The Interplay between the Gut Microbiome and the Immune System in the Context of Infectious Diseases throughout Life and the Role of Nutrition in Optimizing Treatment Strategies. Nutrients 2021, 13, 886. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Song, J.; Lv, Q.; Zhang, H.; Xiang, Q.; Dai, H.; Zheng, H.; Lin, X.; Zhang, W. Alterations in the Gut Microbiome of Young Children with Airway Allergic Disease Revealed by Next-Generation Sequencing. J. Asthma Allergy 2023, 16, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Tohumcu, E.; Raoul, P.; Fiorani, M.; Cintoni, M.; Mele, M.C.; Cammarota, G.; Gasbarrini, A.; Ianiro, G. The role of diet in shaping human gut microbiota. Best Pract. Res. Clin. Gastroenterol. 2023, 62–63, 101828. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, M.; Best, K.; Tang, C.; Liang, X.; Schulz, S.; Gonzalez, E.; White, C.H.; Wyche, T.P.; Kang, J.; Wesseling, H.; et al. Very early life microbiome and metabolome correlates with primary vaccination variability in children. Msystems 2023, 8, e0066123. [Google Scholar] [CrossRef]
- Moroishi, Y.; Gui, J.; Nadeau, K.C.; Morrison, H.G.; Madan, J.; Karagas, M.R. A prospective study of the infant gut microbiome in relation to vaccine response. Pediatr. Res. 2023, 93, 725–731. [Google Scholar] [CrossRef]
- Kumar, M.; James, M.M.; Kumawat, M.; Nabi, B.; Sharma, P.; Pal, N.; Shubham, S.; Tiwari, R.R.; Sarma, D.K.; Nagpal, R. Aging and Microbiome in the Modulation of Vaccine Efficacy. Biomedicines 2022, 10, 1545. [Google Scholar] [CrossRef]
- McCoy, K.D.; Burkhard, R.; Geuking, M.B. The microbiome and immune memory formation. Immunol. Cell Biol. 2019, 97, 625–635. [Google Scholar] [CrossRef]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef]
- Yin, X.; Altman, T.; Rutherford, E.; West, K.A.; Wu, Y.; Choi, J.; Beck, P.L.; Kaplan, G.G.; Dabbagh, K.; DeSantis, T.Z.; et al. A Comparative Evaluation of Tools to Predict Metabolite Profiles From Microbiome Sequencing Data. Front. Microbiol. 2020, 11, 595910. [Google Scholar] [CrossRef]
- Awany, D.; Allali, I.; Dalvie, S.; Hemmings, S.; Mwaikono, K.S.; Thomford, N.E.; Gomez, A.; Mulder, N.; Chimusa, E.R. Host and Microbiome Genome-Wide Association Studies: Current State and Challenges. Front. Genet. 2019, 10, 637. [Google Scholar] [CrossRef]
- Rühlemann, M.C.; Hermes, B.M.; Bang, C.; Doms, S.; Moitinho-Silva, L.; Thingholm, L.B.; Frost, F.; Degenhardt, F.; Wittig, M.; Kässens, J.; et al. Genome-wide association study in 8,956 German individuals identifies influence of ABO histo-blood groups on gut microbiome. Nat. Genet. 2021, 53, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Havulinna, A.S.; Liu, Y.; Jousilahti, P.; Ritchie, S.C.; Tokolyi, A.; Sanders, J.G.; Valsta, L.; Brożyńska, M.; Zhu, Q.; et al. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat. Genet. 2022, 54, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Garay, J.A.R.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Campana, A.M.; Laue, H.E.; Shen, Y.; Shrubsole, M.J.; Baccarelli, A.A. Assessing the role of the gut microbiome at the interface between environmental chemical exposures and human health: Current knowledge and challenges. Environ. Pollut. 2022, 315, 120380. [Google Scholar] [CrossRef]
- Singh, S.; Sharma, P.; Pal, N.; Kumawat, M.; Shubham, S.; Sarma, D.K.; Tiwari, R.R.; Kumar, M.; Nagpal, R. Impact of Environmental Pollutants on Gut Microbiome and Mental Health via the Gut–Brain Axis. Microorganisms 2022, 10, 1457. [Google Scholar] [CrossRef] [PubMed]
- Chiu, K.; Warner, G.; A Nowak, R.; A Flaws, J.; Mei, W. The Impact of Environmental Chemicals on the Gut Microbiome. Toxicol. Sci. 2020, 176, 253–284. [Google Scholar] [CrossRef]
- Salim, S.Y.; Kaplan, G.G.; Madsen, K.L. Air pollution effects on the gut microbiota: A link between exposure and inflammatory disease. Gut Microbes 2013, 5, 215–219. [Google Scholar] [CrossRef]
- Mutlu, E.A.; A Engen, P.; Soberanes, S.; Urich, D.; Forsyth, C.B.; Nigdelioglu, R.; E Chiarella, S.; A Radigan, K.; Gonzalez, A.; Jakate, S.; et al. Particulate matter air pollution causes oxidant-mediated increase in gut permeability in mice. Part. Fibre Toxicol. 2011, 8, 19. [Google Scholar] [CrossRef]
- Gacesa, R.; Kurilshikov, A.; Vila, A.V.; Sinha, T.; Klaassen, M.A.Y.; Bolte, L.A.; Andreu-Sánchez, S.; Chen, L.; Collij, V.; Hu, S.; et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature 2022, 604, 732–739. [Google Scholar] [CrossRef]
- Van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; De Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.W.M.; Tijssen, J.G.P.; et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef]
- Gupta, K.; Tappiti, M.; Nazir, A.M.; Koganti, B.; Memon, M.S.; Zahid, M.B.A.; Kumar, V.S.; A Mostafa, J. Fecal Microbiota Transplant in Recurrent Clostridium Difficile Infections: A Systematic Review. Cureus 2022, 14, e24754. [Google Scholar] [CrossRef] [PubMed]
- Nygren, E.; Li, B.-L.; Holmgren, J.; Attridge, S.R. Establishment of an Adult Mouse Model for Direct Evaluation of the Efficacy of Vaccines against Vibrio cholerae. Infect. Immun. 2009, 77, 3475–3484. [Google Scholar] [CrossRef] [PubMed]
- Sit, B.; Fakoya, B.; Waldor, M.K. Animal models for dissecting Vibrio cholerae intestinal pathogenesis and immunity. Curr. Opin. Microbiol. 2022, 65, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Liu, R.; Macbeth, J.C.; Hsiao, A. The Interface of Vibrio cholerae and the Gut Microbiome. Gut Microbes 2021, 13, 1937015. [Google Scholar] [CrossRef] [PubMed]
- Weil, A.A.; Becker, R.L.; Harris, J.B. Vibrio cholerae at the Intersection of Immunity and the Microbiome. mSphere 2019, 4, e00597-19:1–e00597-19:16. [Google Scholar] [CrossRef] [PubMed]
- Becattini, S.; Littmann, E.R.; Carter, R.A.; Kim, S.G.; Morjaria, S.M.; Ling, L.; Gyaltshen, Y.; Fontana, E.; Taur, Y.; Leiner, I.M.; et al. Commensal microbes provide first line defense against Listeria monocytogenes infection. J. Exp. Med. 2017, 214, 1973–1989. [Google Scholar] [CrossRef]
- Rogers, A.W.L.; Tsolis, R.M.; Bäumler, A.J. Salmonella versus the Microbiome. Microbiol. Mol. Biol. Rev. 2021, 85, e00027-19:1–e00027-19:30. [Google Scholar] [CrossRef]
- Buffie, C.G.; Pamer, E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013, 13, 790–801. [Google Scholar] [CrossRef]
- Kim, S.; Covington, A.; Pamer, E.G. The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens. Immunol. Rev. 2017, 279, 90–105. [Google Scholar] [CrossRef]
- Goodridge, H.S.; Ahmed, S.S.; Curtis, N.; Kollmann, T.R.; Levy, O.; Netea, M.G.; Pollard, A.J.; van Crevel, R.; Wilson, C.B. Harnessing the beneficial heterologous effects of vaccination. Nat. Rev. Immunol. 2016, 16, 392–400. [Google Scholar] [CrossRef]
- Rogala, A.R.; Oka, A.; Sartor, R.B. Strategies to Dissect Host-Microbial Immune Interactions That Determine Mucosal Homeostasis vs. Intestinal Inflammation in Gnotobiotic Mice. Front. Immunol. 2020, 11, 214. [Google Scholar] [CrossRef] [PubMed]
- Lamousé-Smith, E.S.; Tzeng, A.; Starnbach, M.N. The Intestinal Flora Is Required to Support Antibody Responses to Systemic Immunization in Infant and Germ Free Mice. PLoS ONE 2011, 6, e27662. [Google Scholar] [CrossRef] [PubMed]
- Bostick, J.W.; Schonhoff, A.M.; Mazmanian, S.K. Gut microbiome-mediated regulation of neuroinflammation. Curr. Opin. Immunol. 2022, 76, 102177. [Google Scholar] [CrossRef] [PubMed]
- Liwinski, T.; Zheng, D.; Elinav, E. The microbiome and cytosolic innate immune receptors. Immunol. Rev. 2020, 297, 207–224. [Google Scholar] [CrossRef]
- Gul, L.; Modos, D.; Fonseca, S.; Madgwick, M.; Thomas, J.P.; Sudhakar, P.; Booth, C.; Stentz, R.; Carding, S.R.; Korcsmaros, T. Extracellular vesicles produced by the human commensal gut bacterium Bacteroides thetaiotaomicron affect host immune pathways in a cell-type specific manner that are altered in inflammatory bowel disease. J. Extracell. Vesicles 2022, 11, e12189. [Google Scholar] [CrossRef]
- Sorbara, M.T.; Pamer, E.G. Microbiome-based therapeutics. Nat. Rev. Microbiol. 2022, 20, 365–380. [Google Scholar] [CrossRef]
- Rasouli-Saravani, A.; Jahankhani, K.; Moradi, S.; Gorgani, M.; Shafaghat, Z.; Mirsanei, Z.; Mehmandar, A.; Mirzaei, R. Role of microbiota short-chain fatty acids in the pathogenesis of autoimmune diseases. BioMedicine 2023, 162, 114620. [Google Scholar] [CrossRef]
- Tiwari, S.K. Bacteriocin-Producing Probiotic Lactic Acid Bacteria in Controlling Dysbiosis of the Gut Microbiota. Front. Cell. Infect. Microbiol. 2022, 12, 851140. [Google Scholar] [CrossRef]
- Corr, S.C.; Li, Y.; Riedel, C.U.; O’Toole, P.W.; Hill, C.; Gahan, C.G.M. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. USA 2007, 104, 7617–7621. [Google Scholar] [CrossRef]
- Sun, Y.; Wilkinson, B.J.; Standiford, T.J.; Akinbi, H.T.; O’Riordan, M.X.D. Fatty Acids Regulate Stress Resistance and Virulence Factor Production for Listeria monocytogenes. J. Bacteriol. 2012, 194, 5274–5284. [Google Scholar] [CrossRef]
- Costa, G.T.; Vasconcelos, Q.D.J.S.; Aragão, G.F. Fructooligosaccharides on inflammation, immunomodulation, oxidative stress, and gut immune response: A systematic review. Nutr. Rev. 2022, 80, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Wang, H.; Ji, L.; Song, P.; Ma, X. Impacts of Fructose on Intestinal Barrier Function, Inflammation and Microbiota in a Piglet Model. Nutrients 2021, 13, 3515. [Google Scholar] [CrossRef]
- Ducarmon, Q.R.; Zwittink, R.D.; Hornung, B.V.H.; Van Schaik, W.; Young, V.B.; Kuijper, E.J. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 2019, 83, e00007-19. [Google Scholar] [CrossRef]
- Herzog, M.K.-M.; Cazzaniga, M.; Peters, A.; Shayya, N.; Beldi, L.; Hapfelmeier, S.; Heimesaat, M.M.; Bereswill, S.; Frankel, G.; Gahan, C.G.; et al. Mouse models for bacterial enteropathogen infections: Insights into the role of colonization resistance. Gut Microbes 2023, 15, 2172667. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.; Littman, D.R. Segmented filamentous bacteria take the stage. Mucosal Immunol. 2010, 3, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Tannock, G.W.; Miller, J.R.; Savage, D.C. Host specificity of filamentous, segmented microorganisms adherent to the small bowel epithelium in mice and rats. Appl. Environ. Microbiol. 1984, 47, 441–442. [Google Scholar] [CrossRef]
- Li, F.; Ke, H.; Wang, S.; Mao, W.; Fu, C.; Chen, X.; Fu, Q.; Qin, X.; Huang, Y.; Li, B.; et al. Leaky Gut Plays a Critical Role in the Pathophysiology of Autism in Mice by Activating the Lipopolysaccharide-Mediated Toll-Like Receptor 4–Myeloid Differentiation Factor 88–Nuclear Factor Kappa B Signaling Pathway. Neurosci. Bull. 2023, 39, 911–928. [Google Scholar] [CrossRef]
- Kociszewska, D.; Vlajkovic, S.M. The Association of Inflammatory Gut Diseases with Neuroinflammatory and Auditory Disorders. Front. Biosci. 2022, 14, 8. [Google Scholar] [CrossRef]
- Sadagopan, A.; Mahmoud, A.; Begg, M.; Tarhuni, M.; A Gonzalez, N.; Sanivarapu, R.R.; Osman, U.; Kumar, A.L.; Mohammed, L.; Gonzalez, N.A.; et al. Understanding the Role of the Gut Microbiome in Diabetes and Therapeutics Targeting Leaky Gut: A Systematic Review. Cureus 2023, 15, e41559. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Zhao, W.; Hu, Y.; Li, C.; Li, N.; Zhu, S.; Tan, X.; Li, M.; Zhang, Y.; Xu, Z.; Ding, Z.; et al. Transplantation of fecal microbiota from patients with alcoholism induces anxiety/depression behaviors and decreases brain mGluR1/PKC ε levels in mouse. BioFactors 2020, 46, 38–54. [Google Scholar] [CrossRef]
- Hamada, H.; Hiroi, T.; Nishiyama, Y.; Takahashi, H.; Masunaga, Y.; Hachimura, S.; Kaminogawa, S.; Takahashi-Iwanaga, H.; Iwanaga, T.; Kiyono, H.; et al. Identification of Multiple Isolated Lymphoid Follicles on the Antimesenteric Wall of the Mouse Small Intestine. J. Immunol. 2002, 168, 57–64. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Hunziker, L.; McCoy, K.; Lamarre, A. IgA responses in the intestinal mucosa against pathogenic and non-pathogenic microorganisms. Microbes Infect. 2001, 3, 1021–1035. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E.; Santiago-Mateo, K.; Wertz, N.; Sun, X.; Sinkora, M.; Francis, D.L. Antibody repertoire development in fetal and neonatal piglets. XXIV. Hypothesis: The ileal Peyer patches (IPP) are the major source of primary, undiversified IgA antibodies in newborn piglets. Dev. Comp. Immunol. 2016, 65, 340–351. [Google Scholar] [CrossRef] [PubMed]
- McDole, J.R.; Wheeler, L.W.; McDonald, K.G.; Wang, B.; Konjufca, V.; Knoop, K.A.; Newberry, R.D.; Miller, M.J. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 2012, 483, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Knoop, K.; McDonald, K.; McCrate, S.; McDole, J.; Newberry, R. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 2015, 8, 198–210. [Google Scholar] [CrossRef]
- Qiu, X.; Zhang, M.; Yang, X.; Hong, N.; Yu, C. Faecalibacterium prausnitzii upregulates regulatory T cells and anti-inflammatory cytokines in treating TNBS-induced colitis. J. Crohn’s Colitis 2013, 7, e558–e568. [Google Scholar] [CrossRef]
- Lee, Y.; Awasthi, A.; Yosef, N.; Quintana, F.J.; Xiao, S.; Peters, A.; Wu, C.; Kleinewietfeld, M.; Kunder, S.; A Hafler, D.; et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 2012, 13, 991–999. [Google Scholar] [CrossRef]
- Neziraj, T.; Siewert, L.; Pössnecker, E.; Pröbstel, A. Therapeutic targeting of gut-originating regulatory B cells in neuroinflammatory diseases. Eur. J. Immunol. 2023, e2250033. [Google Scholar] [CrossRef]
- Moosavi, S.M.; Sepahi, A.A.; Mousavi, S.F.; Vaziri, F.; Siadat, S.D. The effect of Faecalibacterium prausnitzii and its extracellular vesicles on the permeability of intestinal epithelial cells and expression of PPARs and ANGPTL4 in the Caco-2 cell culture model. J. Diabetes Metab. Disord. 2020, 19, 1061–1069. [Google Scholar] [CrossRef]
- de Castro, M.J.; Pardo-Seco, J.J.; Martinón-Torres, F. Nonspecific (Heterologous) Protection of Neonatal BCG Vaccination Against Hospitalization Due to Respiratory Infection and Sepsis. Clin. Infect. Dis. 2015, 60, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Arts, R.J.; Carvalho, A.; La Rocca, C.; Palma, C.; Rodrigues, F.; Silvestre, R.; Kleinnijenhuis, J.; Lachmandas, E.; Gonçalves, L.G.; Belinha, A.; et al. Immunometabolic Pathways in BCG-Induced Trained Immunity. Cell Rep. 2016, 17, 2562–2571. [Google Scholar] [CrossRef]
- Mitroulis, I.; Ruppova, K.; Wang, B.; Chen, L.S.; Grzybek, M.; Grinenko, T.; Eugster, A.; Troullinaki, M.; Palladini, A.; Kourtzelis, I.; et al. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell 2018, 172, 147–161.e12. [Google Scholar] [CrossRef] [PubMed]
- Rabiei, N.; Badi, S.A.; Marvasti, F.E.; Sattari, T.N.; Vaziri, F.; Siadat, S.D. Induction effects of Faecalibacterium prausnitzii and its extracellular vesicles on toll-like receptor signaling pathway gene expression and cytokine level in human intestinal epithelial cells. Cytokine 2019, 121, 154718. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Rios-Covian, D.; Huillet, E.; Auger, S.; Khazaal, S.; Bermúdez-Humarán, L.G.; Sokol, H.; Chatel, J.-M.; Langella, P. Faecalibacterium: A bacterial genus with promising human health applications. FEMS Microbiol. Rev. 2023, 47, fuad039. [Google Scholar] [CrossRef]
- Chelakkot, C.; Choi, Y.; Kim, D.-K.; Park, H.T.; Ghim, J.; Kwon, Y.; Jeon, J.; Kim, M.-S.; Jee, Y.-K.; Gho, Y.S.; et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018, 50, e450. [Google Scholar] [CrossRef]
- Molina-Tijeras, J.A.; Gálvez, J.; Rodríguez-Cabezas, M.E. The Immunomodulatory Properties of Extracellular Vesicles Derived from Probiotics: A Novel Approach for the Management of Gastrointestinal Diseases. Nutrients 2019, 11, 1038. [Google Scholar] [CrossRef]
- Hori, T.; Matsuda, K.; Oishi, K. Probiotics: A Dietary Factor to Modulate the Gut Microbiome, Host Immune System, and Gut–Brain Interaction. Microorganisms 2020, 8, 1401. [Google Scholar] [CrossRef]
- Bajaj, B.K.; Claes, I.J.; Lebeer, S. Functional mechanisms of probiotics. J. Microbiol. Biotechnol. Food Sci. 2015, 4, 321–327. [Google Scholar] [CrossRef]
- Luu, M.; Pautz, S.; Kohl, V.; Singh, R.; Romero, R.; Lucas, S.; Hofmann, J.; Raifer, H.; Vachharajani, N.; Carrascosa, L.C.; et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun. 2019, 10, 760. [Google Scholar] [CrossRef]
- Kimura, I.; Inoue, D.; Maeda, T.; Hara, T.; Ichimura, A.; Miyauchi, S.; Kobayashi, M.; Hirasawa, A.; Tsujimoto, G. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc. Natl. Acad. Sci. USA 2011, 108, 8030–8035. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Vitetta, L. The Role of Butyrate in Attenuating Pathobiont-Induced Hyperinflammation. Immune Netw. 2020, 20, e15. [Google Scholar] [CrossRef] [PubMed]
- Effendi, R.M.R.A.; Anshory, M.; Kalim, H.; Dwiyana, R.F.; Suwarsa, O.; Pardo, L.M.; Nijsten, T.E.C.; Thio, H.B. Akkermansia muciniphila and Faecalibacterium prausnitzii in Immune-Related Diseases. Microorganisms 2022, 10, 2382. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Jung, D.-H.; Park, C.-S. Resistant starch utilization by Bifidobacterium, the beneficial human gut bacteria. Food Sci. Biotechnol. 2023, 32, 441–452. [Google Scholar] [CrossRef]
- Jacobson, A.; Lam, L.; Rajendram, M.; Tamburini, F.; Honeycutt, J.; Pham, T.; Van Treuren, W.; Pruss, K.; Stabler, S.R.; Lugo, K.; et al. A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe 2018, 24, 296–307.e7. [Google Scholar] [CrossRef]
- Pensinger, D.A.; Fisher, A.T.; Dobrila, H.A.; Van Treuren, W.; Gardner, J.O.; Higginbottom, S.K.; Carter, M.M.; Schumann, B.; Bertozzi, C.R.; Anikst, V.; et al. Butyrate Differentiates Permissiveness to Clostridioides difficile Infection and Influences Growth of Diverse C. difficile Isolates. Infect. Immun. 2023, 91, e0057022. [Google Scholar] [CrossRef]
- Wang, C.; Wei, S.; Liu, B.; Wang, F.; Lu, Z.; Jin, M.; Wang, Y. Maternal consumption of a fermented diet protects offspring against intestinal inflammation by regulating the gut microbiota. Gut Microbes 2022, 14, 2057779. [Google Scholar] [CrossRef]
- Un-Nisa, A.; Khan, A.; Zakria, M.; Siraj, S.; Ullah, S.; Tipu, M.K.; Ikram, M.; Kim, M.O. Updates on the Role of Probiotics against Different Health Issues: Focus on Lactobacillus. Int. J. Mol. Sci. 2023, 24, 142. [Google Scholar] [CrossRef]
- Chudzik, A.; Orzyłowska, A.; Rola, R.; Stanisz, G.J. Probiotics, Prebiotics and Postbiotics on Mitigation of Depression Symptoms: Modulation of the Brain–Gut–Microbiome Axis. Biomolecules 2021, 11, 1000. [Google Scholar] [CrossRef]
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017, 8, 1162. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Gaulke, C.A.; Sharpton, T.J. The Influence of Ethnicity and Geography on human Gut Microbiome Composition. Nat. Med. 2018, 24, 1492–1498. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.; Carding, S.R.; Hall, L.J. The early-life gut microbiome and vaccine efficacy. Lancet Microbe 2022, 3, e787–e794. [Google Scholar] [CrossRef] [PubMed]
- Vaiserman, A.; Romanenko, M.; Piven, L.; Moseiko, V.; Lushchak, O.; Kryzhanovska, N.; Guryanov, V.; Koliada, A. Differences in the gut Firmicutes to Bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol. 2020, 20, 221:1–221:8. [Google Scholar] [CrossRef]
- Houtman, T.A.; Eckermann, H.A.; Smidt, H.; de Weerth, C. Gut microbiota and BMI throughout childhood: The role of firmicutes, bacteroidetes, and short-chain fatty acid producers. Sci. Rep. 2022, 12, 3140. [Google Scholar] [CrossRef]
- Rahat-Rozenbloom, S.; Fernandes, J.; Gloor, G.B.; Wolever, T.M.S. Evidence for greater production of colonic short-chain fatty acids in overweight than lean humans. Int. J. Obes. 2014, 38, 1525–1531. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human Gut Microbes Associated with Obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Desselberger, U. The Mammalian Intestinal Microbiome: Composition, Interaction with the Immune System, Significance for Vaccine Efficacy, and Potential for Disease Therapy. Pathogens 2018, 7, 57. [Google Scholar] [CrossRef]
- Jamieson, A.M. Influence of the microbiome on response to vaccination. Hum. Vaccines Immunother. 2016, 11, 2329–2331. [Google Scholar] [CrossRef]
- Jurburg, S.D.; Bossers, A. Age Matters: Community Assembly in the Pig Fecal Microbiome in the First Month of Life. Front. Microbiol. 2021, 12, 564408. [Google Scholar] [CrossRef] [PubMed]
- La Torre, D.; Verbeke, K.; Dalile, B. Dietary fibre and the gut–brain axis: Microbiota-dependent and independent mechanisms of action. Gut Microbiome 2021, 2, e3:1–e3:39. [Google Scholar] [CrossRef]
- Mavrogeni, M.E.; Asadpoor, M.; Henricks, P.A.J.; Keshavarzian, A.; Folkerts, G.; Braber, S. Direct Action of Non-Digestible Oligosaccharides against a Leaky Gut. Nutrients 2022, 14, 4699. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pasolli, E.; Ercolini, D. Newly Explored Faecalibacterium Diversity Is Connected to Age, Lifestyle, Geography, and Disease. Curr. Biol. 2020, 30, 4932–4943. [Google Scholar] [CrossRef] [PubMed]
- Quévrain, E.; A Maubert, M.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermúdez-Humarán, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liang, R.; Zhang, W.; Tian, K.; Li, J.; Chen, X.; Yu, T.; Chen, Q. Faecalibacterium prausnitzii-derived microbial anti-inflammatory molecule regulates intestinal integrity in diabetes mellitus mice via modulating tight junction protein expression. J. Diabetes 2020, 12, 224–236. [Google Scholar] [CrossRef]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef]
- Chen, H.; Ou, R.; Tang, N.; Su, W.; Yang, R.; Yu, X.; Zhang, G.; Jiao, J.; Zhou, X. Alternation of the gut microbiota in irritable bowel syndrome: An integrated analysis based on multicenter amplicon sequencing data. J. Transl. Med. 2023, 21, 117:1–117:16. [Google Scholar] [CrossRef]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef]
- Lukovac, S.; Belzer, C.; Pellis, L.; Keijser, B.J.; de Vos, W.M.; Montijn, R.C.; Roeselers, G. Differential Modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of Host Peripheral Lipid Metabolism and Histone Acetylation in Mouse Gut Organoids. Mbio 2014, 5, e01438. [Google Scholar] [CrossRef]
- Wang, Z.; Qin, X.; Hu, D.; Huang, J.; Guo, E.; Xiao, R.; Li, W.; Sun, C.; Chen, G. Akkermansia supplementation reverses the tumor-promoting effect of the fecal microbiota transplantation in ovarian cancer. Cell Rep. 2022, 41, 111890. [Google Scholar] [CrossRef] [PubMed]
- Karcher, N.; Nigro, E.; Punčochář, M.; Blanco-Míguez, A.; Ciciani, M.; Manghi, P.; Zolfo, M.; Cumbo, F.; Manara, S.; Golzato, D.; et al. Genomic diversity and ecology of human-associated Akkermansia species in the gut microbiome revealed by extensive metagenomic assembly. Genome Biol. 2021, 22, 209. [Google Scholar] [CrossRef] [PubMed]
- Alavi, S.; Mitchell, J.D.; Cho, J.Y.; Liu, R.; Macbeth, J.C.; Hsiao, A. Interpersonal Gut Microbiome Variation Drives Susceptibility and Resistance to Cholera Infection. Cell 2020, 181, 1533–1546.e13. [Google Scholar] [CrossRef] [PubMed]
- Breen, P.; Winters, A.D.; Theis, K.R.; Withey, J.H. The Vibrio cholerae Type Six Secretion System Is Dispensable for Colonization but Affects Pathogenesis and the Structure of Zebrafish Intestinal Microbiome. Infect. Immun. 2021, 89, e00151. [Google Scholar] [CrossRef]
- Hsiao, A.; Zhu, J. Pathogenicity and virulence regulation of Vibrio cholerae at the interface of host-gut microbiome interactions. Virulence 2020, 11, 1582–1599. [Google Scholar] [CrossRef]
- Chen, H.; Yin, Y.; Wang, Y.; Wang, X.; Xiang, C. Host Specificity of Flagellins from Segmented Filamentous Bacteria Affects Their Patterns of Interaction with Mouse Ileal Mucosal Proteins. Appl. Environ. Microbiol. 2017, 83, e01061-17:1–e01061-17:16. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Uematsu, S.; Akira, S. Immune responses of TLR5+ lamina propria dendritic cells in enterobacterial infection. J. Gastroenterol. 2009, 44, 803–811. [Google Scholar] [CrossRef]
- Schnell, A.; Littman, D.R.; Kuchroo, V.K. TH17 cell heterogeneity and its role in tissue inflammation. Nat. Immunol. 2023, 24, 19–29. [Google Scholar] [CrossRef]
- Yi, J.; Jung, J.; Han, D.; Surh, C.D.; Lee, Y.J. Segmented Filamentous Bacteria Induce Divergent Populations of Antigen-Specific CD4 T Cells in the Small Intestine. Mol. Cells 2019, 42, 228–236. [Google Scholar] [CrossRef]
- Biazzo, M.; Deidda, G. Fecal Microbiota Transplantation as New Therapeutic Avenue for Human Diseases. J. Clin. Med. 2022, 11, 4119. [Google Scholar] [CrossRef]
- Mahmoudi, H.; Hossainpour, H. Application and Development of Fecal Microbiota Transplantation in the Treatment of Gastrointestinal and Metabolic Diseases: A Review. Saudi. J. Gastroenterol. 2023, 29, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Kanlioz, M.; Ekici, U.; Ferhatoğlu, M.F. Total Gastrointestinal Flora Transplantation in the Treatment of Leaky Gut Syndrome and Flora Loss. Cureus 2022, 14, e31071. [Google Scholar] [CrossRef] [PubMed]
- Danne, C.; Rolhion, N.; Sokol, H. Recipient factors in faecal microbiota transplantation: One stool does not fit all. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Taur, Y.; Coyte, K.; Schluter, J.; Robilotti, E.; Figueroa, C.; Gjonbalaj, M.; Littmann, E.R.; Ling, L.; Miller, L.; Gyaltshen, Y.; et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci. Transl. Med. 2018, 10, aap9489. [Google Scholar] [CrossRef] [PubMed]
- Ermolenko, E.; Kotyleva, M.; Kotrova, A.; Tichonov, S.; Lavrenova, N.; Voropaeva, L.; Topalova, Y.; Karaseva, A.; Azarov, D.; Ermolenko, K.; et al. Consortium of Indigenous Fecal Bacteria in the Treatment of Metabolic Syndrome. Microorganisms 2022, 10, 1574. [Google Scholar] [CrossRef]
- Tian, B.; Zhang, Y.; Deng, C.; Guo, C. Efficacy of Probiotic Consortium Transplantation on Experimental Necrotizing Enterocolitis. J. Surg. Res. 2022, 279, 598–610. [Google Scholar] [CrossRef]
- Malik, A.; Malik, M.I. Fecal Microbiota Transplantation in Human Immunodeficiency Virus-Infected Patient Population: A Systematic Review and Meta-Analysis. Gastroenterol. Res. 2023, 16, 209–216. [Google Scholar] [CrossRef]
- Li, Y.; Guo, B.; Wu, Z.; Wang, W.; Li, C.; Liu, G.; Cai, H. Effects of Fermented Soybean Meal Supplementation on the Growth Performance and Cecal Microbiota. Animals 2020, 10, 1098. [Google Scholar] [CrossRef]
- Ciric, A.; Radu, N.; Zaharie, M.G.O.; Neagu, G.; Pirvu, L.C.; Begea, M.; Stefaniu, A. Potential Antitumor Effect of Functional Yogurts Formulated with Prebiotics from Cereals and a Consortium of Probiotic Bacteria. Foods 2023, 12, 1250. [Google Scholar] [CrossRef]
- Tello, L.; Flores, L.; Usca, J.; Moreno, I. Lactobacillus and Its Probiotic Role in the Digestive and Nutritional Processes of Pigs: A Review. ESPOCH Congr. Ecuadorian J. STEAM 2021, 1, 1425–1439. [Google Scholar] [CrossRef]
- Lokapirnasari, W.P.; Pribadi, T.B.; Al Arif, A.; Soeharsono, S.; Hidanah, S.; Harijani, N.; Najwan, R.; Huda, K.; Wardhani, H.C.P.; Rahman, N.F.N.; et al. Potency of probiotics Bifidobacterium spp. and Lactobacillus casei to improve growth performance and business analysis in organic laying hens. Veter-World 2019, 12, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Ramlucken, U.; Lalloo, R.; Roets, Y.; Moonsamy, G.; van Rensburg, C.J.; Thantsha, M. Advantages of Bacillus-based probiotics in poultry production. Livest. Sci. 2020, 241, 104215. [Google Scholar] [CrossRef]
- Ritter, A.C.; Paula, A.; Correa, F.; Fonseca Veras, F.; Brandelli, A. Characterization of Bacillus Subtilis Available as Probiotics. J. Microbiol. Res. 2018, 8, 23–32. [Google Scholar] [CrossRef]
- Vazquez, A.P. Bacillus species are Superior Probiotic Feed-Additives for Poultry. J. Bacteriol. Mycol. Open Access 2016, 2, 23–25. [Google Scholar] [CrossRef]
- Hoa, N.T.; Baccigalupi, L.; Huxham, A.; Smertenko, A.; Van, P.H.; Ammendola, S.; Ricca, E.; Cutting, S.M. Characterization of Bacillus Species Used for Oral Bacteriotherapy and Bacterioprophylaxis of Gastrointestinal Disorders. Appl. Environ. Microbiol. 2000, 66, 5241–5247. [Google Scholar] [CrossRef]
- Cartman, S.T.; La Ragione, R.M.; Woodward, M.J. Bacillus subtilis Spores Germinate in the Chicken Gastrointestinal Tract. Appl. Environ. Microbiol. 2008, 74, 5254–5258. [Google Scholar] [CrossRef]
- Bernardeau, M.; Lehtinen, M.J.; Forssten, S.D.; Nurminen, P. Importance of the gastrointestinal life cycle of Bacillus for probiotic functionality. J. Food Sci. Technol. 2017, 54, 2570–2584. [Google Scholar] [CrossRef]
- Dumitru, M.; Hăbeanu, M.; Sorescu, I.; Tabuc, C. Effects of Bacillus spp. as a supplemental probiotic in diets for weaned piglets. South Afr. J. Anim. Sci. 2021, 51, 578–586. [Google Scholar] [CrossRef]
- Fritts, C.A.; Kersey, J.H.; Motl, M.A.; Kroger, E.C.; Yan, F.; Si, J.; Jiang, Q.; Campos, M.M.; Waldroup, A.L.; Waldroup, P.W. Bacillus subtilis C-3102 (Calsporin) Improves Live Performance and Microbiological Status of Broiler Chickens. J. Appl. Poult. Res. 2000, 9, 149–155. [Google Scholar] [CrossRef]
- Maruta, K.; Miyazaki, H.; Masuda, S.; Takahashi, M.; Marubashi, T.; Tadano, Y.; Takahashi, H. Exclusion of Intestinal Pathogens by Continuous Feeding with Bacillus subtilis C-3102 and its Influence on the Intestinal Microflora in Broilers. Animal Sci. Technol. 1996, 67, 273–280. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, S.; Di, H.; Deng, Z.; Liu, J.; Wang, H. Gut health benefit and application of postbiotics in animal production. J. Anim. Sci. Biotechnol. 2022, 13, 38. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.N.; Kogut, M.H.; Genovese, K.; He, H.; Kazemi, S.; Arsenault, R.J. Administration of a Postbiotic Causes Immunomodulatory Responses in Broiler Gut and Reduces Disease Pathogenesis Following Challenge. Microorganisms 2019, 7, 268. [Google Scholar] [CrossRef]
- Biswas, I.; Das Mohapatra, P.K. Recent advancement in metabiotics: A consortium with bioactive molecules after fermentation by probiotic bacteria with multidisciplinary application potential and future solution in health sector. Bioresour. Technol. Rep. 2023, 23, 101583. [Google Scholar] [CrossRef]
- Elayaraja, S.; Annamalai, N.; Mayavu, P.; Balasubramanian, T. Production, purification and characterization of bacteriocin from Lactobacillus murinus AU06 and its broad antibacterial spectrum. Asian Pac. J. Trop. Biomed. 2014, 4, S305–S311. [Google Scholar] [CrossRef]
- Garsa, A.K.; Kumariya, R.; Sood, S.K.; Kumar, A.; Kapila, S. Bacteriocin Production and Different Strategies for Their Recovery and Purification. Probiotics Antimicrob. Proteins 2014, 6, 47–58. [Google Scholar] [CrossRef]
- Stern, N.J.; Svetoch, E.A.; Eruslanov, B.V.; Kovalev, Y.N.; Volodina, L.I.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Levchuk, V.P. Paenibacillus polymyxa Purified Bacteriocin To Control Campylobacter jejuni in Chickens. J. Food Prot. 2005, 68, 1450–1453. [Google Scholar] [CrossRef]
- Pascale, N.; Gu, F.; Larsen, N.; Jespersen, L.; Respondek, F. The Potential of Pectins to Modulate the Human Gut Microbiota Evaluated by In Vitro Fermentation: A Systematic Review. Nutrients 2022, 14, 3629. [Google Scholar] [CrossRef]
- Furrie, E.; Macfarlane, S.; Kennedy, A.; Cummings, J.H.; Walsh, S.V.; O’neil, D.A.; Macfarlane, G.T. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: A randomised controlled pilot trial. Gut 2005, 54, 242–249. [Google Scholar] [CrossRef]
- Duysburgh, C.; Van den Abbeele, P.; Krishnan, K.; Bayne, T.F.; Marzorati, M. A synbiotic concept containing spore-forming Bacillus strains and a prebiotic fiber blend consistently enhanced metabolic activity by modulation of the gut microbiome in vitro. Int. J. Pharm. X 2019, 1, 100021. [Google Scholar] [CrossRef]
- Barrett, J.S. How to Institute the Low-FODMAP Diet. J. Gastroenter. Hepatol. 2017, 32, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Black, C.J.; Staudacher, H.M.; Ford, A.C. Efficacy of a low FODMAP diet in irritable bowel syndrome: Systematic review and network meta-analysis. Gut 2021, 71, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Vervier, K.; Moss, S.; Kumar, N.; Adoum, A.; Barne, M.; Browne, H.; Kaser, A.; Kiely, C.J.; Neville, B.A.; Powell, N.; et al. Two microbiota subtypes identified in irritable bowel syndrome with distinct responses to the low FODMAP diet. Gut 2023, 71, 1821–1830. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).