Switching Rat Resident Macrophages from M1 to M2 Phenotype by Iba1 Silencing Has Analgesic Effects in SNL-Induced Neuropathic Pain
Abstract
:1. Introduction
2. Results
2.1. SNL-Induced Clusters of Iba1 (+) Macrophages around DRG Neurons Are Maintained Even after Iba1 Silencing
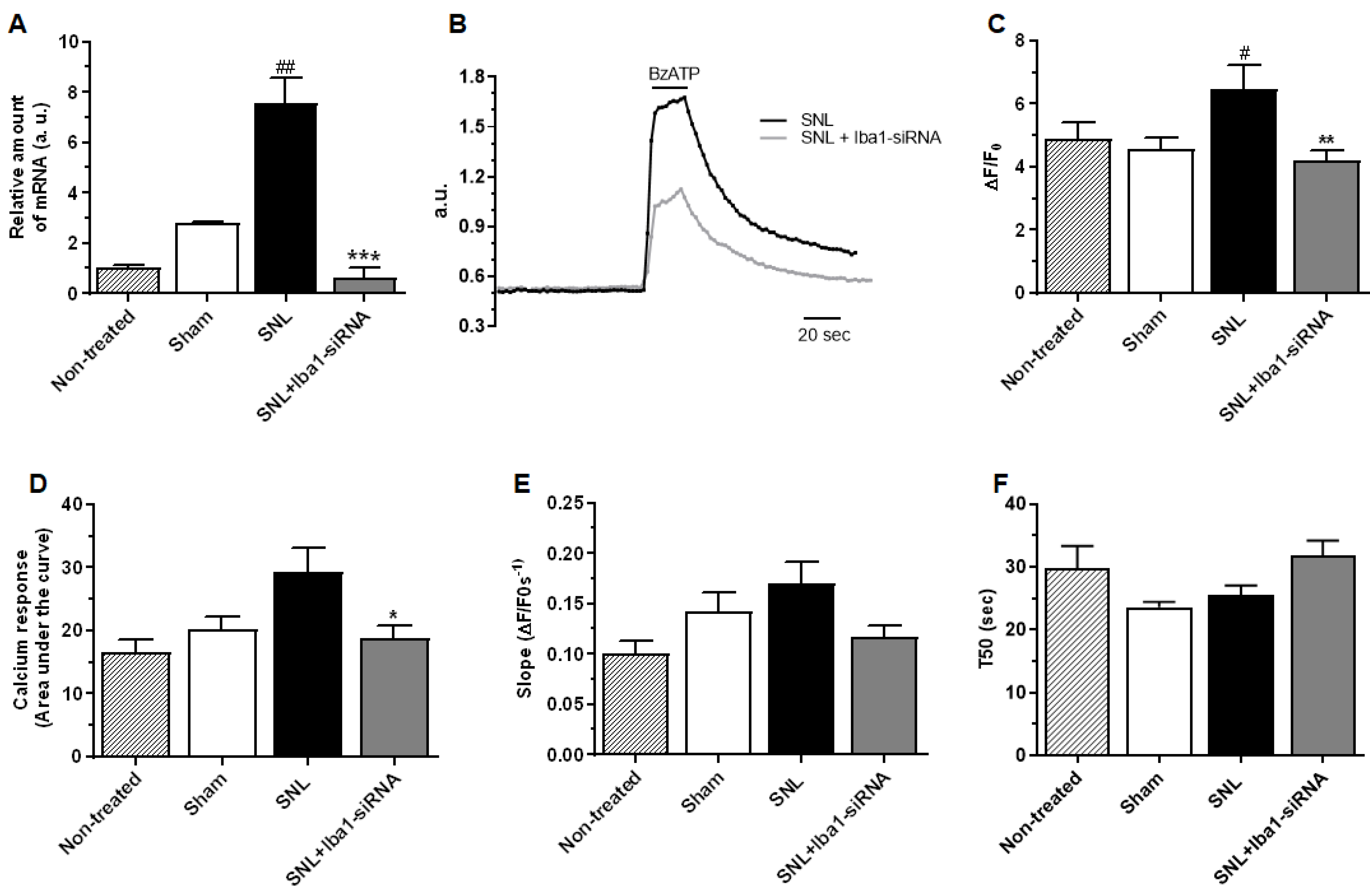
2.2. Intra-Ganglionic Delivery of Iba1-siRNA Significantly Reduced the SNL-Induced Neuropathic Pain, without Reducing Neuronal Excitability
2.3. Iba1 Silencing Is Switching Macrophages from M1 Pro-Inflammatory Phenotype to M2 Anti-Inflammatory Phenotype
3. Discussion
4. Materials and Methods
4.1. Animals and Experimental Design
4.2. SNL Surgery and Intra-Ganglionic Delivery Procedure
4.3. Behavioral Tests
4.3.1. Mechanical Stimulation
4.3.2. Thermal Stimulation: Acetone Test
4.3.3. Thermal Stimulation: Hot Plate Test
4.4. Quantitative Reverse Transcription—Polymerase Chain Reaction (qRT-PCR)
4.5. Western Blotting
4.6. Electron Microscopy (EM)
4.7. Immunohistochemistry (IHC)
4.8. Immunopanning and Cultures of Macrophages
4.9. Migration Transwell Assay
4.10. Electrophysiological Recordings
4.11. Ratiometric Intracellular Ca2+ Imaging
4.12. Neurite Measurement Assay on DRG Explants
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADP100 | Duration of the action potential base |
| ADPos | Duration at the overshoot level |
| AHP | Action potential hyperpolarization amplitude |
| AHPsub | Subtracted voltage value from resting potential to AHP |
| Aif1 | Allograft inflammatory factor 1 |
| ANOVA | Analysis of variance |
| AP | Action potential |
| AP Amp | Action potential total amplitude |
| AP FP | Duration of the AP falling phase |
| AP OS | Action potential overshoot value |
| AP RPh | Duration of the AP rising phase |
| AP Th | Threshold potential |
| AUC | Area under the curve |
| BCA assay | Bicinchoninic acid assay |
| BDNF | Brain derived neurotrophic factor |
| BLAST | Basic Local Alignment Search Tool |
| BzATP | (2′(3′)-O-(4-Benzoylbenzoyl)adenosine-5′-triphosphate) |
| CaCl | Calcium Chloride |
| CCI | Chronic Constriction Injury |
| CCL2 | Chemokine (C-C motif) ligand 2 |
| CD163 | Cluster of Differentiation 163 |
| CD206 | Cluster of Differentiation 206 |
| CD32 | Cluster of Differentiation 32 |
| CD86 | Cluster of Differentiation 86 |
| CD 68 (ED1) | Cluster of Differentiation 68 |
| Cd11b | Cluster of Differentiation 11b |
| CGRP | Calcitonin gene-related peptide |
| CSF1 | Colony stimulating factor 1 |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DNA | Deoxyribonucleic acid |
| DPA | Dynamic plantar aesthesiometer |
| DRG | Dorsal root ganglia |
| EGTA | 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid |
| E11 | Embryonic day 11 |
| EM | Electron microscopy |
| F-actin | Filamentous actin |
| FPharea | Area at the falling phase of action potential |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| HRP | Horseradish peroxidase |
| IB4 | Isolectin B4 |
| Iba1 | Ionized binding adapter protein 1 |
| ICC | Immunocytochemistry |
| IgG | Immunoglobulin G |
| IHC | Immunohistochemistry |
| IL-4 | Interleukin 4 |
| IL-1β | Interleukin 1β |
| IL-6 | Interleukin 6 |
| iNOS | inducible Nitric oxide synthase |
| KCl | Potassium Chloride |
| KOH | Potassium Hydroxide |
| LPS | Lipopolysaccharide |
| MAC1 | Macrophage-1 antigen |
| MAFIA | Macrophage Fas-induced apoptosis |
| MgCl | Magnesium Chloride |
| MHC-II | Major histocompatibility complex class II |
| mRNA | messenger Ribonucleic acid |
| MUSCLE | MUltiple Sequence Comparison by Log-Expectation |
| NaCl | Sodium Chloride |
| NaOH | Sodium hydroxide |
| NCBI | National Center for Biotechnology Information |
| NF200 | Neurofilament 200 kDa |
| NGF | Nerve growth factor |
| NLRP3 | NOD-, LRR- and pyrin domain-containing protein 3 = Nucleotide-Binding Oligomerization Domain, Leucine Rich Repeat and Pyrin Domain Containing protein 3 |
| NO | Nitric oxide |
| NT-3 | Neurotrophin 3 |
| ORF | Open reading frame |
| P2x7R | P2x7 receptor |
| PBS | Phosphate buffer saline |
| PFA | Paraformaldehyde |
| PVDF | Polyvinylidene difluoride |
| qRT-PCR | Quantitative Reverse Transcription–Polymerase Chain Reaction |
| RIPA | Radioimmunoprecipitation assay buffer |
| RNA | Ribonucleic acid |
| RP | Resting potential |
| RPharea | Area at the rising phase of action potential |
| S100A9 | S100 calcium-binding protein A9 |
| SDS-PAGE | Sodium dodecyl sulphate–polyacrylamide gel electrophoresis |
| SEM | Standard error of the mean |
| siRNA | Small interfering RNA |
| SNI | Spared nerve injury |
| SNL | Spinal nerve ligation |
| TNF-α | Tumor necrosis factor-α |
| VSB | Visual Basic for Applications |
References
- Costigan, M.; Scholz, J.; Woolf, C.J. Neuropathic pain: A maladaptive response of the nervous system to damage. Annu. Rev. Neurosci. 2009, 32, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Gangadharan, V.; Zheng, H.; Taberner, F.J.; Landry, J.; Nees, T.A.; Pistolic, J.; Agarwal, N.; Mannich, D.; Benes, V.; Helmstaedter, M.; et al. Neuropathic pain caused by miswiring and abnormal end organ targeting. Nature 2022, 606, 137–145. [Google Scholar] [CrossRef] [PubMed]
- von Hehn, C.A.; Baron, R.; Woolf, C.J. Deconstructing the neuropathic pain phenotype to reveal neural mechanisms. Neuron 2012, 73, 638–652. [Google Scholar] [CrossRef] [PubMed]
- Raoof, R.; Willemen, H.; Eijkelkamp, N. Divergent roles of immune cells and their mediators in pain. Rheumatology 2018, 57, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.S.; Kam, P.C. Neuroimmune mechanisms of pain: Basic science and potential therapeutic modulators. Anaesth. Intensive Care 2020, 48, 167–178. [Google Scholar] [CrossRef]
- Calvo, M.; Dawes, J.M.; Bennett, D.L. The role of the immune system in the generation of neuropathic pain. Lancet Neurol. 2012, 11, 629–642. [Google Scholar] [CrossRef]
- Grace, P.M.; Hutchinson, M.R.; Maier, S.F.; Watkins, L.R. Pathological pain and the neuroimmune interface. Nat. Rev. Immunol. 2014, 14, 217–231. [Google Scholar] [CrossRef]
- Gheorghe, R.O.; Grosu, A.V.; Bica-Popi, M.; Ristoiu, V. The Yin/Yang Balance of Communication between Sensory Neurons and Macrophages in Traumatic Peripheral Neuropathic Pain. Int. J. Mol. Sci. 2022, 23, 12389. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.E.A.; Guimaraes, R.M.; Cunha, T.M. Sensory neuron-associated macrophages as novel modulators of neuropathic pain. Pain Rep. 2021, 6, e873. [Google Scholar] [CrossRef]
- Domoto, R.; Sekiguchi, F.; Tsubota, M.; Kawabata, A. Macrophage as a Peripheral Pain Regulator. Cells 2021, 10, 1881. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zou, W.; Du, J.; Zhao, Y. The origins and homeostasis of monocytes and tissue-resident macrophages in physiological situation. J. Cell. Physiol. 2018, 233, 6425–6439. [Google Scholar] [CrossRef]
- De Logu, F.; Nassini, R.; Materazzi, S.; Carvalho Goncalves, M.; Nosi, D.; Rossi Degl’Innocenti, D.; Marone, I.M.; Ferreira, J.; Li Puma, S.; Benemei, S.; et al. Schwann cell TRPA1 mediates neuroinflammation that sustains macrophage-dependent neuropathic pain in mice. Nat. Commun. 2017, 8, 1887. [Google Scholar] [CrossRef]
- Liu, T.; van Rooijen, N.; Tracey, D.J. Depletion of macrophages reduces axonal degeneration and hyperalgesia following nerve injury. Pain 2000, 86, 25–32. [Google Scholar] [CrossRef]
- Shepherd, A.J.; Mickle, A.D.; Golden, J.P.; Mack, M.R.; Halabi, C.M.; de Kloet, A.D.; Samineni, V.K.; Kim, B.S.; Krause, E.G.; Gereau, R.W.t.; et al. Macrophage angiotensin II type 2 receptor triggers neuropathic pain. Proc. Natl. Acad. Sci. USA 2018, 115, E8057–E8066. [Google Scholar] [CrossRef]
- Yu, X.; Liu, H.; Hamel, K.A.; Morvan, M.G.; Yu, S.; Leff, J.; Guan, Z.; Braz, J.M.; Basbaum, A.I. Dorsal root ganglion macrophages contribute to both the initiation and persistence of neuropathic pain. Nat. Commun. 2020, 11, 264. [Google Scholar] [CrossRef]
- Cobos, E.J.; Nickerson, C.A.; Gao, F.; Chandran, V.; Bravo-Caparros, I.; Gonzalez-Cano, R.; Riva, P.; Andrews, N.A.; Latremoliere, A.; Seehus, C.R.; et al. Mechanistic Differences in Neuropathic Pain Modalities Revealed by Correlating Behavior with Global Expression Profiling. Cell Rep. 2018, 22, 1301–1312. [Google Scholar] [CrossRef]
- Kolter, J.; Kierdorf, K.; Henneke, P. Origin and Differentiation of Nerve-Associated Macrophages. J. Immunol. 2020, 204, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.L.; Yim, A.K.Y.; Kim, K.W.; Avey, D.; Czepielewski, R.S.; Colonna, M.; Milbrandt, J.; Randolph, G.J. Peripheral nerve resident macrophages share tissue-specific programming and features of activated microglia. Nat. Commun. 2020, 11, 2552. [Google Scholar] [CrossRef] [PubMed]
- Ydens, E.; Amann, L.; Asselbergh, B.; Scott, C.L.; Martens, L.; Sichien, D.; Mossad, O.; Blank, T.; De Prijck, S.; Low, D.; et al. Profiling peripheral nerve macrophages reveals two macrophage subsets with distinct localization, transcriptome and response to injury. Nat. Neurosci. 2020, 23, 676–689. [Google Scholar] [CrossRef] [PubMed]
- Ristoiu, V. Contribution of macrophages to peripheral neuropathic pain pathogenesis. Life Sci. 2013, 93, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Zigmond, R.E.; Echevarria, F.D. Macrophage biology in the peripheral nervous system after injury. Prog. Neurobiol. 2019, 173, 102–121. [Google Scholar] [CrossRef]
- Kalinski, A.L.; Yoon, C.; Huffman, L.D.; Duncker, P.C.; Kohen, R.; Passino, R.; Hafner, H.; Johnson, C.; Kawaguchi, R.; Carbajal, K.S.; et al. Analysis of the immune response to sciatic nerve injury identifies efferocytosis as a key mechanism of nerve debridement. eLife 2020, 9, e60223. [Google Scholar] [CrossRef]
- Krishnan, A.; Bhavanam, S.; Zochodne, D. An Intimate Role for Adult Dorsal Root Ganglia Resident Cycling Cells in the Generation of Local Macrophages and Satellite Glial Cells. J. Neuropathol. Exp. Neurol. 2018, 77, 929–941. [Google Scholar] [CrossRef]
- Ton, B.H.; Chen, Q.; Gaina, G.; Tucureanu, C.; Georgescu, A.; Strungaru, C.; Flonta, M.L.; Sah, D.; Ristoiu, V. Activation profile of dorsal root ganglia Iba-1 (+) macrophages varies with the type of lesion in rats. Acta Histochem. 2013, 115, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Vega-Avelaira, D.; Geranton, S.M.; Fitzgerald, M. Differential regulation of immune responses and macrophage/neuron interactions in the dorsal root ganglion in young and adult rats following nerve injury. Mol. Pain 2009, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J.; Kim, J.; Shin, H.; Jeong, S.R.; Kang, Y.M.; Choi, J.Y.; Hwang, D.H.; Kim, B.G. Contribution of macrophages to enhanced regenerative capacity of dorsal root ganglia sensory neurons by conditioning injury. J. Neurosci. 2013, 33, 15095–15108. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J.; Shin, H.Y.; Cui, Y.; Kim, H.; Thi, A.H.; Choi, J.Y.; Kim, E.Y.; Hwang, D.H.; Kim, B.G. CCL2 Mediates Neuron-Macrophage Interactions to Drive Proregenerative Macrophage Activation Following Preconditioning Injury. J. Neurosci. 2015, 35, 15934–15947. [Google Scholar] [CrossRef]
- Rotshenker, S. Wallerian degeneration: The innate-immune response to traumatic nerve injury. J. Neuroinflamm. 2011, 8, 109. [Google Scholar] [CrossRef]
- Dubovy, P.; Jancalek, R.; Klusakova, I.; Svizenska, I.; Pejchalova, K. Intra- and extraneuronal changes of immunofluorescence staining for TNF-alpha and TNFR1 in the dorsal root ganglia of rat peripheral neuropathic pain models. Cell Mol. Neurobiol. 2006, 26, 1205–1217. [Google Scholar] [CrossRef]
- Ajami, B.; Bennett, J.L.; Krieger, C.; McNagny, K.M.; Rossi, F.M. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat. Neurosci. 2011, 14, 1142–1149. [Google Scholar] [CrossRef]
- Varvel, N.H.; Neher, J.J.; Bosch, A.; Wang, W.; Ransohoff, R.M.; Miller, R.J.; Dingledine, R. Infiltrating monocytes promote brain inflammation and exacerbate neuronal damage after status epilepticus. Proc. Natl. Acad. Sci. USA 2016, 113, E5665–E5674. [Google Scholar] [CrossRef]
- Ohsawa, K.; Imai, Y.; Kanazawa, H.; Sasaki, Y.; Kohsaka, S. Involvement of Iba1 in membrane ruffling and phagocytosis of macrophages/microglia. J. Cell Sci. 2000, 113 Pt 17, 3073–3084. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.F.; Ho, D.W.; Lau, C.K.; Lam, C.T.; Lum, C.T.; Poon, R.T.; Fan, S.T. Allograft inflammatory factor-1 (AIF-1) is crucial for the survival and pro-inflammatory activity of macrophages. Int. Immunol. 2005, 17, 1391–1397. [Google Scholar] [CrossRef]
- Kanazawa, H.; Ohsawa, K.; Sasaki, Y.; Kohsaka, S.; Imai, Y. Macrophage/microglia-specific protein Iba1 enhances membrane ruffling and Rac activation via phospholipase C-gamma -dependent pathway. J. Biol. Chem. 2002, 277, 20026–20032. [Google Scholar] [CrossRef]
- Ohsawa, K.; Imai, Y.; Sasaki, Y.; Kohsaka, S. Microglia/macrophage-specific protein Iba1 binds to fimbrin and enhances its actin-bundling activity. J. Neurochem. 2004, 88, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Ohsawa, K.; Kanazawa, H.; Kohsaka, S.; Imai, Y. Iba1 is an actin-cross-linking protein in macrophages/microglia. Biochem. Biophys. Res. Commun. 2001, 286, 292–297. [Google Scholar] [CrossRef]
- Gheorghe, R.O.; Deftu, A.; Filippi, A.; Grosu, A.; Bica-Popi, M.; Chiritoiu, M.; Chiritoiu, G.; Munteanu, C.; Silvestro, L.; Ristoiu, V. Silencing the Cytoskeleton Protein Iba1 (Ionized Calcium Binding Adapter Protein 1) Interferes with BV2 Microglia Functioning. Cell Mol. Neurobiol. 2020, 40, 1011–1027. [Google Scholar] [CrossRef]
- Puljak, L.; Kojundzic, S.L.; Hogan, Q.H.; Sapunar, D. Lidocaine injection into the rat dorsal root ganglion causes neuroinflammation. Anesth. Analg. 2009, 108, 1021–1026. [Google Scholar] [CrossRef]
- Blum, E.; Procacci, P.; Conte, V.; Hanani, M. Systemic inflammation alters satellite glial cell function and structure. A possible contribution to pain. Neuroscience 2014, 274, 209–217. [Google Scholar] [CrossRef]
- Choi, Y.; Yoon, Y.W.; Na, H.S.; Kim, S.H.; Chung, J.M. Behavioral signs of ongoing pain and cold allodynia in a rat model of neuropathic pain. Pain 1994, 59, 369–376. [Google Scholar]
- Kim, S.H.; Chung, J.M. An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain 1992, 50, 355–363. [Google Scholar] [CrossRef]
- Sapunar, D.; Vukojevic, K.; Kostic, S.; Puljak, L. Attenuation of pain-related behavior evoked by injury through blockade of neuropeptide Y Y2 receptor. Pain 2011, 152, 1173–1181. [Google Scholar] [CrossRef]
- Ma, C.; Shu, Y.; Zheng, Z.; Chen, Y.; Yao, H.; Greenquist, K.W.; White, F.A.; LaMotte, R.H. Similar electrophysiological changes in axotomized and neighboring intact dorsal root ganglion neurons. J. Neurophysiol. 2003, 89, 1588–1602. [Google Scholar] [CrossRef] [PubMed]
- Sapunar, D.; Ljubkovic, M.; Lirk, P.; McCallum, J.B.; Hogan, Q.H. Distinct membrane effects of spinal nerve ligation on injured and adjacent dorsal root ganglion neurons in rats. Anesthesiology 2005, 103, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Djouhri, L.; Zeidan, A.; Alzoghaibi, M.; Al Otaibi, M.F.; Abd El-Aleem, S.A. L5 Spinal Nerve Axotomy Induces Distinct Electrophysiological Changes in Axotomized L5- and Adjacent L4-Dorsal Root Ganglion Neurons in Rats In Vivo. J. Neurotrauma 2021, 38, 330–341. [Google Scholar] [CrossRef]
- Schwab, J.M.; Frei, E.; Klusman, I.; Schnell, L.; Schwab, M.E.; Schluesener, H.J. AIF-1 expression defines a proliferating and alert microglial/macrophage phenotype following spinal cord injury in rats. J. Neuroimmunol. 2001, 119, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D. Anatomy of a discovery: M1 and M2 macrophages. Front. Immunol. 2015, 6, 212. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Huang, S.C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef]
- Li, Y.; Ji, A.; Weihe, E.; Schafer, M.K. Cell-specific expression and lipopolysaccharide-induced regulation of tumor necrosis factor alpha (TNFalpha) and TNF receptors in rat dorsal root ganglion. J. Neurosci. 2004, 24, 9623–9631. [Google Scholar] [CrossRef]
- Li, L.R.; Bai, L.Y.; Yang, K.L.; Zhang, J.; Gao, Y.; Jiang, M.J.; Yang, Y.; Zhang, X.; Wang, L.; Wang, X.L.; et al. KDM6B epigenetically regulated-interleukin-6 expression in the dorsal root ganglia and spinal dorsal horn contributes to the development and maintenance of neuropathic pain following peripheral nerve injury in male rats. Brain Behav. Immun. 2021, 98, 265–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Li, L.; Bai, L.; Gao, Y.; Yang, Y.; Wang, L.; Qiao, Y.; Wang, X.; Xu, J.T. Activation of Double-Stranded RNA-Activated Protein Kinase in the Dorsal Root Ganglia and Spinal Dorsal Horn Regulates Neuropathic Pain Following Peripheral Nerve Injury in Rats. Neurother. J. Am. Soc. Exp. NeuroTherapeutics 2022, 19, 1381–1400. [Google Scholar] [CrossRef] [PubMed]
- Li, X.N.; Yang, H.Q.; Ouyang, Q.; Liu, F.T.; Li, J.; Xiang, Z.H.; Yuan, H.B. Enhanced RAGE Expression in the Dorsal Root Ganglion May Contribute to Neuropathic Pain Induced by Spinal Nerve Ligation in Rats. Pain Med. 2016, 17, 803–812. [Google Scholar] [CrossRef]
- Ousingsawat, J.; Wanitchakool, P.; Kmit, A.; Romao, A.M.; Jantarajit, W.; Schreiber, R.; Kunzelmann, K. Anoctamin 6 mediates effects essential for innate immunity downstream of P2x7 receptors in macrophages. Nat. Commun. 2015, 6, 6245. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, J.J.; Pelegrin, P. Assessment of Cell Adhesion After Purinoceptor Activation. Methods Mol. Biol. 2020, 2041, 351–358. [Google Scholar] [CrossRef]
- Di Virgilio, F.; Dal Ben, D.; Sarti, A.C.; Giuliani, A.L.; Falzoni, S. The P2x7 Receptor in Infection and Inflammation. Immunity 2017, 47, 15–31. [Google Scholar] [CrossRef]
- Pelegrin, P.; Surprenant, A. Dynamics of macrophage polarization reveal new mechanism to inhibit IL-1beta release through pyrophosphates. EMBO J. 2009, 28, 2114–2127. [Google Scholar] [CrossRef]
- Franceschini, A.; Capece, M.; Chiozzi, P.; Falzoni, S.; Sanz, J.M.; Sarti, A.C.; Bonora, M.; Pinton, P.; Di Virgilio, F. The P2x7 receptor directly interacts with the NLRP3 inflammasome scaffold protein. FASEB J. 2015, 29, 2450–2461. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.J.; Feng, D.T.; Zhang, X.X.; Xia, C.C.; Zhang, Z.P.; Kang, J.H.; Tan, Z.Y.; Wu, B. Involvement of P2x7 receptors in satellite glial cells of dorsal root ganglia in the BmK I -induced pain model of rats. Gen. Physiol. Biophys. 2019, 38, 407–416. [Google Scholar] [CrossRef] [PubMed]
- de Torre-Minguela, C.; Barbera-Cremades, M.; Gomez, A.I.; Martin-Sanchez, F.; Pelegrin, P. Macrophage activation and polarization modify P2x7 receptor secretome influencing the inflammatory process. Sci. Rep. 2016, 6, 22586. [Google Scholar] [CrossRef]
- Niemi, J.P.; DeFrancesco-Lisowitz, A.; Roldan-Hernandez, L.; Lindborg, J.A.; Mandell, D.; Zigmond, R.E. A critical role for macrophages near axotomized neuronal cell bodies in stimulating nerve regeneration. J. Neurosci. 2013, 33, 16236–16248. [Google Scholar] [CrossRef]
- Caroleo, M.C.; Costa, N.; Tirassa, P.; Aloe, L. Nerve growth factor produced by activated human monocytes/macrophages is severely affected by ethanol. Alcohol 2004, 34, 107–114. [Google Scholar] [CrossRef]
- Barouch, R.; Appel, E.; Kazimirsky, G.; Brodie, C. Macrophages express neurotrophins and neurotrophin receptors. Regulation of nitric oxide production by NT-3. J. Neuroimmunol. 2001, 112, 72–77. [Google Scholar] [CrossRef]
- Barrette, B.; Hebert, M.A.; Filali, M.; Lafortune, K.; Vallieres, N.; Gowing, G.; Julien, J.P.; Lacroix, S. Requirement of myeloid cells for axon regeneration. J. Neurosci. 2008, 28, 9363–9376. [Google Scholar] [CrossRef]
- Hind, L.E.; Lurier, E.B.; Dembo, M.; Spiller, K.L.; Hammer, D.A. Effect of M1-M2 Polarization on the Motility and Traction Stresses of Primary Human Macrophages. Cell Mol. Bioeng. 2016, 9, 455–465. [Google Scholar] [CrossRef]
- Cui, K.; Ardell, C.L.; Podolnikova, N.P.; Yakubenko, V.P. Distinct Migratory Properties of M1, M2, and Resident Macrophages Are Regulated by alpha(D)beta(2) and alpha(M)beta(2) Integrin-Mediated Adhesion. Front. Immunol. 2018, 9, 2650. [Google Scholar] [CrossRef] [PubMed]
- Davis-Taber, R.A.; Scott, V.E.S. Transcriptional profiling of dorsal root ganglia in a neuropathic pain model using microarray and laser capture microdissection. Drug Dev. Res. 2006, 67, 308–330. [Google Scholar] [CrossRef]
- Jeon, S.M.; Lee, K.M.; Cho, H.J. Expression of monocyte chemoattractant protein-1 in rat dorsal root ganglia and spinal cord in experimental models of neuropathic pain. Brain Res. 2009, 1251, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Engle, M.P.; Merrill, M.A.; De Prado, B.M.; Hammond, D.L. Spinal nerve ligation decreases gamma-aminobutyric acid(B) receptors on specific populations of immunohistochemically identified neurons in L5 dorsal root ganglion of the rat. J. Comp. Neurol. 2012, 520, 1663–1677. [Google Scholar] [CrossRef] [PubMed]
- Hammond, D.L.; Ackerman, L.; Holdsworth, R.; Elzey, B. Effects of spinal nerve ligation on immunohistochemically identified neurons in the L4 and L5 dorsal root ganglia of the rat. J. Comp. Neurol. 2004, 475, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Iwai, H.; Ataka, K.; Suzuki, H.; Dhar, A.; Kuramoto, E.; Yamanaka, A.; Goto, T. Tissue-resident M2 macrophages directly contact primary sensory neurons in the sensory ganglia after nerve injury. J. Neuroinflamm. 2021, 18, 227. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Hayashi, S.; Ibuki, T.; Tanaka, M.; Ibata, Y. Morphological and Functional Roles of Macrophages and Postganglionic Sympathetic Fibers in Dorsal Root Ganglions of a Rat Neuropathic Pain Model. Acta Histochem. Cytochem. 1999, 32, 85–93. [Google Scholar] [CrossRef]
- Bowen, S.; Ateh, D.D.; Deinhardt, K.; Bird, M.M.; Price, K.M.; Baker, C.S.; Robson, J.C.; Swash, M.; Shamsuddin, W.; Kawar, S.; et al. The phagocytic capacity of neurones. Eur. J. Neurosci. 2007, 25, 2947–2955. [Google Scholar] [CrossRef] [PubMed]
- Abram, S.E.; Yi, J.; Fuchs, A.; Hogan, Q.H. Permeability of injured and intact peripheral nerves and dorsal root ganglia. Anesthesiology 2006, 105, 146–153. [Google Scholar] [CrossRef]
- Rastogi, V.; Sharma, R.; Misra, S.R.; Yadav, L.; Sharma, V. Emperipolesis—A review. J. Clin. Diagn. Res. JCDR 2014, 8, ZM01–ZM02. [Google Scholar] [CrossRef]
- Ng, Y.K.; Ling, E.A. Emperipolesis of lymphoid cells in vagal efferent neurons following an intraneural injection of ricin into the vagus nerve in rats. Neurosci. Lett. 1999, 270, 153–156. [Google Scholar] [CrossRef]
- Fais, S.; Overholtzer, M. Cell-in-cell phenomena in cancer. Nat. Rev. Cancer 2018, 18, 758–766. [Google Scholar] [CrossRef]
- Borensztejn, K.; Tyrna, P.; Gawel, A.M.; Dziuba, I.; Wojcik, C.; Bialy, L.P.; Mlynarczuk-Bialy, I. Classification of Cell-in-Cell Structures: Different Phenomena with Similar Appearance. Cells 2021, 10, 2569. [Google Scholar] [CrossRef]
- Kramer, P.R.; Puri, J.; Bellinger, L.L. Knockdown of Fcgamma receptor III in an arthritic temporomandibular joint reduces the nociceptive response in rats. Arthritis Rheum. 2010, 62, 3109–3118. [Google Scholar] [CrossRef]
- Celik, M.O.; Labuz, D.; Keye, J.; Glauben, R.; Machelska, H. IL-4 induces M2 macrophages to produce sustained analgesia via opioids. JCI Insight 2020, 5, e133093. [Google Scholar] [CrossRef]
- Kiguchi, N.; Kobayashi, Y.; Saika, F.; Sakaguchi, H.; Maeda, T.; Kishioka, S. Peripheral interleukin-4 ameliorates inflammatory macrophage-dependent neuropathic pain. Pain 2015, 156, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Gaojian, T.; Dingfei, Q.; Linwei, L.; Xiaowei, W.; Zheng, Z.; Wei, L.; Tong, Z.; Benxiang, N.; Yanning, Q.; Wei, Z.; et al. Parthenolide promotes the repair of spinal cord injury by modulating M1/M2 polarization via the NF-kappa B and STAT 1/3 signaling pathway. Cell Death Discov. 2020, 6, 97. [Google Scholar] [CrossRef] [PubMed]
- McWhorter, F.Y.; Wang, T.T.; Nguyen, P.; Chung, T.; Liu, W.F. Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. USA 2013, 110, 17253–17258. [Google Scholar] [CrossRef]
- Kim, M.; Jiang, L.H.; Wilson, H.L.; North, R.A.; Surprenant, A. Proteomic and functional evidence for a P2x7 receptor signalling complex. EMBO J. 2001, 20, 6347–6358. [Google Scholar] [CrossRef]
- Vogel, D.Y.S.; Heijnen, P.D.A.M.; Breur, M.; de Vries, H.E.; Tool, A.T.J.; Amor, S.; Dijkstra, C.D. Macrophages migrate in an activation-dependent manner to chemokines involved in neuroinflammation. J. Neuroinflamm. 2014, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Lever, I.J.; Bradbury, E.J.; Cunningham, J.R.; Adelson, D.W.; Jones, M.G.; McMahon, S.B.; Marvizon, J.C.G.; Malcangio, M. Brain-derived neurotrophic factor is released in the dorsal horn by distinctive patterns of afferent fiber stimulation. J. Neurosci. 2001, 21, 4469–4477. [Google Scholar] [CrossRef]
- Sikandar, S.; Minett, M.S.; Millet, Q.; Santana-Varela, S.; Lau, J.; Wood, J.N.; Zhao, J. Brain-derived neurotrophic factor derived from sensory neurons plays a critical role in chronic pain. Brain 2018, 141, 1028–1039. [Google Scholar] [CrossRef]
- Barker, P.A.; Mantyh, P.; Arendt-Nielsen, L.; Viktrup, L.; Tive, L. Nerve Growth Factor Signaling and Its Contribution to Pain. J. Pain. Res. 2020, 13, 1223–1241. [Google Scholar] [CrossRef]
- Eaton, M.J.; Blits, B.; Ruitenberg, M.J.; Verhaagen, J.; Oudega, M. Amelioration of chronic neuropathic pain after partial nerve injury by adeno-associated viral (AAV) vector-mediated over-expression of BDNF in the rat spinal cord. Gene Ther. 2002, 9, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Cejas, P.J.; Martinez, M.; Karmally, S.; McKillop, M.; McKillop, J.; Plunkett, J.A.; Oudega, M.; Eaton, M.J. Lumbar transplant of neurons genetically modified to secrete brain-derived neurotrophic factor attenuates allodynia and hyperalgesia after sciatic nerve constriction. Pain 2000, 86, 195–210. [Google Scholar] [CrossRef]
- Pezet, S.; Cunningham, J.; Patel, J.; Grist, J.; Gavazzi, I.; Lever, I.J.; Malcangio, M. BDNF modulates sensory neuron synaptic activity by a facilitation of GABA transmission in the dorsal horn. Mol. Cell Neurosci. 2002, 21, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, K.I.; Clayton, B.A.; Johnson, J.E.; Eisenach, J.C. Brain derived nerve growth factor induces spinal noradrenergic fiber sprouting and enhances clonidine analgesia following nerve injury in rats. Pain 2008, 136, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Mogil, J.S. Qualitative sex differences in pain processing: Emerging evidence of a biased literature. Nat. Rev. Neurosci. 2020, 21, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Niemi, J.P.; DeFrancesco-Lisowitz, A.; Cregg, J.M.; Howarth, M.; Zigmond, R.E. Overexpression of the monocyte chemokine CCL2 in dorsal root ganglion neurons causes a conditioning-like increase in neurite outgrowth and does so via a STAT3 dependent mechanism. Exp. Neurol. 2016, 275 Pt 1, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Lindborg, J.A.; Niemi, J.P.; Howarth, M.A.; Liu, K.W.; Moore, C.Z.; Mahajan, D.; Zigmond, R.E. Molecular and cellular identification of the immune response in peripheral ganglia following nerve injury. J. Neuroinflamm. 2018, 15, 192. [Google Scholar] [CrossRef]
- Komori, T.; Morikawa, Y.; Inada, T.; Hisaoka, T.; Senba, E. Site-specific subtypes of macrophages recruited after peripheral nerve injury. Neuroreport 2011, 22, 911–917. [Google Scholar] [CrossRef]
- Decosterd, I.; Woolf, C.J. Spared nerve injury: An animal model of persistent peripheral neuropathic pain. Pain 2000, 87, 149–158. [Google Scholar] [CrossRef]
- Lindenlaub, T.; Sommer, C. Partial sciatic nerve transection as a model of neuropathic pain: A qualitative and quantitative neuropathological study. Pain 2000, 89, 97–106. [Google Scholar] [CrossRef]
- Puljak, L.; Kojundzic, S.L.; Hogan, Q.H.; Sapunar, D. Targeted delivery of pharmacological agents into rat dorsal root ganglion. J. Neurosci. Methods 2009, 177, 397–402. [Google Scholar] [CrossRef]
- Fischer, G.; Kostic, S.; Nakai, H.; Park, F.; Sapunar, D.; Yu, H.; Hogan, Q. Direct injection into the dorsal root ganglion: Technical, behavioral, and histological observations. J. Neurosci. Methods 2011, 199, 43–55. [Google Scholar] [CrossRef]
- Sotocinal, S.G.; Sorge, R.E.; Zaloum, A.; Tuttle, A.H.; Martin, L.J.; Wieskopf, J.S.; Mapplebeck, J.C.; Wei, P.; Zhan, S.; Zhang, S.; et al. The Rat Grimace Scale: A partially automated method for quantifying pain in the laboratory rat via facial expressions. Mol. Pain 2011, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Nirogi, R.; Goura, V.; Shanmuganathan, D.; Jayarajan, P.; Abraham, R. Comparison of manual and automated filaments for evaluation of neuropathic pain behavior in rats. J. Pharmacol. Toxicol. Methods 2012, 66, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Moriarty, O.; Roche, M.; McGuire, B.E.; Finn, D.P. Validation of an air-puff passive-avoidance paradigm for assessment of aversive learning and memory in rat models of chronic pain. J. Neurosci. Methods 2012, 204, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Xu, Y.; Han, F.; Sun, D.; Zhang, H.; Li, X.; Yao, X.; Wang, H. Quercetin alleviates thermal and cold hyperalgesia in a rat neuropathic pain model by inhibiting Toll-like receptor signaling. Biomed. Pharmacother. 2017, 94, 652–658. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Mattfeldt, T.; Mall, G.; Gharehbaghi, H.; Moller, P. Estimation of surface area and length with the orientator. J. Microsc. 1990, 159, 301–317. [Google Scholar] [CrossRef]
- A Simple Python Interface for Axon Binary Format (ABF) Files, H., SW. pyABF 2.3.5. Available online: https://pypi.org/project/pyabf (accessed on 19 September 2022).
- Eyo, U.B.; Miner, S.A.; Ahlers, K.E.; Wu, L.J.; Dailey, M.E. P2x7 receptor activation regulates microglial cell death during oxygen-glucose deprivation. Neuropharmacology 2013, 73, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Meijering, E.; Jacob, M.; Sarria, J.C.F.; Steiner, P.; Hirling, H.; Unser, M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytom. Part A 2004, 58A, 167–176. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gheorghe, R.-O.; Grosu, A.V.; Magercu, M.; Ghenghea, M.-S.; Zbarcea, C.E.; Tanase, A.; Negres, S.; Filippi, A.; Chiritoiu, G.; Gherghiceanu, M.; et al. Switching Rat Resident Macrophages from M1 to M2 Phenotype by Iba1 Silencing Has Analgesic Effects in SNL-Induced Neuropathic Pain. Int. J. Mol. Sci. 2023, 24, 15831. https://doi.org/10.3390/ijms242115831
Gheorghe R-O, Grosu AV, Magercu M, Ghenghea M-S, Zbarcea CE, Tanase A, Negres S, Filippi A, Chiritoiu G, Gherghiceanu M, et al. Switching Rat Resident Macrophages from M1 to M2 Phenotype by Iba1 Silencing Has Analgesic Effects in SNL-Induced Neuropathic Pain. International Journal of Molecular Sciences. 2023; 24(21):15831. https://doi.org/10.3390/ijms242115831
Chicago/Turabian StyleGheorghe, Roxana-Olimpia, Andreea Violeta Grosu, Melania Magercu, Mihail-Sebastian Ghenghea, Cristina Elena Zbarcea, Alexandra Tanase, Simona Negres, Alexandru Filippi, Gabriela Chiritoiu, Mihaela Gherghiceanu, and et al. 2023. "Switching Rat Resident Macrophages from M1 to M2 Phenotype by Iba1 Silencing Has Analgesic Effects in SNL-Induced Neuropathic Pain" International Journal of Molecular Sciences 24, no. 21: 15831. https://doi.org/10.3390/ijms242115831





