Label-Free Quantitative Proteomics Reveal the Mechanisms of Young Wheat (Triticum aestivum L.) Ears’ Response to Spring Freezing
Abstract
:1. Introduction
2. Results
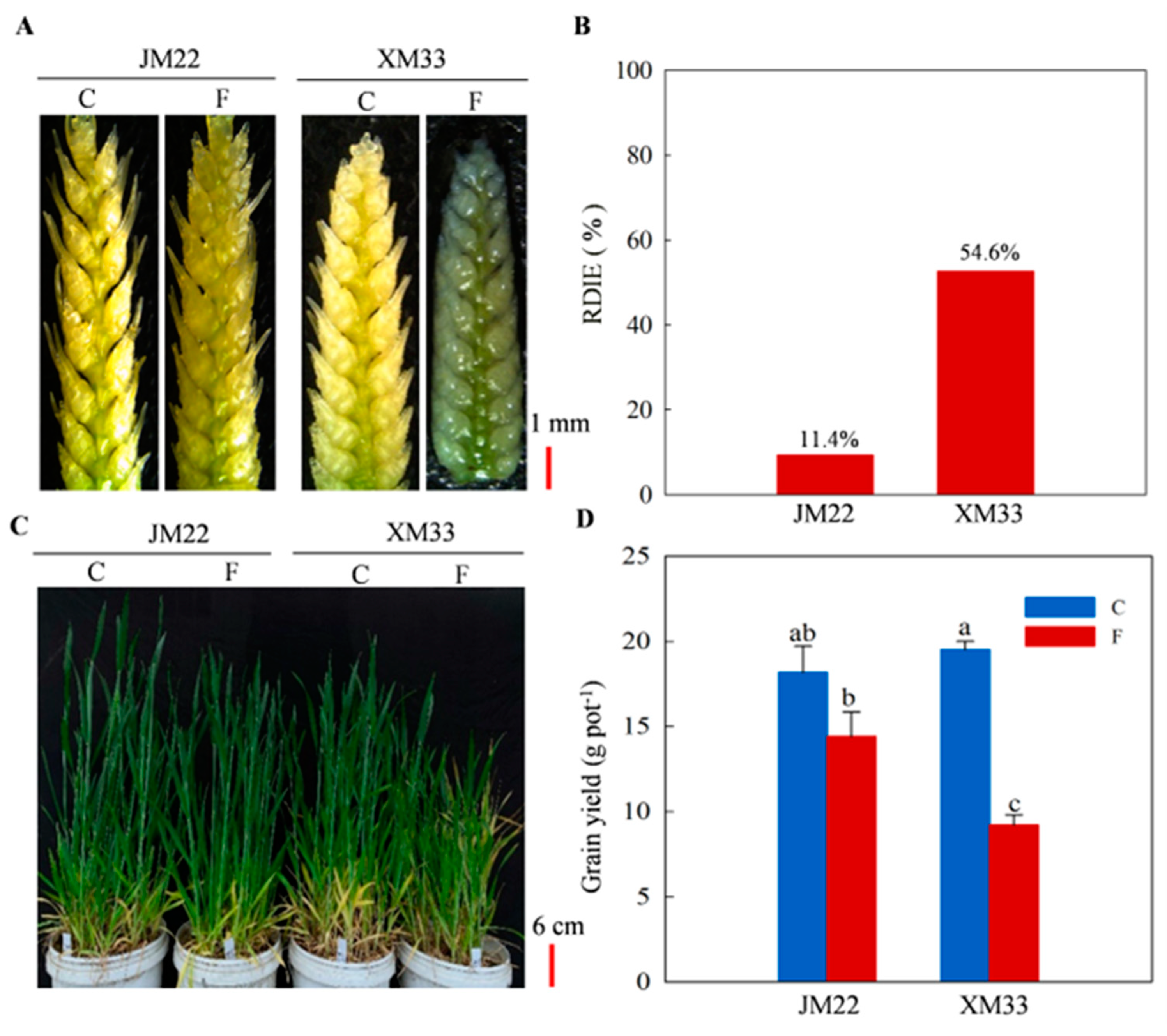
2.1. Phenotypic Differences between JM22 and XM33 under Freezing Stress
2.2. Physiological Response of Young Ears to Freezing Stress
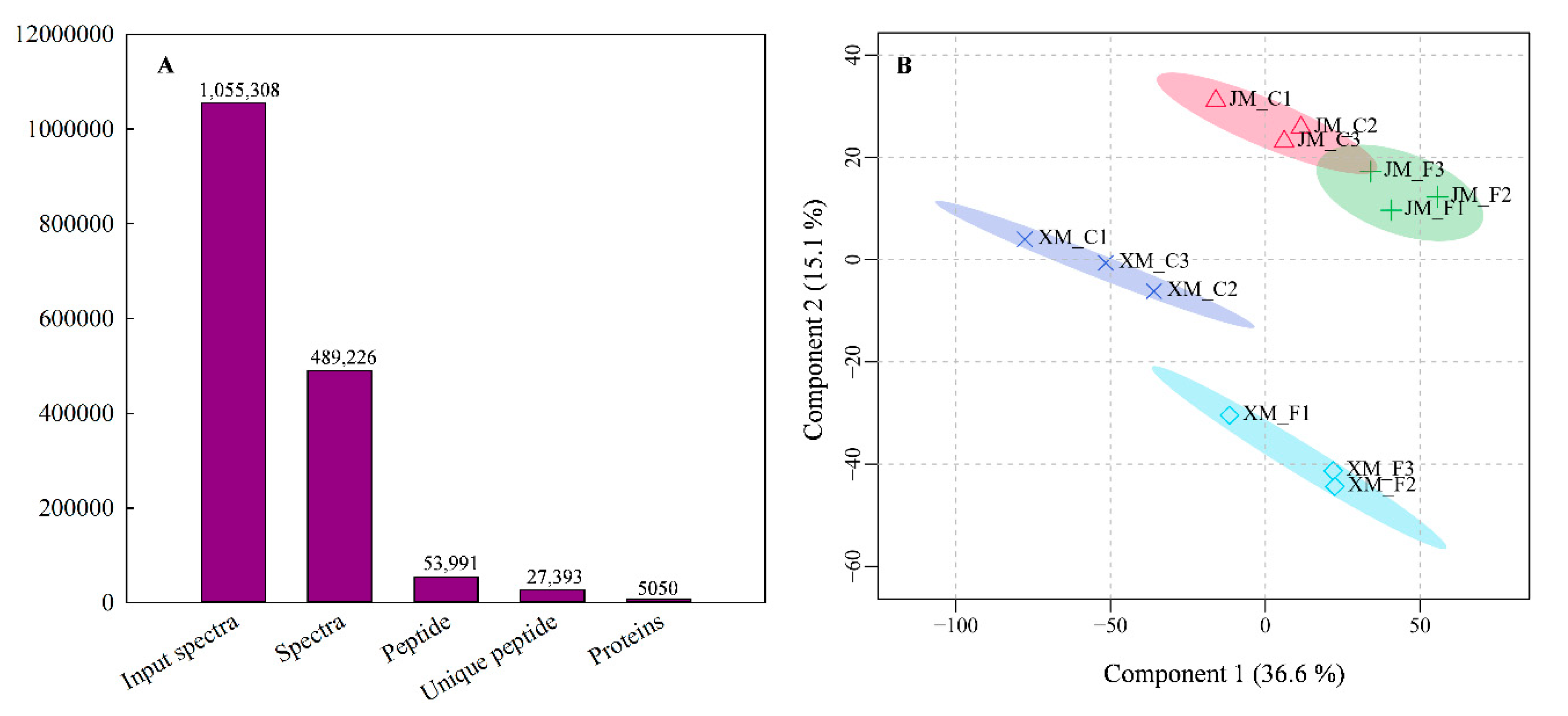
2.3. Protein Identification and Quantification
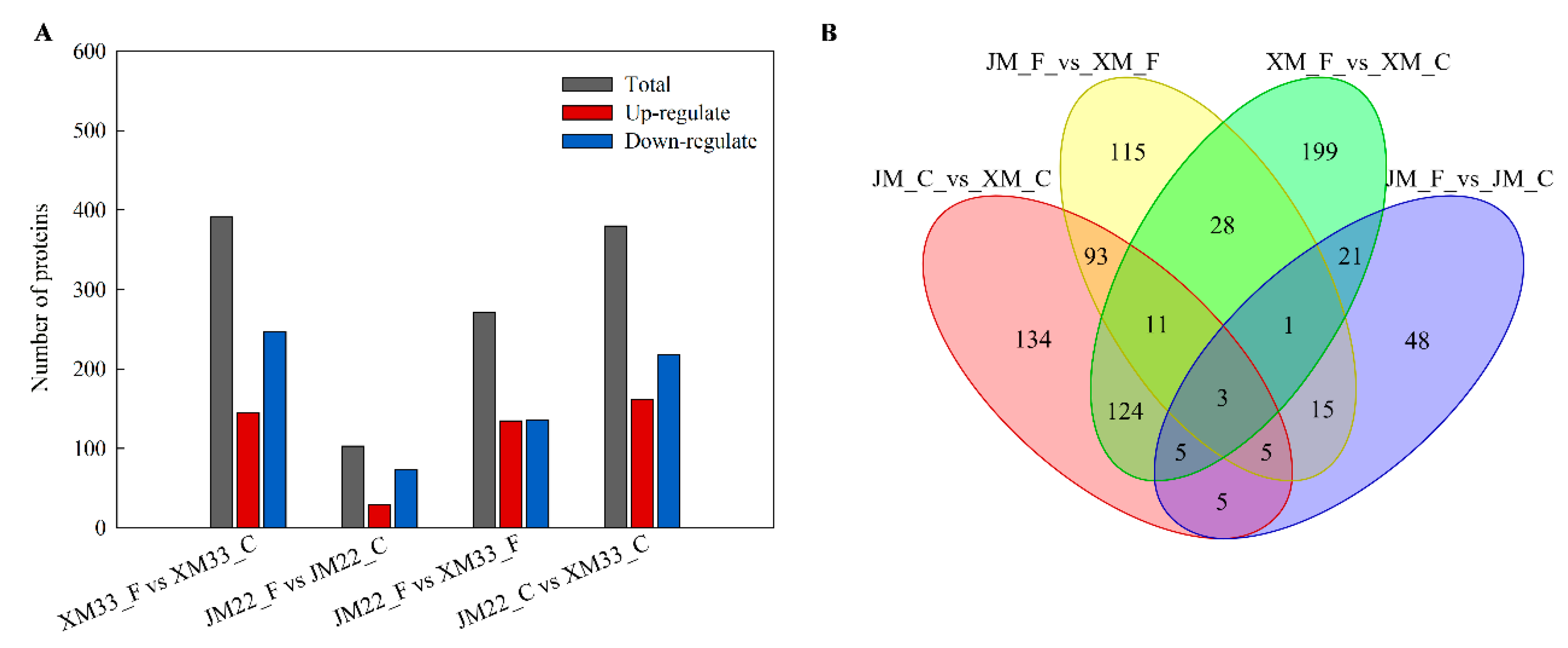
2.4. Identification and Analysis of Differently Expressed Proteins
2.5. Functional Analysis of the DEPs in Different Comparison Groups
2.6. Key Proteins Associated with Freezing Stress Response in the Young Ears
2.6.1. Antioxidant-Related Proteins
2.6.2. Heat Shock Proteins
2.6.3. Cell Wall−Modifying Related Proteins
3. Discussion
3.1. Enhancing Antioxidant Capacity Was Beneficial for Young Wheat Ears’ Freezing Tolerance
3.2. Heat Shock Proteins Were Involved in Young Wheat Ears Coping with Freezing Stress
3.3. Changes in Cell Wall Traits Participated in the Response of Young Wheat Ears to Freezing Stress
3.4. Other Proteins May Contribute to Enhance Freezing Tolerance in Young Wheat Ears
3.5. Maintaining Transcriptional Activity Was a Crucial Biological Basis for Freezing Tolerance in Young Wheat Ears
4. Materials and Methods
4.1. Plant Materials, Growing Conditions, and Temperature Treatments
4.2. Young Ears’ Freezing Tolerance Determination
4.3. Physiological Parameters Measurement
4.4. Gene Expression Analysis
4.5. Total Protein Extraction and Digestion
4.6. LC–MS/MS Analysis
4.7. Sequence Database Search
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.; Jiao, G.; Sun, Y.; Chen, J.; Zhong, Y.; Yan, L.; Jiang, D.; Ma, Y.; Xia, L. Modification of starch composition, structure and properties through editing of TaSBEIIa in both winter and spring wheat varieties by CRISPR/Cas9. Plant Biotechnol. J. 2021, 19, 937–951. [Google Scholar] [CrossRef]
- Holman, J.D.; Schlegel, A.J.; Thompson, C.R.; Lingenfelser, J.E. Influence of precipitation, temperature, and 56 years on winter wheat yields in western Kansas. Crop Manag. 2011, 10, 1–10. [Google Scholar] [CrossRef]
- Zohner, C.M.; Mo, L.; Renner, S.S.; Svenning, J.C.; Vitasse, Y.; Benito, B.M.; Ordonez, A.; Baumgarten, F.; Sebald, V.; Reich, P.B. Late-spring frost risk between 1959 and 2017 decreased in North America but increased in Europe and Asia. Proc. Natl. Acad. Sci. USA 2020, 117, 12192–12200. [Google Scholar] [CrossRef]
- Onyemaobi, O.; Sangma, H.; Garg, G.; Wallace, X.; Kleven, S.; Dolferus, R. Transcriptome profiling of the chilling response in wheat spikes: I, acclimation response to long-term chilling treatment. Curr. Plant Biol. 2022, 31, 100255. [Google Scholar]
- Lin, F.; Li, C.; Xu, B.; Chen, J.; Chen, A.; Hassan, M.A.; Liu, B.; Xu, H.; Chen, X.; Sun, J.; et al. Late spring cold reduces grain number at various spike positions by regulating spike growth and assimilate distribution in winter wheat. Crop J. 2023, 11, 1272–1278. [Google Scholar] [CrossRef]
- Wang, W.; Wang, X.; Zhang, J.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. Salicylic acid and cold priming induce late-spring freezing tolerance by maintaining cellular redox homeostasis and protecting photosynthetic apparatus in wheat. Plant Growth Regul. 2021, 90, 109–121. [Google Scholar] [CrossRef]
- Hassan, M.A.; Xiang, C.; Farooq, M.; Muhammad, N.; Yan, Z.; Hui, X.; Yuanyuan, K.; Bruno, A.K.; Lele, Z.; Li, J.C. Cold stress in wheat: Plant acclimation responses and management strategies. Front. Plant Sci. 2021, 12, 676884. [Google Scholar] [CrossRef]
- Zhong, X.; Mei, X.; Li, Y.; Yoshida, H.; Zhao, P.; Wang, X.; Han, L.; Hu, X.; Huang, S.; Huang, J. Changes in frost resistance of wheat young ears with development during jointing stage. J. Agron. Crop Sci. 2008, 194, 343–349. [Google Scholar] [CrossRef]
- Jiang, G.; Hassan, M.A.; Muhammad, N.; Arshad, M.; Chen, X.; Xu, Y.; Xu, H.; Ni, Q.; Liu, B.; Yang, W.; et al. Comparative physiology and transcriptome analysis of young spikes in response to late spring coldness in wheat (Triticum aestivum L.). Front. Plant Sci. 2022, 13, 811884. [Google Scholar] [CrossRef]
- Yue, Y.J.; Zhou, Y.; Wang, J.; Ye, X.Y. Assessing wheat frost risk with the support of GIS: An approach coupling a growing season meteorological index and a hybrid fuzzy neural network model. Sustainability 2016, 8, 1308. [Google Scholar] [CrossRef]
- Xiao, L.; Liu, L.; Asseng, S.; Xia, Y.; Tang, L.; Liu, B.; Cao, W.; Zhu, Y. Estimating spring frost and its impact on yield across winter wheat in China. Agric. Forest Meteorol. 2018, 260, 154–164. [Google Scholar]
- Li, X.; Pu, H.; Liu, F.; Zhou, Q.; Cai, J.; Dai, T.; Cao, W.; Jiang, D. Winter wheat photosynthesis and grain yield responses to spring freeze. Agron. J. 2015, 107, 1002–1010. [Google Scholar]
- Zhang, W.; Wang, J.; Huang, Z.; Mi, L.; Jiang, D. Effects of low temperature at booting stage on sucrose metabolism and endogenous hormone contents in winter wheat spikelet. Front. Plant Sci. 2019, 10, 498. [Google Scholar] [CrossRef] [PubMed]
- Cheong, B.E.; Onyemaobi, O.; Ho, W.W.H.; Biddulph, T.B.; Rupasinghe, T.W.T.; Roessner, U.; Dolferus, R. Phenotyping the chilling and freezing responses of young microspore stage wheat spikes using targeted metabolome and lipidome profiling. Cells 2020, 9, 1309. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Qu, L.; Hu, Y.; Lu, W.; Lu, D. Proteomics reveals the effects of drought stress on the kernel development and starch formation of waxy maize. BMC Plant Biol. 2021, 21, 434. [Google Scholar]
- Han, Q.; Kang, G.; Guo, T. Proteomic analysis of spring freeze-stress responsive proteins in leaves of bread wheat (Triticum aestivum L.). Plant Physiol. Biochem. 2013, 63, 236–244. [Google Scholar]
- Zhang, S.; Song, G.; Li, Y.; Gao, J.; Wang, J.; Chen, G.; Li, H.; Li, G.; Zhao, Z. Comparative proteomic analysis of cold responsive proteins in two wheat cultivars with different tolerance to spring radiation frost. Front. Agric. Sci. Eng. 2014, 1, 37–45. [Google Scholar]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Breusegem, F.V. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Li, X.; Cai, J.; Liu, F.; Dai, T.; Cao, W.; Jiang, D. Cold priming drives the sub-cellular antioxidant systems to protect photosynthetic electron transport against subsequent low temperature stress in winter wheat. Plant Physiol. Biochem. 2014, 82, 34–43. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 1360–1385. [Google Scholar]
- Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.X.; Zhang, H.X.; Wei, A.M.; Gong, Z.H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar]
- Ruellan, E.; Vaultier, M.; Zachowski, A.; Hurry, V. Cold signalling and cold acclimation in plants. Adv. Bot. Res. 2009, 49, 35–150. [Google Scholar]
- Panter, P.E.; Panter, J.R.; Knight, H. Impact of cell-wall structure and composition on plant freezing tolerance. Annu. Plant Rev. 2003, 3, 607–642. [Google Scholar]
- Baldwin, L.; Domon, J.M.; Klimek, J.F.; Fournet, F.; Sellier, H.; Gillet, F.; Pelloux, J.; Lejeune-Hénaut, I.; Carpita, N.C.; Rayon, C. Structural alteration of cell wall pectins accompanies pea development in response to cold. Phytochemistry 2014, 104, 37–47. [Google Scholar] [PubMed]
- Ji, H.; Wang, Y.; Cloix, C.; Li, K.; Jenkins, G.I.; Wang, S.; Shang, Z.; Shi, Y.; Yang, S.; Li, X. The Arabidopsis RCC1 family protein TCF1 regulates freezing tolerance and cold acclimation through modulating lignin biosynthesis. PLoS Genet. 2015, 11, e1005471. [Google Scholar]
- Xue, G.P.; McIntyre, C.L.; Jenkins, C.L.; Glassop, D.; van Herwaarden, A.F.; Shorter, R. Molecular dissection of variation in carbohydrate metabolism related to water-soluble carbohydrate accumulation in stems of wheat. Plant Physiol. 2008, 146, 441–454. [Google Scholar] [PubMed]
- Solecka, D.; Żebrowski, J.; Kacperska, A. Are pectins involved in cold acclimation and de-acclimation of winter oil-seed rape plants? Ann. Bot. 2008, 101, 521–530. [Google Scholar]
- Srivastava, S.; Vishwakarma, R.K.; Arafat, Y.A.; Gupta, S.K.; Khan, B.M. Abiotic stress induces change in Cinnamoyl CoA Reductase (CCR) protein abundance and lignin deposition in developing seedlings of Leucaena leucocephala. Physiol. Mol. Biol. Plants 2015, 21, 197–205. [Google Scholar]
- Liu, W.; Jin, Y.; Li, M.; Dong, L.; Guo, D.; Lu, C.; Qi, H. Analysis of CmCADs and three lignifying enzymes in oriental melon (‘CaiHong7’) seedlings in response to three abiotic stresses. Sci. Hortic. 2018, 237, 257–268. [Google Scholar]
- Lin, J.S.; Huang, X.X.; Li, Q.; Cao, Y.; Bao, Y.; Meng, X.F.; Li, Y.J.; Fu, C.; Hou, B.K. UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J. 2016, 88, 26–42. [Google Scholar] [CrossRef]
- Willick, I.R.; Takahashi, D.; Fowler, D.B.; Uemura, M.; Tanino, K.K. Tissue-specific changes in apoplastic proteins and cell wall structure during cold acclimation of winter wheat crowns. J. Exp. Bot. 2018, 69, 1221–1234. [Google Scholar] [PubMed]
- Miedes, E.; Lorences, E.P. Xyloglucan endotransglucosylase/hydrolases (XTHs) during tomato fruit growth and ripening. J. Plant Physiol. 2009, 166, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Johnson, K.L.; Hao, P.; Tuong, T.; Erban, A.; Sampathkumar, A.; Bacic, A.; Livingston, D.P., III; Kopka, J.; Kuroha, T. Cell wall modification by the xyloglucan endotransglucosylase/hydrolase XTH19 influences freezing tolerance after cold and sub-zero acclimation. Plant Cell Environ. 2021, 44, 915–930. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, X.; Wang, X.; Wang, B.; Li, H.; Feng, J.; Wu, A. Mutagenesis of UDP-xylose epimerase and xylan arabinosyl-transferase decreases arabinose content and improves saccharification of rice straw. Plant Biotechnol. J. 2021, 19, 863–865. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Griffith, M.; Wiseman, S. Ethylene induces antifreeze activity in winter rye leaves. Plant Physiol. 2001, 126, 1232–1240. [Google Scholar] [PubMed]
- Isobe, A.; Kuwabara, C.; Koike, M.; Sutoh, K.; Sasaki, K.; Imai, R. An apoplastic defensin of wheat elicits the production of extracellular polysaccharides in snow mold. Plants 2021, 10, 1607. [Google Scholar] [CrossRef] [PubMed]
- Hiilovaara-Teijo, M.; Hannukkala, A.; Griffith, M.; Yu, X.; Pihakaski-Maunsbach, K. Snow-mold-induced apoplastic proteins in winter rye leaves lack antifreeze activity. Plant Physiol. 1999, 121, 665–673. [Google Scholar]
- Guo, T.; Weber, H.C.E.; Niemann, M.; Theisl, L.; Leonte, G.; Novák, O.; Werner, T. Arabidopsis HIPP proteins regulate endoplasmic reticulum-associated degradation of CKX proteins and cytokinin responses. Mol. Plant 2021, 14, 1918–1934. [Google Scholar]
- Zhang, X.; Feng, H.; Feng, C.; Xu, H.; Huang, X.; Wang, Q.; Duan, X.; Wang, X.; Wei, G.; Huang, L.; et al. Isolation and characterisation of cDNA encoding a wheat heavy metal-associated isoprenylated protein involved in stress responses. Plant Biol. 2015, 17, 1176–1186. [Google Scholar]
- de Abreu-Neto, J.B.; Turchetto-Zolet, A.C.; de Oliveira, L.F.V.; Bodanese Zanettini, M.H.; Margis-Pinheiro, M. Heavy metal-associated isoprenylated plant protein (HIPP): Characterization of a family of proteins exclusive to plants. FEBS J. 2013, 280, 1604–1616. [Google Scholar] [CrossRef]
- Barth, O.; Vogt, S.; Uhlemann, R.; Zschiesche, W.; Humbeck, K. Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29. Plant Mol. Biol. 2009, 69, 213–226. [Google Scholar] [CrossRef]
- Kurepin, L.V.; Dahal, K.P.; Savitch, L.V.; Singh, J.; Bode, R.; Ivanov, A.G.; Hurry, V.; Hüner, N.P.A. Role of CBFs as integrators of chloroplast redox, phytochrome and plant hormone signaling during cold acclimation. Int. J. Mol. Sci. 2012, 14, 12729–12763. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, X.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Cao, W.; Jiang, D. Hydrogen peroxide and abscisic acid mediate salicylic acid-induced freezing tolerance in wheat. Front. Plant Sci. 2018, 9, 1137. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]



| Protein ID | Protein Description | XM33_F vs. XM33_C | JM22_F vs. JM22_C |
|---|---|---|---|
| A0A3B6HTB1 | Heavy metal−associated isoprenylated plant protein 33−like | up | up |
| W5B397 | Heavy metal−associated isoprenylated plant protein 1 | up | up |
| A0A3B6TCM5 | NUC153 domain−containing protein | up | up |
| A0A3B6KR91 | GLTP domain−containing protein | up | up |
| A0A3B6TGN2 | Polygalacturonase inhibitor−like | up | up |
| W5BGU5 | Mediator of RNA polymerase II transcription subunit 7 | up | up |
| A0A3B6C0Y8 | DIBOA−glucoside dioxygenase BX6−like | up | up |
| A0A3B6FJ71 | Lactamase_B domain−containing protein | up | up |
| A0A3B6CBP0 | Coatomer subunit epsilon | down | down |
| A0A3B5YVU9 | Methyltransferase | down | down |
| A0A3B6TGT6 | Curvature thylakoid 1A, chloroplastic−like protein | down | down |
| A0A3B6RGZ0 | Serinethreonine−protein phosphatase 4 regulatory subunit 2−A−like | down | down |
| W4ZSG9 | Thioredoxin−like protein YLS8 | down | down |
| A0A1D6RNK4 | E3 ubiquitin−protein ligase ARI8 | down | down |
| A0A3B6H1K4 | Coatomer subunit zeta | down | down |
| Q36813 | NAD(P)H dehydrogenase subunit H | down | down |
| Q76ME3 | ADP−ribosylation factor | down | down |
| A0A3B5Y6Q2 | SAC domain−containing protein | down | down |
| A0A3B6TKQ8 | COP9 signalosome complex subunit 3−like isoform X2 | down | down |
| Q6KCK6 | Putative calcium−dependent protein kinase | down | down |
| W5EJ68 | Protein kinase domain-containing protein | down | down |
| A0A3B6JPZ5 | Uncharacterized protein | down | down |
| A0A3B6IPS7 | Uncharacterized protein | down | down |
| A0A3B6N1N4 | Uncharacterized protein | down | down |
| A0A3B6KJM2 | Uncharacterized protein | down | down |
| A0A077RWW5 | Uncharacterized protein | down | down |
| A0A3B5XX28 | Uncharacterized protein | down | down |
| A0A3B6LPS5 | Peroxidase | up | down |
| W5AMD3 | Defensin PDF10 | down | up |
| A0A3B6R8W9 | Uncharacterized protein | down | up |
| Groups | Protein ID | Protein Description | Fold-Change |
|---|---|---|---|
| XM33_F vs. XM33_C | A0A3B6IUJ0 | GST N−terminal domain−containing protein | 1.887 |
| A0A3B6LPS5 | Peroxidase | 1.853 | |
| A0A3B5XVD9 | Probable glutathione S−transferase−cytosolic | 1.839 | |
| A0A3B5YRG4 | Probable glutathione S−transferase DHAR10, cytosolic | 1.754 | |
| A0A3B6JN31 | Glutathione transferase | 1.713 | |
| A0A3B6B868 | Thioredoxin domain−containing protein | 1.705 | |
| A0A3B6ED41 | 2−alkenal reductase (NADP(+) dependent) −like | 1.599 | |
| A0A3B6DBD5 | Thioredoxin | 1.588 | |
| A0A3B6GVH3 | Glutaredoxin−dependent peroxiredoxin | 1.584 | |
| A0A3B6SJF8 | Glutaredoxin−dependent peroxiredoxin | 1.547 | |
| W4ZSG9 | Thioredoxin−like protein YLS8 | 0.495 | |
| A0A3B5YYX5 | Glutathione transferase | 0.342 | |
| JM22_F vs. JM22_C | A0A3B6DL50 | Thioredoxin domain−containing protein | 0.635 |
| A0A3B6SS74 | L−ascorbate oxidase homolog | 0.601 | |
| W4ZSG9 | Thioredoxin−like protein YLS8 | 0.593 | |
| A0A3B6LPS5 | Peroxidase | 0.243 | |
| JM22_F vs. XM33_F | A0A3B5YT02 | Peroxidase | 4.156 |
| A0A3B6CJF7 | Peroxidase | 2.589 | |
| A0A3B5ZRA7 | Peroxidase | 2.275 | |
| A0A3B6LUE2 | Glutathione transferase | 0.611 | |
| A0A3B6SS74 | L−ascorbate oxidase homolog | 0.515 | |
| A0A3B6JN31 | Glutathione transferase | 0.492 | |
| F1DKC1 | Catalase | 0.445 | |
| A0A3B6TWE0 | Peroxidase | 0.365 | |
| A0A3B6QC91 | Peroxidase | 0.250 | |
| JM22_C vs. XM33_C | A0A3B5YT02 | Peroxidase | 4.789 |
| A0A3B6LPS5 | Peroxidase | 3.526 | |
| A0A3B5ZRA7 | Peroxidase | 3.427 | |
| A0A3B6TQJ1 | Thioredoxin domain−containing protein | 3.021 | |
| A0A3B6CJF7 | Peroxidase | 2.500 | |
| A0A3B6IRX6 | L−ascorbate peroxidase | 2.222 | |
| A0A3B5YRG4 | Probable glutathione S−transferase DHAR1, cytosolic | 1.965 | |
| A0A3B6JL78 | L−ascorbate peroxidase | 1.701 | |
| A0A3B6AYZ6 | L−ascorbate peroxidase | 1.536 | |
| A0A1D6D173 | Glutaredoxin domain−containing protein | 1.522 | |
| A0A3B6DNN1 | Peroxidase | 0.597 | |
| F1DKC1 | Catalase | 0.537 | |
| A0A3B6KNG3 | Thioredoxin−like fold domain−containing protein | 0.290 | |
| A0A3B6QC91 | Peroxidase | 0.281 |
| Groups | Protein ID | Protein Description | Fold-Change |
|---|---|---|---|
| XM33_F vs. XM33_C | A0A3B6LMB4 | DEHY | 2.498 |
| A0A3B6PT83 | 18.6 kDa class III heat shock protein | 2.472 | |
| A0A3B5ZYT4 | Heat shock protein 101 | 2.133 | |
| A0A3B6RDZ6 | 26.2 kDa heat shock protein−mitochondrial−like | 2.127 | |
| A0A3B6CA96 | Hsp70−Hsp90 organizing protein−like | 2.093 | |
| A0A3B6JIR3 | Heat shock cognate 70 kDa protein 2−like | 1.571 | |
| A0A3B6KKG9 | Heat shock 70 kDa protein, mitochondrial−like | 1.547 | |
| F4Y594 | Heat shock protein 90 | 0.348 | |
| JM22_F vs. XM33_F | A0A3B6TG72 | BOBBER 1−like protein | 5.145 |
| A0A3B5XUY5 | DnaJ (HSP40) homolog subfamily B member 4−like | 2.125 | |
| A0A3B6GQT8 | DnaJ (HSP40) homolog subfamily B member 4−like | 1.502 | |
| A0A3B5ZYT4 | Heat shock protein 101 | 0.422 | |
| A0A3B6RDZ6 | 26.2 kDa heat shock protein, mitochondrial−like | 0.190 | |
| JM22_C vs. XM33_C | A0A3B6TFV1 | 26.2 kDa heat shock protein, mitochondrial−like | 6.016 |
| A0A3B6LMB4 | SHSP domain−containing protein | 1.894 | |
| A0A3B6RHD3 | 26.2 kDa heat shock protein, mitochondrial−like | 1.758 | |
| Q9ZP24 | 23.6 kDa heat shock protein | 1.587 | |
| A0A3B6RDZ6 | 26.2 kDa heat shock protein, mitochondrial−like | 0.235 |
| Groups | Protein ID | Protein Description | Fold-Change |
|---|---|---|---|
| XM33_F vs. XM33_C | A0A3B6TGN2 | Polygalacturonase inhibitor−like | 2.222 |
| A0A3B6H1P4 | Germin−like protein | 1.955 | |
| A0A1D5V3A3 | Pectinesterase inhibitor 7−like | 1.852 | |
| A0A3B6TWG3 | Xyloglucan endotransglucosylase/hydrolase | 1.833 | |
| A0A3B6CBN0 | Cinnamyl−alcohol dehydrogenase 7−like | 1.506 | |
| A0A3B6TI25 | Alpha−1,3−arabinosyltransferase XAT3 | 0.434 | |
| JM22_F vs. JM22_C | A0A3B6TGN2 | Polygalacturonase inhibitor−like | 1.779 |
| A0A3B6RK39 | Cinnamoyl−CoA reductase 1−like | 0.653 | |
| A0A3B6BYY2 | Probable cinnamyl−alcohol dehydrogenase 6 | 0.509 | |
| JM22_F vs. XM33_F | A0A3B6TGN2 | Polygalacturonase inhibitor−like | 1.790 |
| A0A3B6KU54 | Expansin−B4−like | 1.657 | |
| A0A3B6IKN1 | Endoglucanase | 0.653 | |
| A0A3B6TGM9 | Probable polygalacturonase isoform X1 protein | 0.610 | |
| Q56TP1 | Xyloglucan endotransglucosylase/hydrolase | 0.605 | |
| A0A3B6TWG3 | Xyloglucan endotransglucosylase/hydrolase | 0.587 | |
| A0A3B6CBN0 | Cinnamyl−alcohol dehydrogenase 7−like | 0.556 | |
| A0A3B5Y507 | Endoglucanase | 0.251 | |
| JM22_C vs. XM33_C | A0A3B6B5P1 | UDP−arabinopyranose mutase | 1.503 |
| A0A3B6DED8 | Xyloglucan endotransglucosylasehydrolase | 0.505 | |
| A0A3B6TI25 | Alpha−1,3−arabinosyltransferase XAT3−like | 0.436 | |
| A0A3B5Y507 | Endoglucanase | 0.091 | |
| A0A3B6DMR3 | Beta−D−xylosidase 4−like | 0.052 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Zhang, Y.; Liu, C.; Dong, Y.; Jiang, X.; Zhao, C.; Li, G.; Xu, K.; Huo, Z. Label-Free Quantitative Proteomics Reveal the Mechanisms of Young Wheat (Triticum aestivum L.) Ears’ Response to Spring Freezing. Int. J. Mol. Sci. 2023, 24, 15892. https://doi.org/10.3390/ijms242115892
Wang W, Zhang Y, Liu C, Dong Y, Jiang X, Zhao C, Li G, Xu K, Huo Z. Label-Free Quantitative Proteomics Reveal the Mechanisms of Young Wheat (Triticum aestivum L.) Ears’ Response to Spring Freezing. International Journal of Molecular Sciences. 2023; 24(21):15892. https://doi.org/10.3390/ijms242115892
Chicago/Turabian StyleWang, Weiling, Yuting Zhang, Chang Liu, Yongwen Dong, Xue Jiang, Can Zhao, Guohui Li, Ke Xu, and Zhongyang Huo. 2023. "Label-Free Quantitative Proteomics Reveal the Mechanisms of Young Wheat (Triticum aestivum L.) Ears’ Response to Spring Freezing" International Journal of Molecular Sciences 24, no. 21: 15892. https://doi.org/10.3390/ijms242115892








