Next-Generation Sequencing for Evaluating the Soil Nematode Diversity and Its Role in Composting Processes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Compost Properties
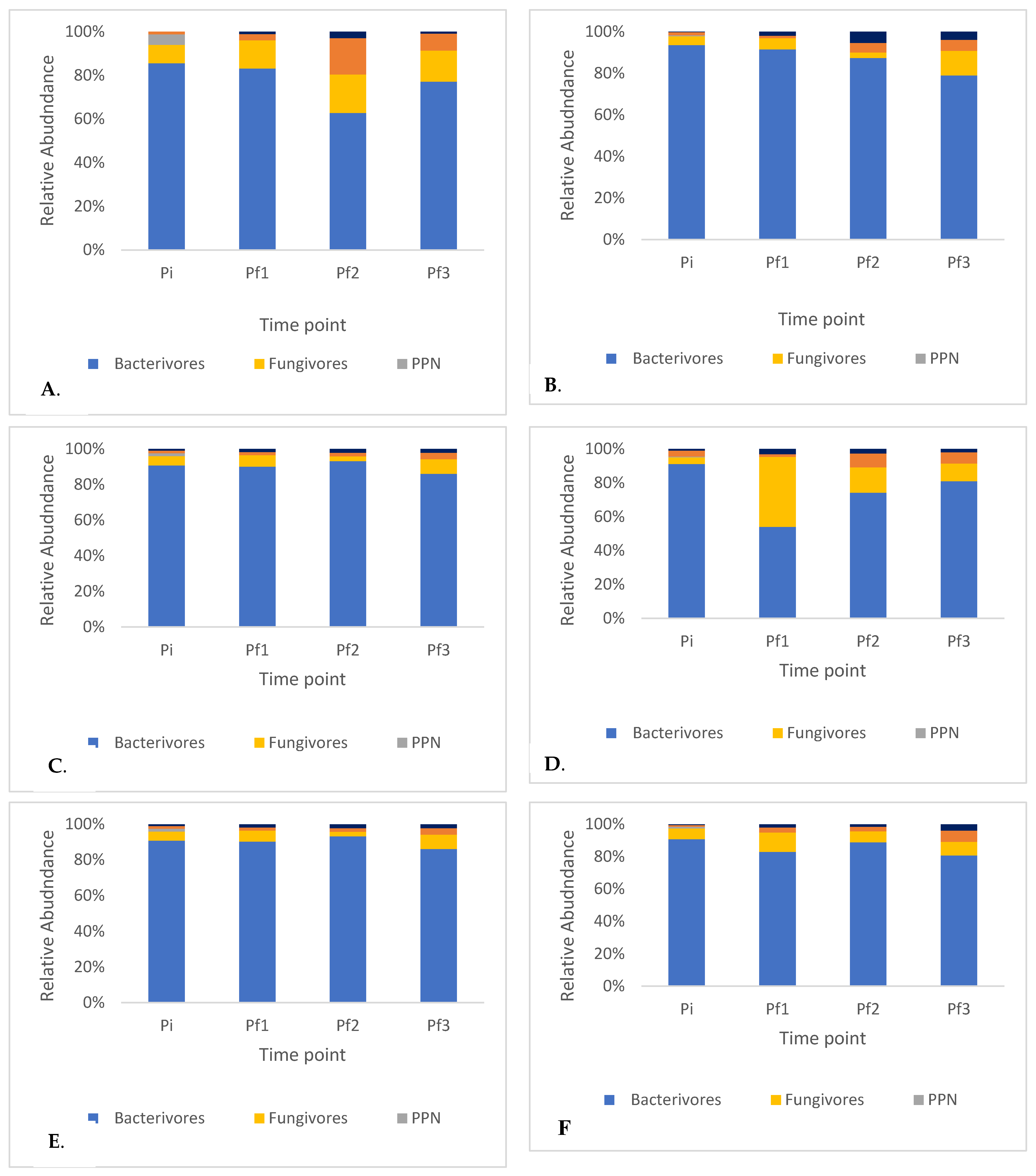
2.2. Nematode Succession
3. Materials and Methods
3.1. Study Site and Compost Preparation Procedure
- C1.
- Sewage sludge (80%) + Sawdust (20%);
- C2.
- Sewage sludge (40%) + Sawdust (10%) + Biodegradable garden and park waste (50%);
- C3.
- Biodegradable garden and park waste (90%) + Sawdust (10%);
- C4.
- Sewage sludge (80%) + Sawdust (20%) + Eisenia fetida;
- C5.
- Sewage sludge (40%) + Sawdust10%) + Biodegradable garden and park waste (50%) + Eisenia fetida;
- C6.
- Biodegradable garden and park waste (90%) + Sawdust (10%) + Eisenia fetida.
3.2. Chemical Analysis of Compost
3.3. Morphological Identification of Nematodes
3.4. DNA Extraction
3.5. Libraries Preparation
3.6. Processing and Analysis of Sequencing Data
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tognetti, C.; Mazzarino, M.; Laos, F. Compost of municipal organic waste: Effects of different management practices on degradability and nutrient release capacity. Soil Biol. Biochem. 2008, 40, 2290–2296. [Google Scholar] [CrossRef]
- Oka, Y. Mechanisms of nematode suppression by organic soil amendments—A review. Appl. Soil Ecol. 2010, 44, 101–115. [Google Scholar] [CrossRef]
- Mehta, C.M.; Palni, U.; Franke-Whittle, I.H.; Sharma, A.K. Compost: Its role, mechanism and impact on reducing soil-borne plant diseases. Waste Manag. 2014, 34, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Yeates, G.W.; Bongers, T.; De Goede, R.G.; Freckman, D.W.; Georgieva, S. Feeding habits in soil nematode families and gene-ra-An outline for soil ecologists. J. Nematol. 1993, 25, 315–331. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2619405/pdf/315.pdf (accessed on 30 September 2023). [PubMed]
- Steel, H.; Moens, T.; Vandecasteele, B.; Hendrickx, F.; De Neve, S.; Neher, D.; Bert, W. Factors influencing the nematode community during composting and nematode-based criteria for compost maturity. Ecol. Indic. 2018, 85, 409–421. [Google Scholar] [CrossRef]
- Steel, H.; de la Peña, E.; Fonderie, P.; Willekens, K.; Borgonie, G.; Bert, W. Nematode succession during composting and the potential of the nematode community as an indicator of compost maturity. Pedobiologia 2010, 53, 181–190. [Google Scholar] [CrossRef]
- Sapkota, R.; Nicolaisen, M. High-throughput sequencing of nematode communities from total soil DNA extractions. BMC Ecol. 2015, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Bogale, M.; Baniya, A.; DiGennaro, P. Nematode Identification Techniques and Recent Advances. Plants 2020, 9, 1260. [Google Scholar] [CrossRef]
- Nisa, R.U.; Tantray, A.Y.; Shah, A.A. Shift from morphological to recent advanced molecular approaches for the identification of nematodes. Genomics 2022, 114, 110295. [Google Scholar] [CrossRef]
- Treonis, A.M.; Unangst, S.K.; Kepler, R.M.; Buyer, J.S.; Cavigelli, M.A.; Mirsky, S.B.; Maul, J.E. Characterization of soil nematode communities in three cropping systems through morphological and DNA metabarcoding approaches. Sci. Rep. 2018, 8, 2004. [Google Scholar] [CrossRef]
- Ahmed, M.; Back, M.A.; Prior, T.; Karssen, G.; Lawson, R.; Adams, I.; Sapp, M. Metabarcoding of soil nematodes: The importance of taxonomic coverage and availability of reference sequences in choosing suitable marker(s). Metabarcoding Metagenomics 2019, 3, 77–99. [Google Scholar] [CrossRef]
- Kawanobe, M.; Toyota, K.; Ritz, K. Development and application of a DNA metabarcoding method for comprehensive analysis of soil nematode communities. Appl. Soil Ecol. 2021, 166, 103974. [Google Scholar] [CrossRef]
- Porazinska, D.L.; Giblin-Davis, R.M.; Faller, L.; Farmerie, W.; Kanzaki, N.; Morris, K.; Powers, T.O.; Tucker, A.E.; Sung, W.; Thomas, W.K. Evaluating high-throughput sequencing as a method for metagenomic analysis of nematode diversity. Mol. Ecol. Resour. 2009, 9, 1439–1450. [Google Scholar] [CrossRef] [PubMed]
- Porazinska, D.L.; Sung, W.; Giblin-Davis, R.M.; Thomas, W.K. Reproducibility of read numbers in high-throughput sequencing analysis of nematode community composition and structure. Mol. Ecol. Resour. 2010, 10, 666–676. [Google Scholar] [CrossRef]
- Holovachov, O.; Haenel, Q.; Bourlat, S.J.; Jondelius, U. Taxonomy assignment approach determines the efficiency of identification of OTUs in marine nematodes. R. Soc. Open Sci. 2017, 4, 170315. [Google Scholar] [CrossRef]
- Steel, H.; Bert, W. Biodiversity of compost mesofauna and its potential as an indicator of the composting process status. Dy-Namic Soil Dyn. Plant. 2011, 2, 45–50. [Google Scholar]
- Steel, H.; Buchan, D.; De Neve, S.; Couvreur, M.; Moens, T.; Bert, W. Nematode and microbial communities in a rapidly changing compost environment: How nematode assemblages reflect composting phases. Eur. J. Soil Biol. 2013, 56, 1–10. [Google Scholar] [CrossRef]
- Wilski, A. Nicienie–Szkodniki Roślin Uprawnych. PWRiL. 1967. 336. Available online: https://scholar.google.com/scholar?q=Wilski%20A.%201967.%20Nicienie%20Szkodniki%20Ro%C5%9Blin%20Uprawnych.%20PWRiL,%20Warszawa,%20336%20pp (accessed on 30 September 2023).
- D’Addabbo, T.; Sasanelli, N.; Greco, N.; Stea, V.; Brandonisio, A. Effect of Water, Soil Temperatures, and Exposure Times on the Survival of the Sugar Beet Cyst Nematode, Heterodera Schachtii. Phytopathology 2005, 95, 339–344. [Google Scholar] [CrossRef]
- Tsai, B.Y. Effect of temperature on the survival of Meloidogyne incognito. Plant Pathol. Bull. 2008, 17, 203–208. Available online: https://www.taiwanphytopath.org/uploads/publication/3e446fc1dd1bad5877b435d63694b06b.pdf (accessed on 30 September 2023).
- Abbassi, B. Elimination of Nematodes from the Soil by Microwave an Dry Heat. University of Central Florida. Stars. Retro-Spective Theses and Dissertations 1979. Available online: https://stars.library.ucf.edu/cgi/viewcontent.cgi?referer=&httpsredir=1&article=1390&context=rtd (accessed on 30 September 2023).
- Veenman, H.; Zonen, N.V. Climatic Influence on the Distribution Pattern of Plant Parasitic and Soil Inhabiting Nematodes. Proefschrift Door Dederico Dao D., 1970. Available online: https://edepot.wur.nl/192442 (accessed on 30 September 2023).
- Sudhaus, W.; Lieven von, A.F. A phylogenetic classification of the Diplogastridae (Secernentea, Nematoda). J. Nema-Tode Morphol. Syst. 2003, 6, 43–90. [Google Scholar]
- Ahlawat, S.; Tahseen, Q. Description and developmental biology of the predatory diplogastrid Acrostichus nudicapitatus (Steiner, 1914) Massey, 1962 (Nematoda: Rhabditida). Helminthologia 2016, 53, 142–154. [Google Scholar] [CrossRef]
- Steel, H.; Vandecasteele, B.; Willekens, K.; Sabbe, K.; Moens, T.; Bert, W. Nematode communities and macronutrients in com-posts and compost-amended soils as affected by feedstock composition. Appl. Soil Ecol. 2012, 61, 100–112. [Google Scholar] [CrossRef]
- Borgonie, G.; Linage-Alvarez, B.; Ojo, A.O.; Mundle, S.; Freese, L.B.; Van Rooyen, C.; Kuloyo, O.; Albertyn, J.; Pohl, C.; Cason, E.D.; et al. Eukaryotic opportunists dominate the deep-subsurface biosphere in South Africa. Nat. Commun. 2015, 6, 8952. [Google Scholar] [CrossRef]
- Hermosilla, C.; Coumbe, K.M.; Habershon-Butcher, J.; Schöniger, S. Fatal equine meningoencephalitis in the United Kingdom caused by the panagrolaimid nematode Halicephalobus gingivalis: Case report and review of the literature. Equine Vet. J. 2011, 43, 759–763. [Google Scholar] [CrossRef] [PubMed]
- Onyiche, T.E.; Okute, T.O.; Oseni, O.S.; Okoro, D.O.; Biu, A.A.; Mbaya, A.W. Parasitic and zoonotic meningoencephalitis in humans and equids: Current knowledge and the role of Halicephalobus gingivalis. Parasite Epidemiol. Control. 2017, 3, 36–42. [Google Scholar] [CrossRef]
- Pillai, V.V.; Mudd, L.J.; Sola, M.F. Disseminated Halicephalobus gingivalis infection in a horse. J. Vet. Diagn. Investig. 2022, 35, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Act of 14 December 2012. On waste. Journal of Laws 2013, item 21. In polish: Dz. U. 2013 poz. 21—Ustawa o Odpadach z Dnia 14 Grudnia 2012. Available online: https://isap.sejm.gov.pl/isap.nsf/download.xsp/WDU20130000021/T/D20130021L.pdf (accessed on 30 July 2023).
- Regulation of the Minister of the Environment of 1 August 2002 on Municipal Sewage Sludge. J. Laws 2002, item. 1140. In polish: Dz. U. 2002 Nr 134 Poz. 1140—Rozporządzenie Ministra Środowiska z Dnia 1 Sierpnia 2002 r. w Sprawie Komunalnych Osadów Ściekowych. Available online: https://isap.sejm.gov.pl/isap.nsf/download.xsp/WDU20021341140/O/D20021140.pdf (accessed on 30 July 2023).
- Zapałowska, A.; Matłok, N.; Piechowiak, T.; Szostek, M.; Puchalski, C.; Balawejder, M. Physiological and Morphological Implications of Using Composts with Different Compositions in the Production of Cucumber Seedlings. Int. J. Mol. Sci. 2023, 24, 14400. [Google Scholar] [CrossRef]
- Zapałowska, A.; Skwiercz, A.T. Populations of parasitic nematodes colonizing Jerusalem artichoke (Helianthus tuberosus L.). Acta Soc. Bot. Pol. 2018, 87, 3578. [Google Scholar] [CrossRef]
- Chałańska, A.; Bogumił, A.; Malewski, T.; Kowalewska, K. The effect of two fixation methods (TAF and DESS) on morphometric parameters of Aphelenchoides ritzemabosi. Zootaxa 2016, 4083, 297–300. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Marcelino, V.R.; Clausen, P.T.L.C.; Buchmann, J.P.; Wille, M.; Iredell, J.R.; Meyer, W.; Lund, O.; Sorrell, T.C.; Holmes, E.C. CCMetagen: Comprehensive and accurate identification of eukaryotes and prokaryotes in metagenomic data. Genome Biol. 2021, 21, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 103. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; Szöke, S.; Cardinali, G.; Eberhardt, U.; Stielow, B.; de Vries, M.; Verkleij, G.J.M.; Crous, P.W.; Boekhout, T.; et al. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016, 85, 91–105. [Google Scholar] [CrossRef] [PubMed]


| Content | Compost Variant | Composting Duration (Days) | |||
|---|---|---|---|---|---|
| 0 | 30 | 60 | 90 | ||
| N (%) | C1 | 3.80 ± 0.09 | 3.2 ± 0.08 | 2.96 ± 0.23 | 4.02 ± 0.32 |
| C2 | 3.72 ± 0.08 | 1.67 ± 0.05 | 2.77 ± 0.22 | 1.82 ± 0.14 | |
| C3 | 1.38 ± 0.04 | 1.32 ± 0.10 | 1.11 ± 0.08 | 1.01 ± 0.08 | |
| C4 | 3.5 ± 0.07 | 3.13 ± 0.25 | 3.01 ± 0.24 | 2.59 ± 0.20 | |
| C5 | 3.54 ± 0.07 | 2.46 ± 0.19 | 2.34 ± 0.18 | 1.86 ± 0.14 | |
| C6 | 1.53 ± 0.03 | 1.47 ± 0.11 | 2.07 ± 0.16 | 1.56 ± 0.12 | |
| P2O5 (g·kg−1 s.m.) | C1 | 12.00 ± 0.96 | 10.82 ± 0.86 | 12.61 ± 1.00 | 13.00 ± 1.04 |
| C2 | 5.81 ± 0.46 | 8.33 ± 0.66 | 6.80 ± 0.54 | 1.09 ± 0.01 | |
| C3 | 1.93 ± 0.15 | 1.72 ± 0.13 | 2.06 ± 0.16 | 2.01 ± 0.16 | |
| C4 | 9.27 ± 0.74 | 8.88 ± 0.71 | 10.06 ± 0.80 | 9.42 ± 0.75 | |
| C5 | 10.22 ± 0.81 | 6.87 ± 0.53 | 7.58 ± 0.60 | 8.89 ± 0.71 | |
| C6 | 2.38 ± 0.19 | 2.76 ± 0.22 | 5,98 ± 0.47 | 3.64 ± 0.29 | |
| K2O5 (g·kg−1 s.m.) | C1 | 4.11 ± 0.39 | 5.23 ± 0.49 | 6.00 ± 0.49 | 4.93 ± 0.30 |
| C2 | 1.00 ± 0.09 | 10.08 ± 0.99 | 8.40 ± 0.69 | 6.88 ± 0.55 | |
| C3 | 11.76 ± 1.63 | 10.02 ± 0.97 | 5.82 ± 0.34 | 4.63 ± 0.37 | |
| C4 | 3.66 ± 0.25 | 4.91 ± 0.48 | 6.06 ± 0.50 | 5.09 ± 0.40 | |
| C5 | 4.10 ± 0.39 | 8.58 ± 0.81 | 9.18 ± 0.76 | 7.63 ± 0.61 | |
| C6 | 5.36 ± 0.49 | 10.08 ±0.10 | 10.62 ± 1.09 | 7.28 ± 0.58 | |
| C:N (%) | C1 | 19.81 ± 0.19 | 24.19 ± 0.30 | 26.14 ± 0.26 | 19.25 ± 0.91 |
| C2 | 12.79 ± 0.23 | 25.74 ± 0.73 | 12.74 ± 0.21 | 23.57 ± 0.86 | |
| C3 | 38.55 ± 0.15 | 35.07 ± 0.20 | 41.44 ± 0.19 | 37.72 ± 0.54 | |
| C4 | 22.8 ± 0.28 | 23.06 ±0.75 | 23.68 ± 0.27 | 28.06 ± 0.42 | |
| C5 | 15.16 ± 0.17 | 17.64 ± 0.18 | 21.02 ± 0.26 | 21.29 ± 0.38 | |
| C6 | 30.45 ± 0.34 | 32.38 ±0.27 | 23.91 ± 0.21 | 30.32 ± 0.19 | |
| Total Metal Concentration | Compost Variant | Composting Duration (Days) | |||
|---|---|---|---|---|---|
| 0 | 30 | 60 | 90 | ||
| Pb (mg∙kg−1 s.m) | C1 | 11.0 ± 0.88 | 15.0 ± 1.2 | 8.5 ± 0.68 | 10.0 ± 0.8 |
| C2 | 10.0 ± 0.80 | <8.0 | <8.0 | <8.0 | |
| C3 | <8.0 | 9.8 ± 0.78 | <8.0 | <8.0 | |
| C4 | <8.0 | <8.0 | <8.0 | <8.0 | |
| C5 | 8.20 ± 0.65 | <8.0 | <8.0 | 8.0 | |
| C6 | 6.0 ± 0.48 | <8.0 | <8.0 | <8.0 | |
| Chr (mg∙kg−1 s.m) | C1 | <10 | <10 | <10 | <10 |
| C2 | <10 | <10 | <10 | <10 | |
| C3 | <10 | <10 | <10 | 12.0 ± 0.96 | |
| C4 | 11.0 ± 0.91 | <10 | <10 | 12.0 ± 0.93 | |
| C5 | 11.0 ± 0.89 | <10 | <10 | 11.0 ± 0.90 | |
| C6 | 16.0 ± 1.28 | 12.0 ± 0.96 | 13.0 ± 1.04 | 23.0 ± 1.84 | |
| Cu (mg∙kg−1 s.m) | C1 | 77.0 ± 6.14 | 65.0 ± 5.2 | 55.0 ± 4.01 | 70.0 ± 5.6 |
| C2 | 31.0 ± 2.48 | 13.0 ± 1.04 | 39.0 ± 3.12 | 58.0 ± 4.64 | |
| C3 | 6.90 ± 0.52 | 9.2 ± 0.76 | 5.2 ± 0.41 | 16.0 ± 1.28 | |
| C4 | 88.0 ± 7.01 | 67.0 ± 5.36 | 81.0 ± 6.48 | 84.0 ± 6.72 | |
| C5 | 78 ± 6.01 | 55.0 ± 4,39 | 42.0 ± 3.36 | 45.0 ± 3.6 | |
| C6 | 9.2 ± 0.73 | 11.0 ± 0.88 | 14.0 ± 1.12 | 16.0 ± 1.28 | |
| Ni (mg∙kg−1 s.m) | C1 | 5.7 ± 0.45 | <5.0 | <5.0 | < 5.0 |
| C2 | <5.0 | <5.0 | <5.0 | < 5.0 | |
| C3 | <5.0 | <5.0 | <5.0 | 5.3 ± 0.42 | |
| C4 | 6.3 ± 0.50 | <5.0 | 5,7 ± | < 5.0 | |
| C5 | 6.8 ± 0.54 | 6.4 ± 0.51 | <5.0 | 5.1 ± 0.40 | |
| C6 | 11.0 ± 0.88 | 7.1 ± 0.65 | 7.7 ± 0.61 | 11.0 ± 0.88 | |
| Cd (mg∙kg−1 s.m) | C1 | 0.473 ± 0.03 | 0.467 ± 0.03 | 0.311 ± 0.02 | 0,44 ± 0.03 |
| C2 | 0.30 ± 0.02 | <0.3 | 0.32 ± 0.02 | 0.42 ± 0.03 | |
| C3 | <0.3 | <0.3 | <0.3 | 0.34 ± 0.03 | |
| C4 | 0.568 ± 0.04 | 0.428 ± 0.03 | 0.392 ± 0.03 | 0.5 ± 0.05 | |
| C5 | 0.456 ± 0.03 | 0.607 ± 0.04 | <0.3 | 0.38 ± 0.03 | |
| C6 | <0,3 | 0.371 ± 0.02 | <0.3 | 0.35 ± 0.03 | |
| Zn (mg∙kg−1 s.m) | C1 | 302 ± 24.1 | 225 ± 18.0 | 241 ± 18.9 | 253 ± 20,2 |
| C2 | 168 ± 13.4 | 54.0 ± 4.32 | 157 ± 12.1 | 223 ± 17.8 | |
| C3 | 42.0 ± 3.06 | 46.0 ± 3.68 | 28.0 ± 2.24 | 91.0 ± 7.28 | |
| C4 | 336 ± 25.9 | 288 ± 18.8 | 274 ± 21.9 | 274 ± 21.91 | |
| C5 | 313 ± 24.9 | 248 ± 19.0 | 147 ± 11.7 | 232 ± 18.56 | |
| C6 | 51.0 ± 4.08 | 57.0 ± 4.02 | 63 ± 5.25 | 78.0 ± 6.24 | |
| Hg (mg∙kg−1 s.m) | C1 | 0.243 ± 0.01 | 0.176 ± 0.01 | 0.133 ± 0.01 | 0.132 ± 0.01 |
| C2 | 0.057 ±0.01 | 0.039 ± 0.00 | 0.047 ± 0.00 | 0.104 ± 0.00 | |
| C3 | 0.035 ±0.00 | 0.032 ± 0.00 | 0.034 ± 0.00 | 0.026 ± 0.00 | |
| C4 | 0.134 ±0.01 | 0.168 ± 0.01 | 0.172 ± 0.01 | 0.18 ± 0.01 | |
| C5 | 0.134 ± 0.01 | 0.074 ± 0.00 | 0.076 ± 0.00 | 0.066 ± 0.00 | |
| C6 | 0.037 ± 0.00 | 0.041 ± 0.00 | 0.023 ± 0.00 | 0.042 ± 0.00 | |
| Species | Compost Variant | Composting Duration (Days) | ||
|---|---|---|---|---|
| 30 | 60 | 90 | ||
| Diplogastridae | C1 | <0.1 | <0.1 | 29.1 ± 5.23 |
| C2 | <0.1 | <0.1 | 27.7 ±± 4.03 | |
| C3 | <0.1 | 25.0 ± 3.19 | 23.2 ± 3.41 | |
| C4 | 28.0 ± 4.21 | 21.7 ± 3.19 | 37.2 ± 4.14 | |
| C5 | 28.0 ± 3.67 | 21.7 ± 4.15 | 36.1 ± 4.96 | |
| C6 | 11.0 ± 3.11 | 4.3 ± 1.48 | 21.8 ± 4.13 | |
| Acrostichus nudicapitatus | C1 | 0.1 ± 0.04 | <0.1 | <0.1 |
| C2 | <0.1 | <0.1 | <0.1 | |
| C3 | <0.1 | <0.1 | <0.1 | |
| C4 | <0.1 | <0.1 | 0.1 ± 0.09 | |
| C5 | <0.1 | <0.1 | 0.1 ± 0.08 | |
| C6 | <0.1 | 6.2 ± 1.42 | <0.1 | |
| Acrostichus sp. | C1 | 59.2 ± 5.11 | 41.2 ± 9.16 | 46.9 ± 4.31 |
| C2 | 72.9 ± 2.13 | 40.6 ± 2.41 | 49.9 ±4.06 | |
| C3 | 1.9 ± 0.47 | 40.1 ± 3.44 | 25.6 ± 3.24 | |
| C4 | 49.8 ± 6.52 | 37.6 ± 4.16 | 54.8 ± 6.32 | |
| C5 | 50.1 ± 7.32 | 37.9 ± 5.42 | 54.9 ± 7.38 | |
| C6 | 10.7 ± 2.02 | 25.4 ± 4.62 | 27.9 ± 3.43 | |
| Acrostichus floridensis | C1 | <0.1 | <0.1 | <0.1 |
| C2 | 4.3 ± 0.27 | 16.9 ± 1.55 | 10.3 ±0.21 | |
| C3 | 16.9 ± 2.72 | 10.4 ± 2.09 | 30.5 ± 4.68 | |
| C4 | <0.1 | 2.2 ± 0.33 | 0.3 ± 0.11 | |
| C5 | <0.1 | 2.1 ± 0.41 | 0.2 ± 0.04 | |
| C6 | <0.1 | 5.9 ± 1.22 | 2.1 ± 0.61 | |
| Mononchoides sp. | C1 | 0.1 ± 0.02 | 32.3 ± 0.58 | 20.1 ± 3.81 |
| C2 | <0.1 | 1.1 ± 0.21 | <0.1 | |
| C3 | <0.1 | 1.1 ± 0.21 | <0.1 | |
| C4 | 0.5 ± 0.21 | 31.3 ± 3.17 | 1.9 ± 0.08 | |
| C5 | 0.5 ± 0.16 | 31.3 ± 4.67 | 1.9 ± 0.33 | |
| C6 | 1.3 ± 0.46 | 14.0 ± 2.61 | 2.0 ± 0.36 | |
| Ektaphelenchus sp. | C1 | <0.1 | <0.1 | <0.1 |
| C2 | <0.1 | <0.1 | <0.1 | |
| C3 | <0.1 | 1.3 ± 0.62 | <0.1 | |
| C4 | <0.1 | <0.1 | 0.1 ± 0.04 | |
| C5 | <0.1 | <0.1 | 0.1 ± 0.05 | |
| C6 | <0.1 | <0.1 | <0.1 | |
| Halicephalobus sp. | C1 | 0.1 ± 0.06 | <0.1 | <0.1 |
| C2 | <0.1 | <0.1 | <0.1 | |
| C3 | <0.1 | <0.1 | <0.1 | |
| C4 | <0.1 | <0.1 | <0.1 | |
| C5 | <0.1 | <0.1 | <0.1 | |
| C6 | 0.3 ± 0.11 | <0.1 | 6.9 ± 0.94 | |
| Halicephalobus gingivalis | C1 | 7.9 ± 1.35 | 1.9 ± 0.42 | <0.1 |
| C2 | 3.9 ± 0.16 | 5.9 ± 1,64 | <0.1 | |
| C3 | 16.4 ± 3.91 | 1.2 ± 0.31 | 6.4 ± 1.16 | |
| C4 | 3.5 ± 0.52 | 0.1 ± 0.07 | <0.1 | |
| C5 | 3.4 ± 1.22 | 0.1 ± 0.04 | <0.1 | |
| C6 | 16.8 ± 2.02 | 6.4 ± 1.41 | <0.1 | |
| Halicephalobus cf. gingivalis | C1 | 30.3 ± 2.21 | 8.9 ± 1.06 | <0.1 |
| C2 | 16.7 ± 1.42 | 23.5 ± 2.46 | <0.1 | |
| C3 | 60.9 ± 3.47 | 1.3 ± 0.43 | 1.9 ± 0.43 | |
| C4 | 14.9 ± 2.11 | 1.2 ± 0.11 | <0.1 | |
| C5 | 15.0 ± 2.47 | 1.1 ± 0.32 | <0.1 | |
| C6 | 52.8 ± 7.43 | 24.1 ± 3.68 | 2.1 ± 0.56 | |
| Oscheius onirici | C1 | <0.1 | <0.1 | <0.1 |
| C2 | <0.1 | <0.1 | <0.1 | |
| C3 | <0.1 | <0.1 | <0.1 | |
| C4 | 0.8 ± 0.21 | <0.1 | 1.2 ± 0.70 | |
| C5 | 0.9 ± 0.16 | <0.1 | 0.9 ± 0.11 | |
| C6 | 0.1 ± 0.04 | <0.1 | <0.1 | |
| Panagrellus redivivus | C1 | <0.1 | <0.1 | <0.1 |
| C2 | <0.1 | 0.2 ± 0.12 | 0.1 ± 0.04 | |
| C3 | <0.1 | <0.1 | 0.2 ± 0.15 | |
| C4 | <0.1 | <0.1 | <0.1 | |
| C5 | <0.1 | <0.1 | <0.1 | |
| C6 | <0.1 | 1.0 ± 0.04 | 0.6 ± 0.04 | |
| Nematoda environmental sample | C1 | 0.1 ± 0.02 | 10.7 ± 2.51 | 1.3 ± 0.32 |
| C2 | 0.1 ± 0.12 | 8.1 ± 0.51 | 9.9 ± 2.49 | |
| C3 | 0.3 ± 0.28 | 1.5 ± 0.32 | <0.1 | |
| C4 | 0.4 ± 0.16 | 2.9 ± 0.85 | 3.4 ± 0.79 | |
| C5 | 0.3 ± 0.07 | 2.8 ± 0.81 | 3.2 ± 0.17 | |
| C6 | 1.4 ± 0.11 | 1.3± 0.12 | 2.6 ± 0.15 | |
| Other nematoda | C1 | 1.2 ± 0.06 | 1.4 ± 0.49 | 2.3 ± 0.79 |
| C2 | 0.7 ± 0.19 | 2.6 ± 0.19 | 1.6 ±0.12 | |
| C3 | 1.4 ± 0.72 | 1.3 ± 0.69 | 1.1 ± 0.17 | |
| C4 | 1.3 ± 0.41 | 1.5 ±0.13 | 2.9 ± 0.39 | |
| C5 | 1.5 ± 0.15 | 1.6 ± 0.48 | 3.1 ± 0.24 | |
| C6 | 2.1 ± 0.16 | 1.7 ± 0.14 | 3.6 ± 0.12 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zapałowska, A.; Skwiercz, A.; Tereba, A.; Puchalski, C.; Malewski, T. Next-Generation Sequencing for Evaluating the Soil Nematode Diversity and Its Role in Composting Processes. Int. J. Mol. Sci. 2023, 24, 15749. https://doi.org/10.3390/ijms242115749
Zapałowska A, Skwiercz A, Tereba A, Puchalski C, Malewski T. Next-Generation Sequencing for Evaluating the Soil Nematode Diversity and Its Role in Composting Processes. International Journal of Molecular Sciences. 2023; 24(21):15749. https://doi.org/10.3390/ijms242115749
Chicago/Turabian StyleZapałowska, Anita, Andrzej Skwiercz, Anna Tereba, Czesław Puchalski, and Tadeusz Malewski. 2023. "Next-Generation Sequencing for Evaluating the Soil Nematode Diversity and Its Role in Composting Processes" International Journal of Molecular Sciences 24, no. 21: 15749. https://doi.org/10.3390/ijms242115749
APA StyleZapałowska, A., Skwiercz, A., Tereba, A., Puchalski, C., & Malewski, T. (2023). Next-Generation Sequencing for Evaluating the Soil Nematode Diversity and Its Role in Composting Processes. International Journal of Molecular Sciences, 24(21), 15749. https://doi.org/10.3390/ijms242115749







