Silencing the CsSnRK2.11 Gene Decreases Drought Tolerance of Cucumis sativus L.
Abstract
:1. Introduction
2. Results
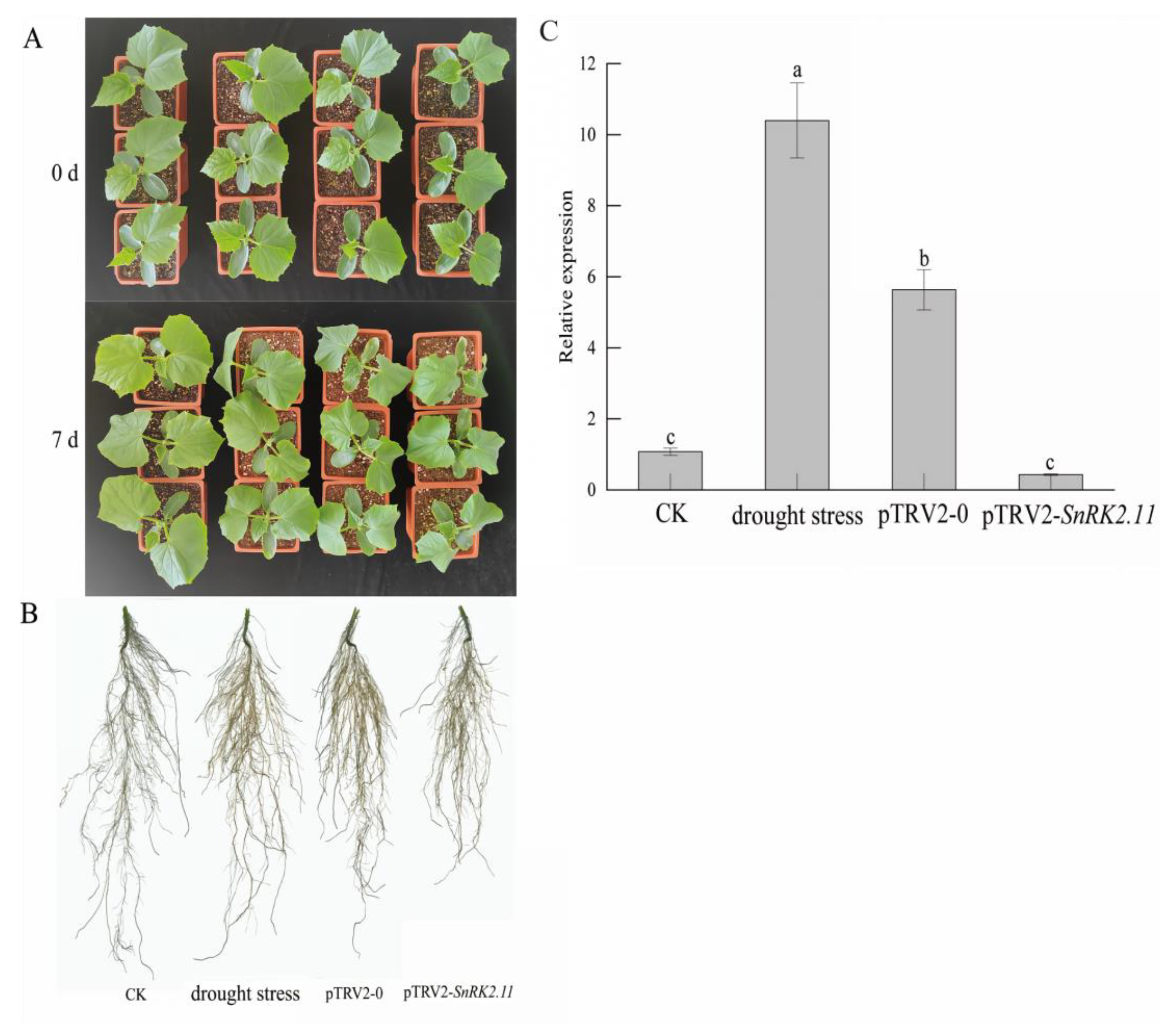
2.1. Plant Phenotype and Relative Expression Level of CsSnRK2.11 under Drought Stress
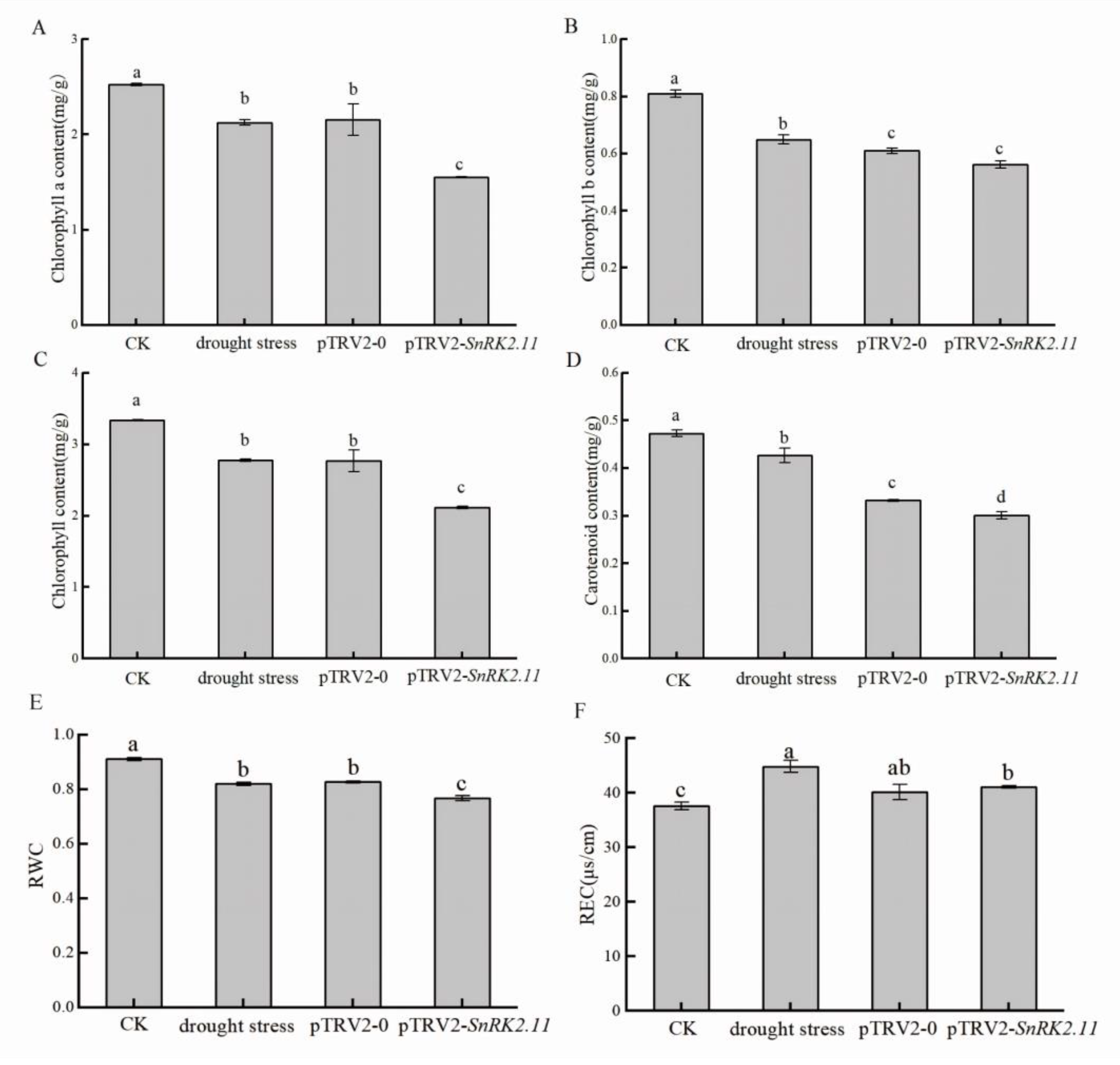
2.2. Effect of Drought Stress on Chlorophyll Content, Relative Water Content, and Relative Electrical Conductivity of Plants
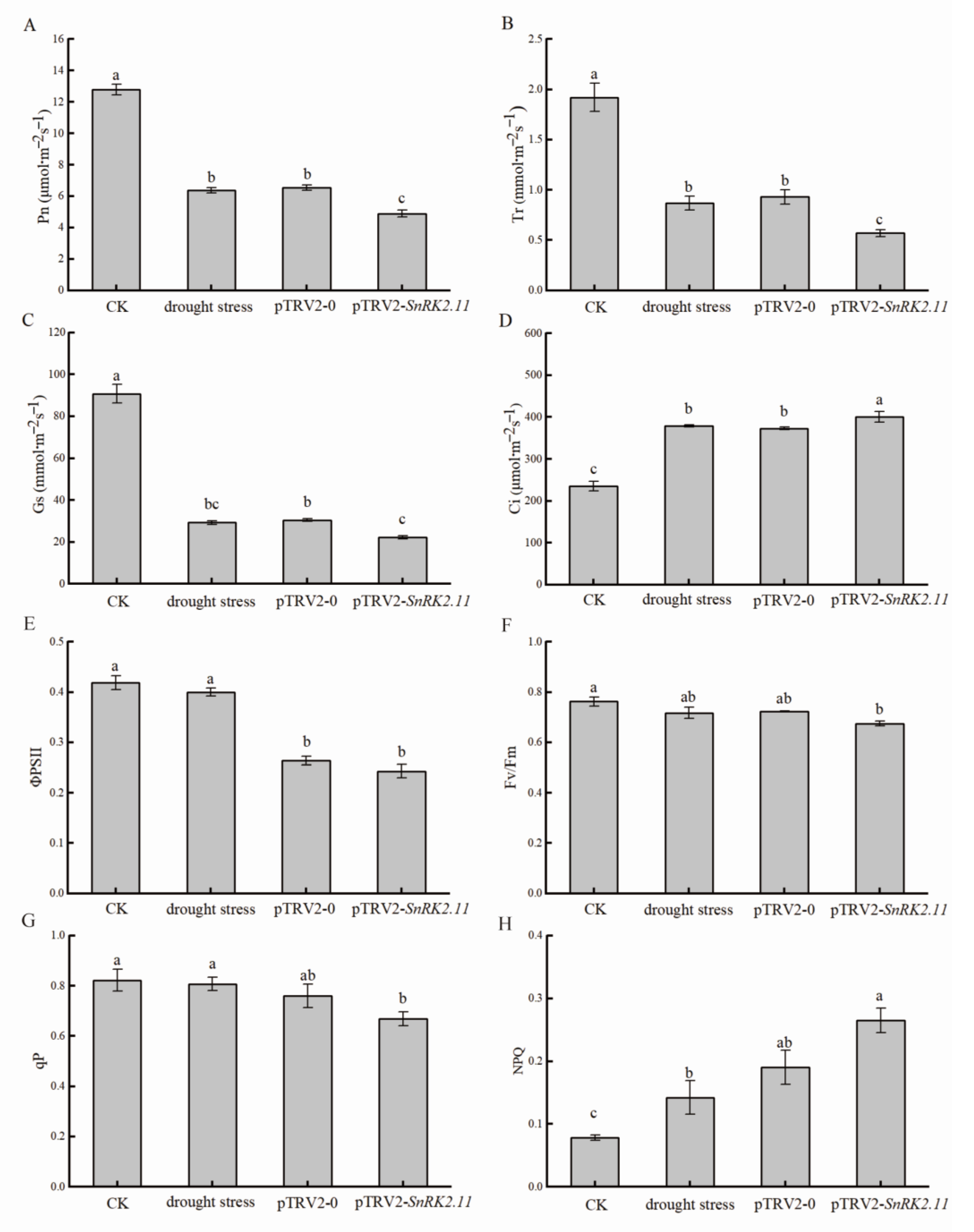
2.3. Effect of Drought Stress on Gas Exchange Parameters and Chlorophyll Fluorescence Parameters of Plant Leaves
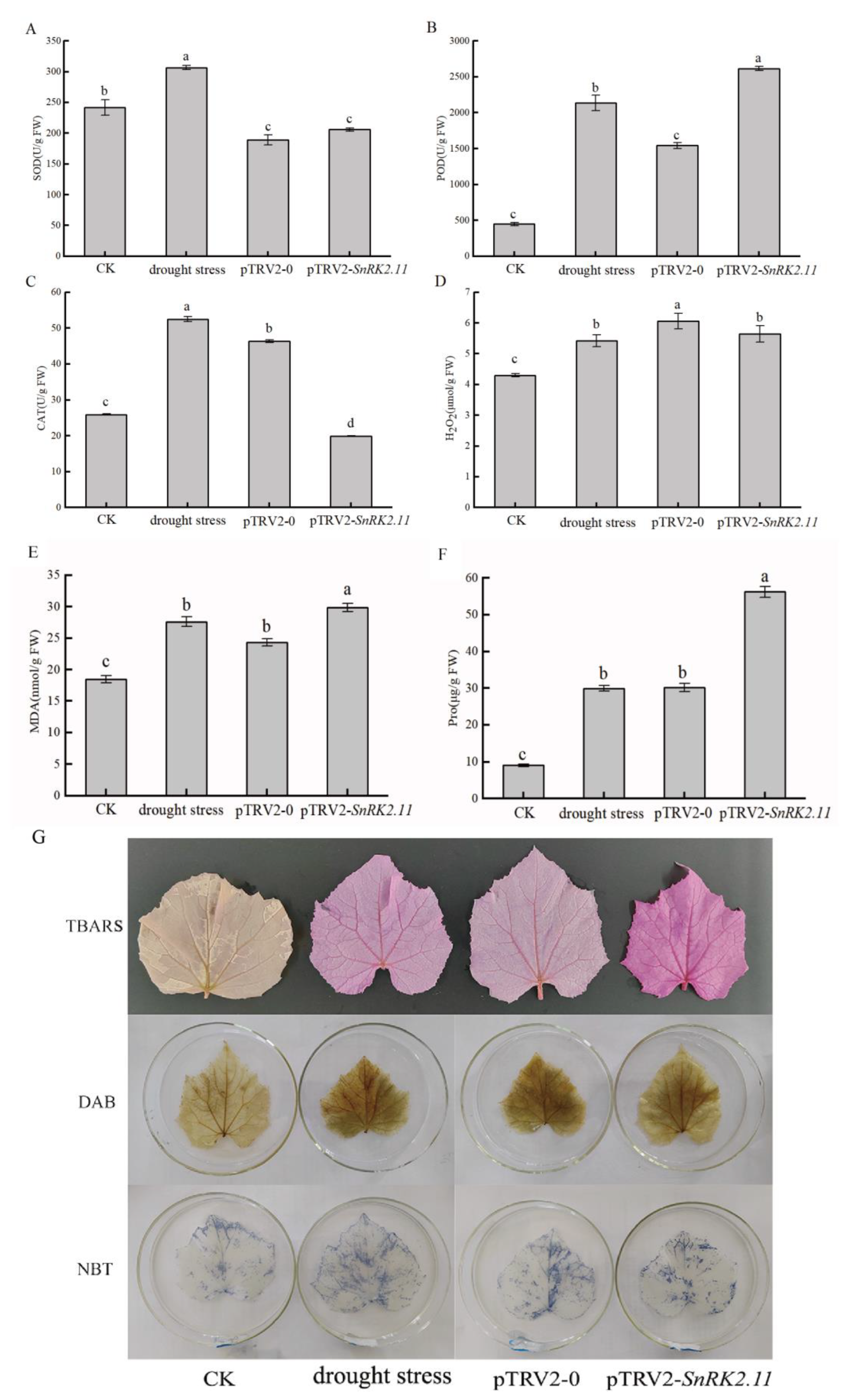
2.4. Effects of Drought Stress on Oxidative Damage and Antioxidant Activity of Plants
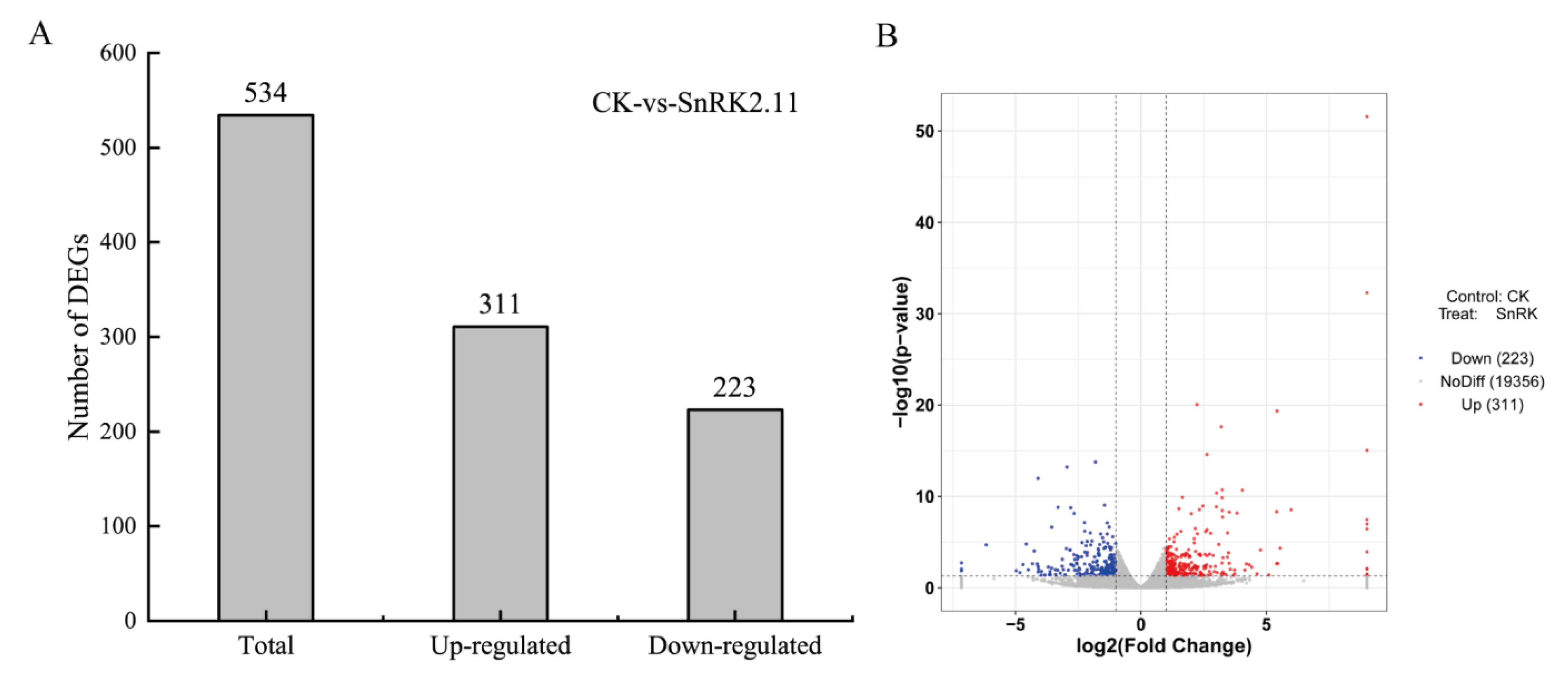
2.5. Differentially Expressed Gene Analysis
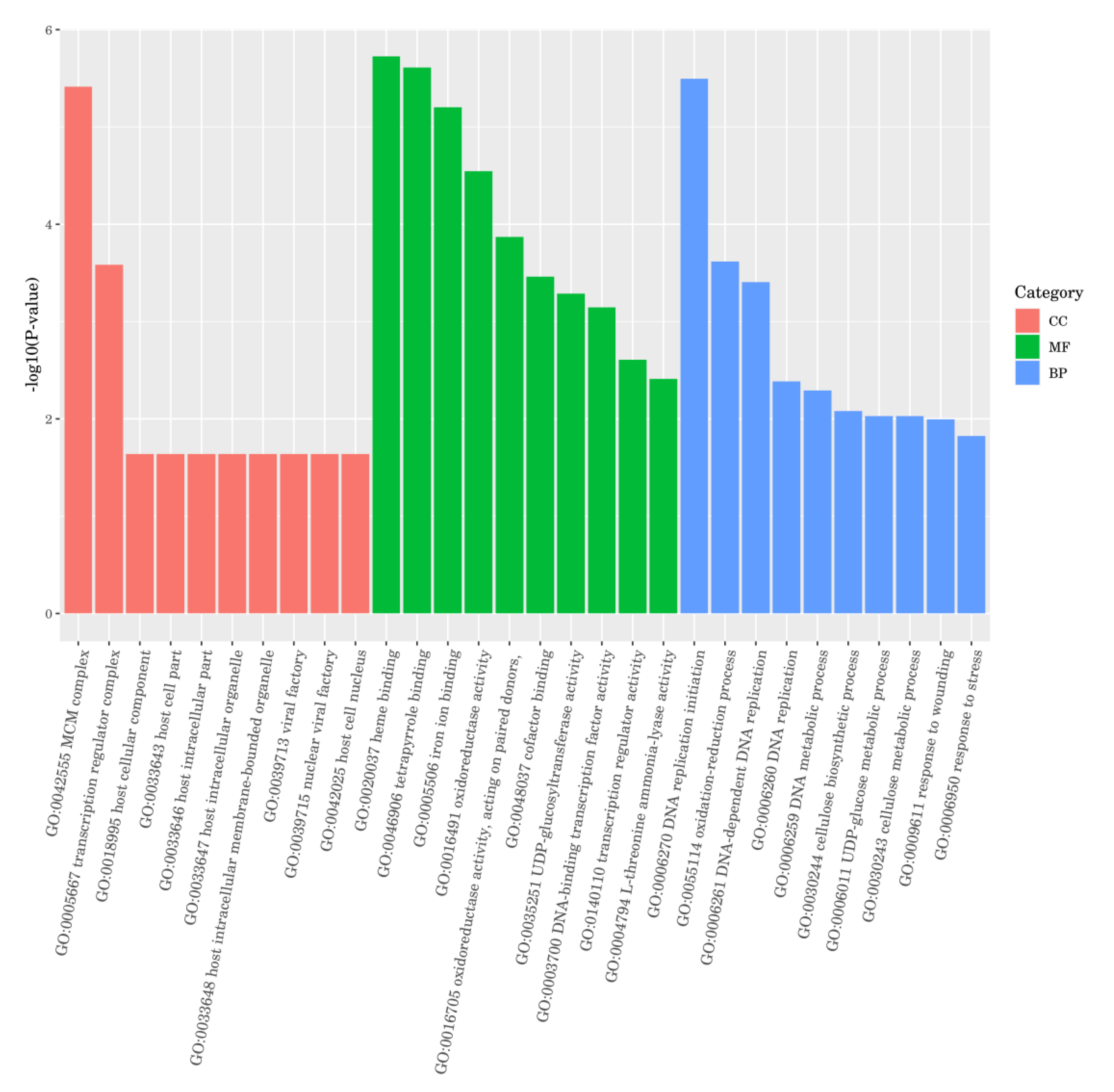
2.6. Differential Gene GO Functional Enrichment Analysis
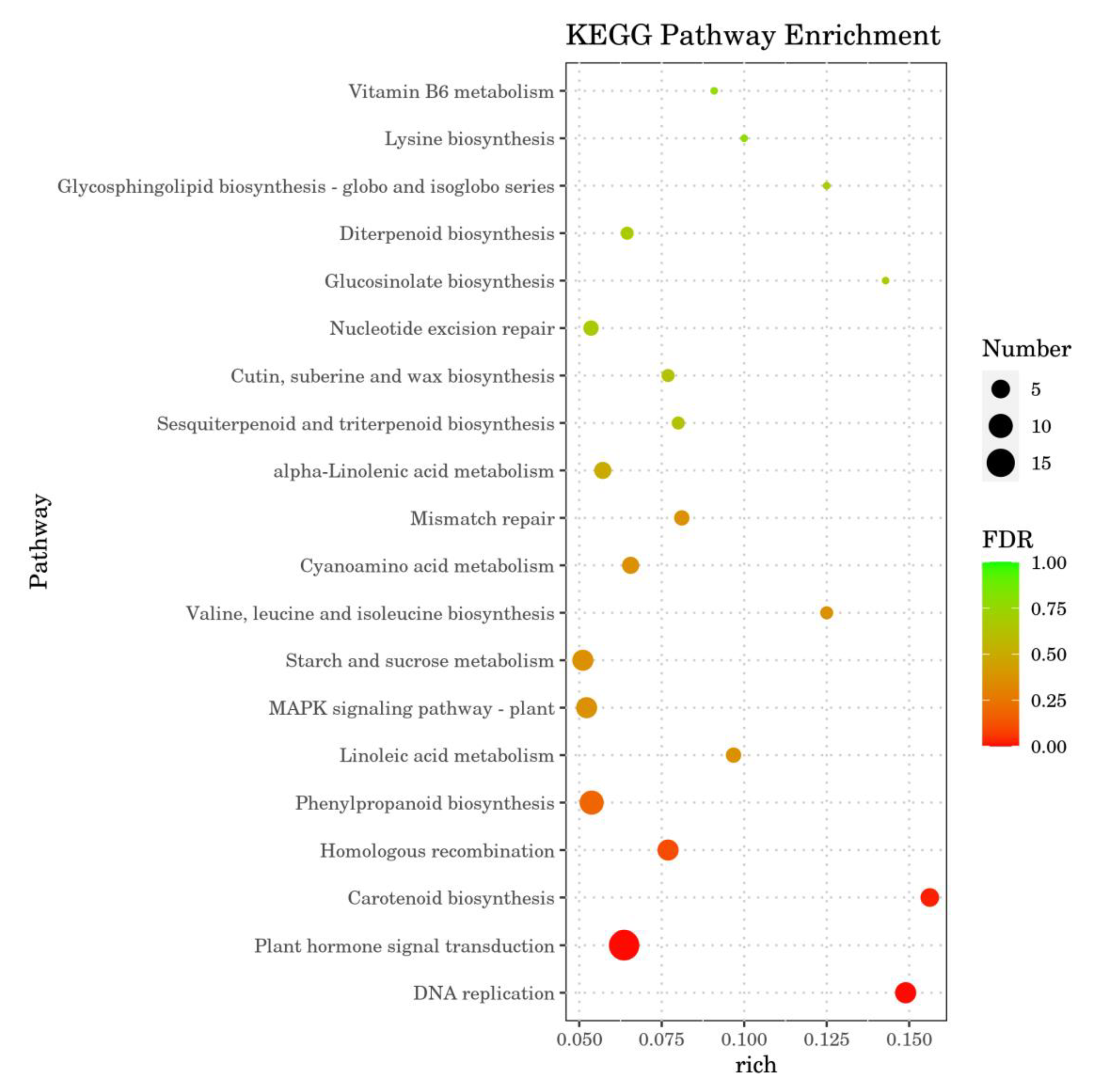
2.7. Annotation Analysis of Differential Gene KEGG
2.8. Analysis of DEGs in the MAPK Signalling Pathway, Plant Hormone Signalling Pathways, and Carotenoid Biosynthetic Pathway
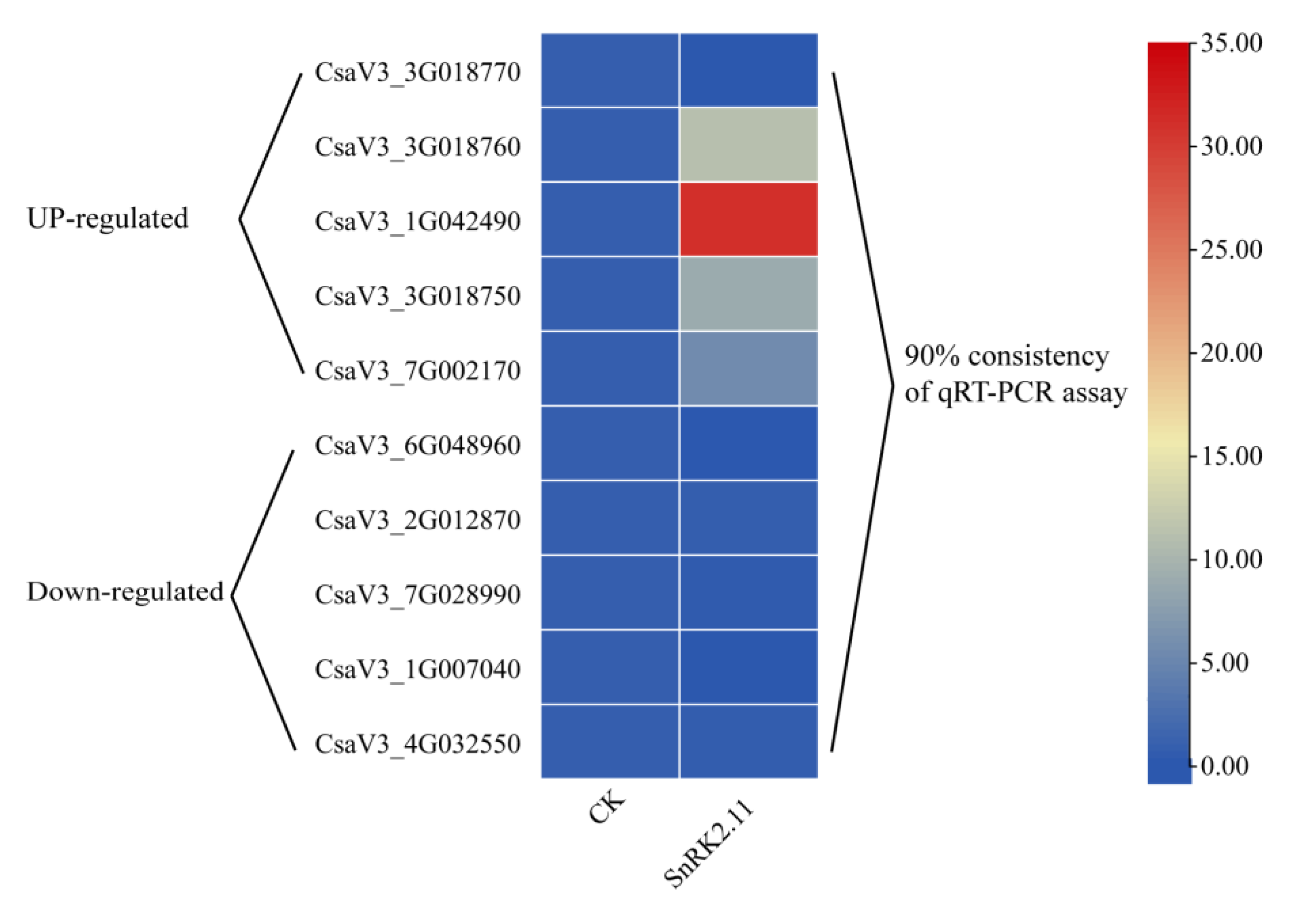
2.9. qRT-PCR Validation of DEGs
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. VIGS Vector Construction for CsSnRK2.11 Gene and Preparation of Agrobacterium Suspension
4.3. Plant Infection
4.4. Silencing Efficiency Assay and Seedling Drought Treatment
4.5. Determination of Chlorophyll Content
4.6. Determination of Leaf Moisture Content and Relative Conductivity
4.7. Measurement of Photosynthetic Parameters and Chlorophyll Fluorescence Parameters
4.8. Determination of Physiological Indexes (SOD, POD, CAT, H2O2, Proline, and MDA)
4.9. Malondialdehyde, Hydrogen Peroxide, Superoxide Anion Staining Observation
4.10. RNA Extraction, Detection, Library Construction, and Transcriptome Sequencing
4.11. qRT-PCR Validation of DEGs
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ajithkumar, I.P.; Panneerselvam, R. ROS scavenging system, osmotic maintenance, pigment and growth status of Panicum sumatrense roth. under drought stress. Cell Biochem. Biophys. 2014, 68, 587–595. [Google Scholar] [CrossRef]
- Wang, W.-B.; Kim, Y.-H.; Lee, H.-S.; Kim, K.-Y.; Deng, X.-P.; Kwak, S.-S. Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses. Plant Physiol. Biochem. 2009, 47, 570–577. [Google Scholar] [CrossRef]
- Li, R.; Shi, F.; Fukuda, K.; Yang, Y. Effects of salt and alkali stresses on germination, growth, photosynthesis and ion accumulation in alfalfa (Medicago sativa L.). Soil Sci. Plant Nutr. 2010, 56, 725–733. [Google Scholar] [CrossRef]
- Yu, Q.; An, L.; Li, W. The CBL–CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014, 33, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.-T.; Ma, S.-L.; Bai, L.-P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.-F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 protein kinases—Key regulators of plant response to abiotic stresses. Omics A J. Integr. Biol. 2011, 15, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Verslues, P.E.; Zhu, J.-K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Mao, X.; Zhang, H.; Chen, S.; Zhai, C.; Yang, S.; Jing, R. Cloning and characterization of TaSnRK2. 3, a novel SnRK2 gene in common wheat. J. Exp. Bot. 2013, 64, 2063–2080. [Google Scholar] [CrossRef]
- Dey, A.; Samanta, M.K.; Gayen, S.; Maiti, M.K. The sucrose non-fermenting 1-related kinase 2 gene SAPK9 improves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression. BMC Plant Biol. 2016, 16, 158. [Google Scholar] [CrossRef]
- Wei, H.; Movahedi, A.; Xu, C.; Wang, P.; Sun, W.; Yin, T.; Zhuge, Q. Heterologous overexpression of the Arabidopsis SnRK2. 8 gene enhances drought and salt tolerance in Populus× euramericana cv ‘Nanlin895’. Plant Biotechnol. Rep. 2019, 13, 245–261. [Google Scholar] [CrossRef]
- Phan, T.-T.; Sun, B.; Niu, J.-Q.; Tan, Q.-L.; Li, J.; Yang, L.-T.; Li, Y.-R. Overexpression of sugarcane gene SoSnRK2. 1 confers drought tolerance in transgenic tobacco. Plant Cell Rep. 2016, 35, 1891–1905. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, H.; Wang, X.; Kang, J.; Lv, B.; Dong, Q.; Li, C.; Chen, H.; Wu, Q. Tartary buckwheat transcription factor FtbZIP5, regulated by FtSnRK2. 6, can improve salt/drought resistance in transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1123. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, S.; Masuda, G.; Bak, H.; Shinozawa, A.; Kamiyama, Y.; Umezawa, T.; Takezawa, D.; Yotsui, I.; Taji, T.; Sakata, Y. Arabidopsis Raf-like kinases act as positive regulators of subclass III SnRK2 in osmostress signaling. Plant J. 2020, 103, 634–644. [Google Scholar] [CrossRef] [PubMed]
- Benedito, V.A.; Visser, P.B.; Angenent, G.C.; Krens, F.A. The potential of virus-induced gene silencing for speeding up functional characterization of plant genes. Genet. Mol. Res. 2004, 3, 323–341. [Google Scholar] [PubMed]
- Igarashi, A.; Yamagata, K.; Sugai, T.; Takahashi, Y.; Sugawara, E.; Tamura, A.; Yaegashi, H.; Yamagishi, N.; Takahashi, T.; Isogai, M. Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology 2009, 386, 407–416. [Google Scholar] [CrossRef]
- Bu, R.; Wang, R.; Wei, Q.; Hu, H.; Sun, H.; Song, P.; Yu, Y.; Liu, Q.; Zheng, Z.; Li, T. Silencing of glycerol-3-phosphate acyltransferase 6 (GPAT6) gene using a newly established virus induced gene silencing (VIGS) system in cucumber alleviates autotoxicity mimicked by cinnamic acid (CA). Plant Soil 2019, 438, 329–346. [Google Scholar] [CrossRef]
- Wan, Z.; Luo, S.; Zhang, Z.; Liu, Z.; Qiao, Y.; Gao, X.; Yu, J.; Zhang, G. Identification and expression profile analysis of the SnRK2 gene family in cucumber. PeerJ 2022, 10, e13994. [Google Scholar] [CrossRef]
- Sharma, A.; Zheng, B. Melatonin mediated regulation of drought stress: Physiological and molecular aspects. Plants 2019, 8, 190. [Google Scholar] [CrossRef]
- Korkmaz, A.; Korkmaz, Y.; Demirkıran, A.R. Enhancing chilling stress tolerance of pepper seedlings by exogenous application of 5-aminolevulinic acid. Environ. Exp. Bot. 2010, 67, 495–501. [Google Scholar] [CrossRef]
- Alexander, R.D.; Wendelboe-Nelson, C.; Morris, P.C. The barley transcription factor HvMYB1 is a positive regulator of drought tolerance. Plant Physiol. Biochem. 2019, 142, 246–253. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, X.; Wang, C.; Jing, R. Overexpression of a common wheat gene TaSnRK2. 8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS ONE 2010, 5, e16041. [Google Scholar] [CrossRef] [PubMed]
- Keyvan, S. The effects of drought stress on yield, relative water content, proline, soluble carbohydrates and chlorophyll of bread wheat cultivars. J. Anim. Plant Sci. 2010, 8, 1051–1060. [Google Scholar]
- Liu, Y.; Xi, M.; Li, Y.; Cheng, Z.; Wang, S.; Kong, F. Improvement in salt tolerance of Iris pseudacorus L. in constructed wetland by exogenous application of salicylic acid and calcium chloride. J. Environ. Manag. 2021, 300, 113703. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.-A.A.; Mekki, B.; Abd El-Sadek, M.E.; El Lateef, E.E. Effect of L-Ornithine application on improving drought tolerance in sugar beet plants. Heliyon 2019, 5, e02631. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, H.; Tadayon, M.R.; Nadeem, M.; Cheema, M.; Razmjoo, J. Proline-mediated changes in antioxidant enzymatic activities and the physiology of sugar beet under drought stress. Acta Physiol. Plant. 2019, 41, 23. [Google Scholar] [CrossRef]
- Shao, Y.; Zhang, X.; van Nocker, S.; Gong, X.; Ma, F. Overexpression of a protein kinase gene MpSnRK2. 10 from Malus prunifolia confers tolerance to drought stress in transgenic Arabidopsis thaliana and apple. Gene 2019, 692, 26–34. [Google Scholar] [CrossRef]
- Khan, T.A.; Fariduddin, Q.; Yusuf, M. Low-temperature stress: Is phytohormones application a remedy? Environ. Sci. Pollut. Res. 2017, 24, 21574–21590. [Google Scholar] [CrossRef]
- Judycki, J.; Jaskula, P.; Dolzycki, B.; Pszczola, M.; Jaczewski, M.; Rys, D.; Stienss, M. Investigation of low-temperature cracking in newly constructed high-modulus asphalt concrete base course of a motorway pavement. Road Mater. Pavement Des. 2015, 16, 362–388. [Google Scholar] [CrossRef]
- Yu, X.-Z.; Yang, L.; Feng, Y.-X. Comparative response of SOD in different plants against cadmium and drought stress at the molecular level. Appl. Environ. Biotechnol. 2020, 5, 14–27. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Bao, Y.-F.; Tong, P.; Wu, T.-R.; Xu, D.; He, M.-X.; Yue, W.; Liu, Q.-F.; Yang, H.-H.; Jiang, J.-B. Silencing the SLB3 transcription factor gene decreases drought stress tolerance in tomato. J. Integr. Agric. 2020, 19, 2699–2708. [Google Scholar] [CrossRef]
- Jingjiang, H.; Zhenyu, G.; Jianlei, W.; Shuqing, W. Effect of water stress on membrane lipid peroxidation in maple. J. Northwest For. Coll. 1999, 14, 7–11. [Google Scholar]
- Yang, X.; Yang, X. The effect of water stress on cell protective enzyme activity and membrane lipid peroxidation in broccoli (Brassica oleracea L. var. italica P.). Adv. Hortic. 1998, 2, 567–571. [Google Scholar]
- Kohli, S.K.; Khanna, K.; Bhardwaj, R.; Abd_Allah, E.F.; Ahmad, P.; Corpas, F.J. Assessment of subcellular ROS and NO metabolism in higher plants: Multifunctional signaling molecules. Antioxidants 2019, 8, 641. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Plant peroxisomes: A factory of reactive species. Front. Plant Sci. 2020, 11, 853. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Li, H.; Liu, C.; Fan, H. Understanding of the postgerminative development response to salinity and drought stresses in cucumber seeds by integrated proteomics and transcriptomics analysis. J. Proteom. 2021, 232, 104062. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-C.; Yang, Z.-R.; Wang, M.; Li, W.; Li, S.-C. High-throughput sequencing technology and its application. China Biotechnol. 2012, 32, 109–114. [Google Scholar]
- Khan, R.; Zhou, P.; Ma, X.; Zhou, L.; Wu, Y.; Ullah, Z.; Wang, S. Transcriptome profiling, biochemical and physiological analyses provide new insights towards drought tolerance in Nicotiana tabacum L. Genes 2019, 10, 1041. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, W.; Lv, Y.; Bai, J.; Li, T.; Yang, X.; Liu, L.; Zhou, H. Comparative transcriptomics reveals new insights into melatonin-enhanced drought tolerance in naked oat seedlings. PeerJ 2022, 10, e13669. [Google Scholar] [CrossRef]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant–pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef]
- Jung, C.; Nguyen, N.H.; Cheong, J.-J. Transcriptional regulation of protein phosphatase 2C genes to modulate abscisic acid signaling. Int. J. Mol. Sci. 2020, 21, 9517. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sola, M.Á.; Rodríguez-Concepción, M. Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arab. Book/Am. Soc. Plant Biol. 2012, 10, e0158. [Google Scholar] [CrossRef] [PubMed]
- Chernys, J.T.; Zeevaart, J.A. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 2000, 124, 343–354. [Google Scholar] [CrossRef]
- Arraes, F.B.M.; Beneventi, M.A.; Lisei de Sa, M.E.; Paixao, J.F.R.; Albuquerque, E.V.S.; Marin, S.R.R.; Purgatto, E.; Nepomuceno, A.L.; Grossi-de-Sa, M.F. Implications of ethylene biosynthesis and signaling in soybean drought stress tolerance. BMC Plant Biol. 2015, 15, 213. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of Total Carotenoids and Chlorophylls a and b of Leaf Extracts in Different Solvents; Portland Press Ltd.: London, UK, 1983. [Google Scholar]
- Turk, H.; Erdal, S. Melatonin alleviates cold-induced oxidative damage in maize seedlings by up-regulating mineral elements and enhancing antioxidant activity. J. Plant Nutr. Soil Sci. 2015, 178, 433–439. [Google Scholar] [CrossRef]
- Gao, J. Plant Physiology Laboratory Guide; Higher Education Press: Beijing, China, 2006. [Google Scholar]
- Liu, J.; Zhang, R.; Xu, X.; Fowler, J.C.; Miller, T.E.; Dong, T. Effect of summer warming on growth, photosynthesis and water status in female and male Populus cathayana: Implications for sex-specific drought and heat tolerances. Tree Physiol. 2020, 40, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Kramer, D.M.; Johnson, G.; Kiirats, O.; Edwards, G.E. New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth. Res. 2004, 79, 209–218. [Google Scholar] [CrossRef]
- Lu, T.; Yu, H.; Li, Q.; Chai, L.; Jiang, W. Improving plant growth and alleviating photosynthetic inhibition and oxidative stress from low-light stress with exogenous GR24 in tomato (Solanum lycopersicum L.) seedlings. Front. Plant Sci. 2019, 10, 490. [Google Scholar] [CrossRef]
- Cang, J.; Zhao, H. Experimental Course of Plant Physiology; Higher Education Press: Beijing, China, 2013. [Google Scholar]
- Kong, F.; Deng, Y.; Zhou, B.; Wang, G.; Wang, Y.; Meng, Q. A chloroplast-targeted DnaJ protein contributes to maintenance of photosystem II under chilling stress. J. Exp. Bot. 2014, 65, 143–158. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, J.; Chen, X.; Gao, Z.; Xuan, W.; Xu, S.; Ding, X.; Shen, W. Carbon monoxide alleviates cadmium-induced oxidative damage by modulating glutathione metabolism in the roots of Medicago sativa. New Phytol. 2008, 177, 155–166. [Google Scholar] [CrossRef]
- Grellet Bournonville, C.F.; Díaz-Ricci, J.C. Quantitative determination of superoxide in plant leaves using a modified NBT staining method. Phytochem. Anal. 2011, 22, 268–271. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Rahman, A.; Ansary, M.M.U.; Watanabe, A.; Fujita, M.; Tran, L.-S.P. Hydrogen sulfide modulates cadmium-induced physiological and biochemical responses to alleviate cadmium toxicity in rice. Sci. Rep. 2015, 5, 14078. [Google Scholar] [CrossRef]
- Luo, P.; Shen, Y.; Jin, S.; Huang, S.; Cheng, X.; Wang, Z.; Li, P.; Zhao, J.; Bao, M.; Ning, G. Overexpression of Rosa rugosa anthocyanidin reductase enhances tobacco tolerance to abiotic stress through increased ROS scavenging and modulation of ABA signaling. Plant Sci. 2016, 245, 35–49. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Wan, Z.; Luo, S.; Wei, H.; Zhao, J.; Wang, G.; Yu, J.; Zhang, G. Silencing the CsSnRK2.11 Gene Decreases Drought Tolerance of Cucumis sativus L. Int. J. Mol. Sci. 2023, 24, 15761. https://doi.org/10.3390/ijms242115761
Wang P, Wan Z, Luo S, Wei H, Zhao J, Wang G, Yu J, Zhang G. Silencing the CsSnRK2.11 Gene Decreases Drought Tolerance of Cucumis sativus L. International Journal of Molecular Sciences. 2023; 24(21):15761. https://doi.org/10.3390/ijms242115761
Chicago/Turabian StyleWang, Peng, Zilong Wan, Shilei Luo, Haotai Wei, Jianuo Zhao, Guoshuai Wang, Jihua Yu, and Guobin Zhang. 2023. "Silencing the CsSnRK2.11 Gene Decreases Drought Tolerance of Cucumis sativus L." International Journal of Molecular Sciences 24, no. 21: 15761. https://doi.org/10.3390/ijms242115761
APA StyleWang, P., Wan, Z., Luo, S., Wei, H., Zhao, J., Wang, G., Yu, J., & Zhang, G. (2023). Silencing the CsSnRK2.11 Gene Decreases Drought Tolerance of Cucumis sativus L. International Journal of Molecular Sciences, 24(21), 15761. https://doi.org/10.3390/ijms242115761




