Identification and Characterization of the HbPP2C Gene Family and Its Expression in Response to Biotic and Abiotic Stresses in Rubber Tree
Abstract
:1. Introduction
2. Results
2.1. Characterization and Analysis of PP2C Gene Family Members in Rubber Tree
| Gene Name | Gene ID | Length of CDS (bp) | Number of Exons | Predicted Protein | ||
|---|---|---|---|---|---|---|
| Siza (aa) | MW (Da) | PI | ||||
| HbPP2C1 | 110650351 | 1236 | 4 | 411 | 43,539.76 | 5.92 |
| HbPP2C2 | 110664056 | 681 | 4 | 226 | 25,130.49 | 5.41 |
| HbPP2C3 | 110644406 | 1311 | 3 | 436 | 36,712.72 | 5.47 |
| HbPP2C4 | 110640824 | 2001 | 5 | 666 | 65,573.51 | 5.39 |
| HbPP2C5 | 110666131 | 1287 | 7 | 428 | 46,120.9 | 6.40 |
| HbPP2C6 | 110670175 | 1251 | 4 | 416 | 45,312.93 | 5.21 |
| HbPP2C7 | 110659418 | 1506 | 4 | 501 | 54,241.09 | 4.62 |
| HbPP2C8 | 110653766 | 1086 | 6 | 361 | 39,711.86 | 6.21 |
| HbPP2C9 | 110667550 | 882 | 5 | 293 | 26,712.08 | 8.31 |
| HbPP2C10 | 110672253 | 846 | 5 | 281 | 30,766.68 | 8.70 |
| HbPP2C11 | 110634536 | 990 | 8 | 329 | 30,162.58 | 4.79 |
| HbPP2C12 | 110664354 | 1275 | 8 | 424 | 46,211.85 | 5.62 |
| HbPP2C13 | 110651406 | 861 | 1 | 286 | 18,178.68 | 5.63 |
| HbPP2C14 | 110639512 | 999 | 4 | 332 | 33,255.94 | 7.96 |
| HbPP2C15 | 110666808 | 1539 | 5 | 512 | 30,078.9 | 8.72 |
| HbPP2C16 | 110641533 | 1638 | 4 | 545 | 58,926.58 | 4.71 |
| HbPP2C17 | 110652402 | 1032 | 10 | 343 | 39,528.75 | 5.39 |
| HbPP2C18 | 110638740 | 1515 | 6 | 504 | 56,592.87 | 5.49 |
| HbPP2C19 | 110651937 | 3285 | 16 | 1094 | 121,034.7 | 5.05 |
| HbPP2C20 | 110665418 | 849 | 4 | 282 | 28,091.82 | 6.17 |
| HbPP2C21 | 110657379 | 1032 | 9 | 343 | 31,444.84 | 6.09 |
| HbPP2C22 | 110637316 | 1140 | 3 | 379 | 31,413.62 | 5.15 |
| HbPP2C23 | 110659164 | 1170 | 2 | 389 | 27,827.86 | 4.79 |
| HbPP2C24 | 110636987 | 1305 | 3 | 434 | 36,716.07 | 8.19 |
| HbPP2C25 | 110647724 | 1125 | 4 | 374 | 40,501.11 | 7.06 |
| HbPP2C26 | 110666575 | 915 | 12 | 304 | 32,822.4 | 5.09 |
| HbPP2C27 | 110669236 | 1149 | 4 | 382 | 41,895.72 | 5.24 |
| HbPP2C28 | 110653172 | 1230 | 2 | 409 | 14,735.89 | 5.79 |
| HbPP2C29 | 110657742 | 2364 | 4 | 787 | 86,792.4 | 5.40 |
| HbPP2C30 | 110639138 | 1158 | 4 | 385 | 42,130.4 | 4.88 |
| HbPP2C31 | 110669048 | 1236 | 5 | 411 | 26,258.97 | 4.99 |
| HbPP2C31 | 110661701 | 2700 | 3 | 899 | 87,750.35 | 6.81 |
| HbPP2C33 | 110630051 | 1539 | 5 | 512 | 28,981.91 | 8.83 |
| HbPP2C34 | 110639297 | 1074 | 3 | 357 | 39,813.2 | 6.36 |
| HbPP2C35 | 110664833 | 1602 | 4 | 533 | 51,567.47 | 5.54 |
| HbPP2C36 | 110655005 | 852 | 4 | 283 | 26,636.45 | 6.45 |
| HbPP2C37 | 110643225 | 1278 | 3 | 425 | 46,328.04 | 5.12 |
| HbPP2C38 | 110650239 | 1230 | 3 | 409 | 44,117.94 | 8.54 |
| HbPP2C39 | 110666277 | 855 | 3 | 284 | 19,352.03 | 5.94 |
| HbPP2C40 | 110631453 | 1593 | 4 | 530 | 57,315.53 | 5.34 |
| HbPP2C41 | 110667158 | 1194 | 4 | 397 | 44,117.94 | 8.54 |
| HbPP2C42 | 110669425 | 777 | 4 | 258 | 26,124.6 | 5.63 |
| HbPP2C43 | 110646201 | 1134 | 5 | 377 | 43,622.84 | 7.23 |
| HbPP2C44 | 110635324 | 852 | 8 | 283 | 32,449.25 | 9.76 |
| HbPP2C45 | 110637488 | 912 | 2 | 303 | 38,238.89 | 5.63 |
| HbPP2C46 | 110642221 | 687 | 4 | 229 | 27,451.31 | 6.30 |
| HbPP2C47 | 110672864 | 1158 | 4 | 385 | 41,882.79 | 5.96 |
| HbPP2C48 | 110651368 | 1158 | 4 | 385 | 42,961.3 | 8.29 |
| HbPP2C49 | 110640918 | 1185 | 5 | 394 | 43,697.35 | 4.93 |
| HbPP2C50 | 110672178 | 1050 | 4 | 349 | 40,916.22 | 4.85 |
| HbPP2C51 | 110645615 | 3072 | 23 | 1023 | 110,551.67 | 5.44 |
| HbPP2C52 | 110668244 | 948 | 4 | 315 | 34,981.15 | 4.67 |
| HbPP2C53 | 110646945 | 1647 | 3 | 548 | 59,173.58 | 4.68 |
| HbPP2C54 | 110646864 | 1107 | 4 | 368 | 40,969.80 | 8.44 |
| HbPP2C55 | 110655808 | 1548 | 4 | 515 | 55,811.2 | 6.24 |
| HbPP2C56 | 110642429 | 1632 | 11 | 543 | 59,251.99 | 4.90 |
| HbPP2C57 | 110650667 | 1164 | 5 | 387 | 42,380.07 | 4.97 |
| HbPP2C58 | 110671158 | 876 | 8 | 291 | 32,120.55 | 8.70 |
| HbPP2C59 | 110657354 | 891 | 10 | 296 | 31,721.07 | 4.77 |
| HbPP2C60 | 110646895 | 1110 | 3 | 369 | 40,325.37 | 5.23 |
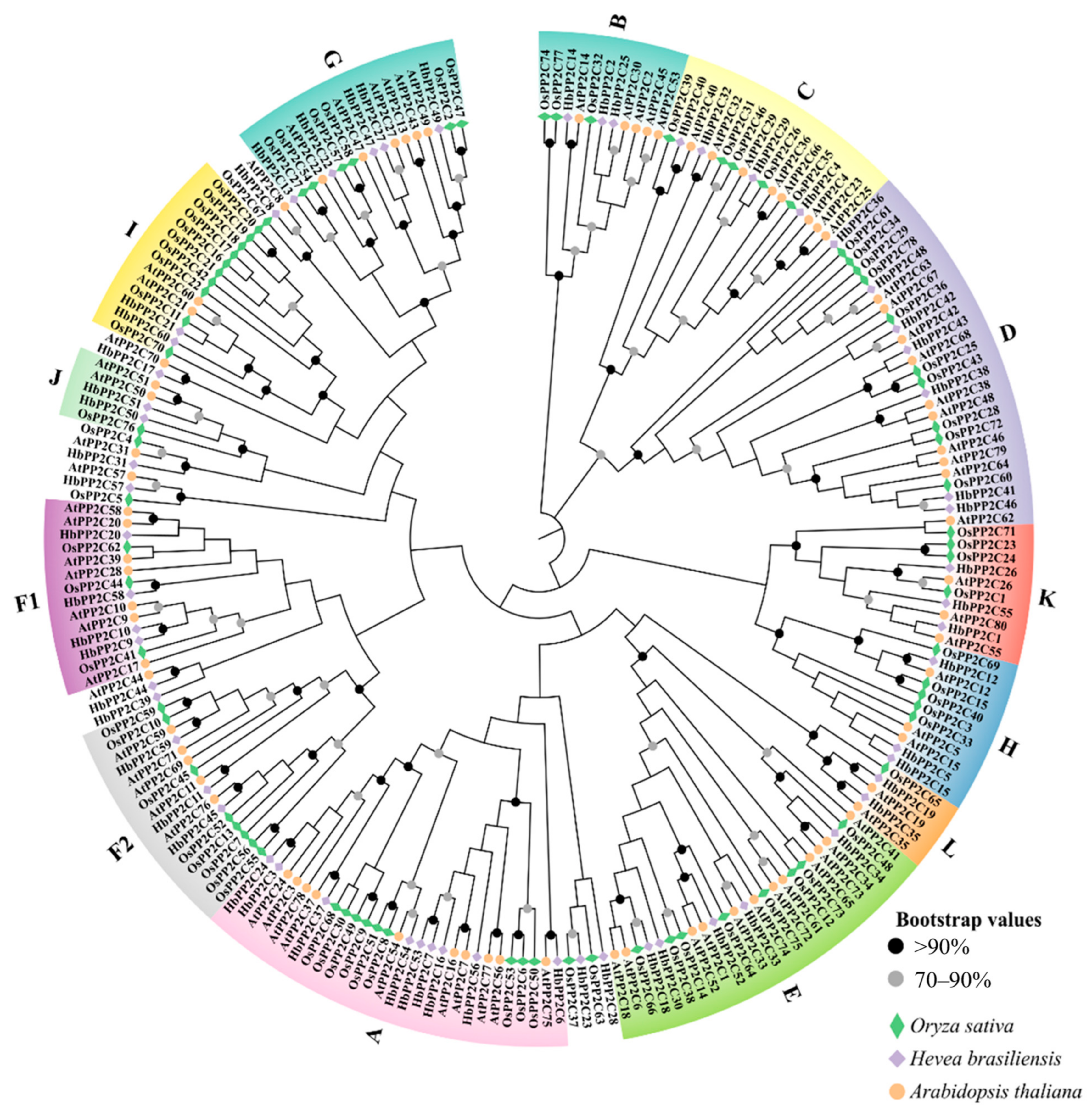
2.2. Phylogenetic Analysis of HbPP2Cs
2.3. Conserved Domain and Motifs of HbPP2C Proteins in Rubber Tree
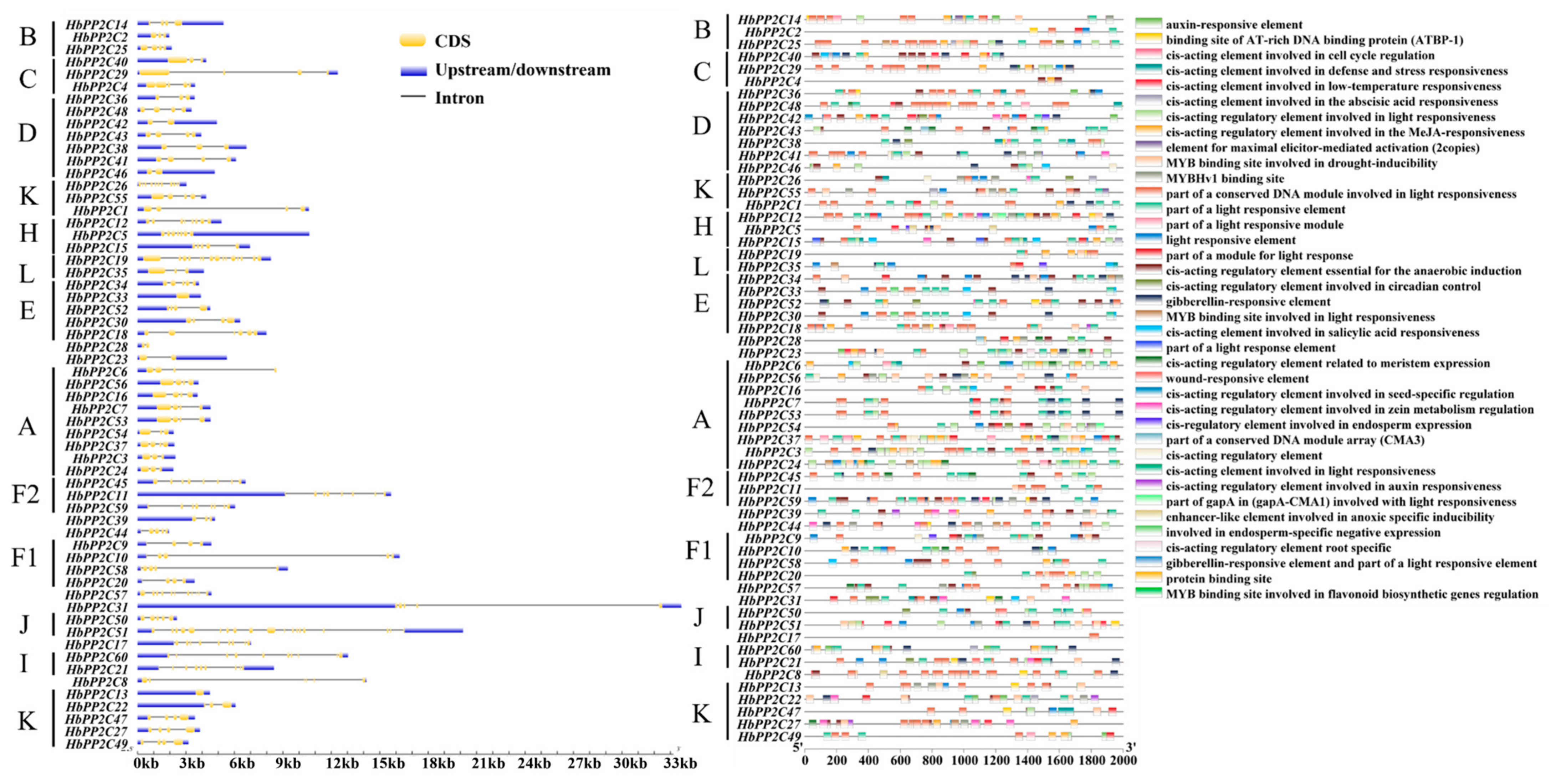
2.4. Structure of PP2C Genes in Rubber Tree
2.5. Cis-Acting Element Analysis of the Promoters of the HbPP2C Genes
2.6. Expression Profiles of HbPP2Cs in Different Tissues
2.7. Expression of HbPP2C Genes under Abiotic Stress Conditions

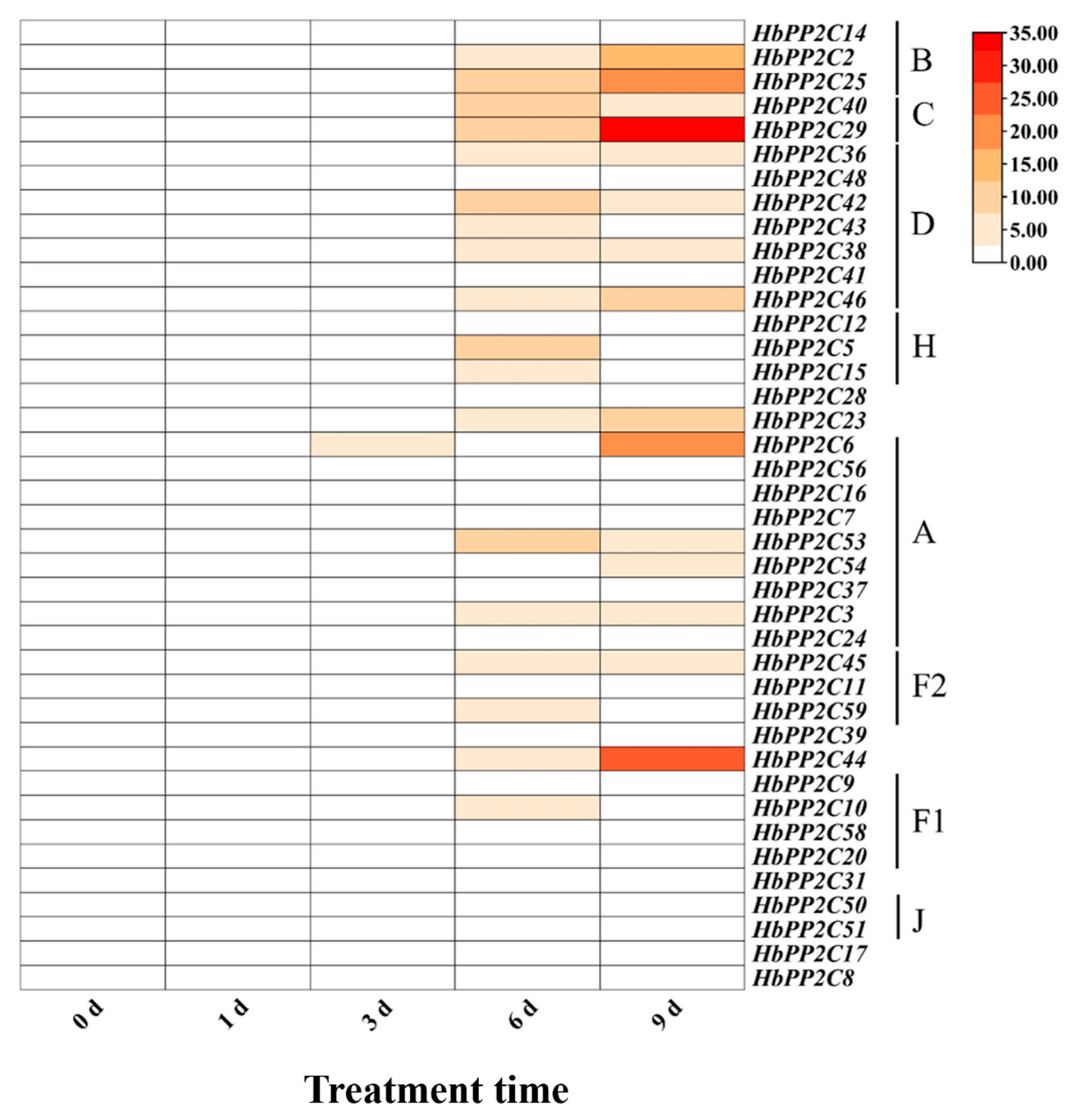
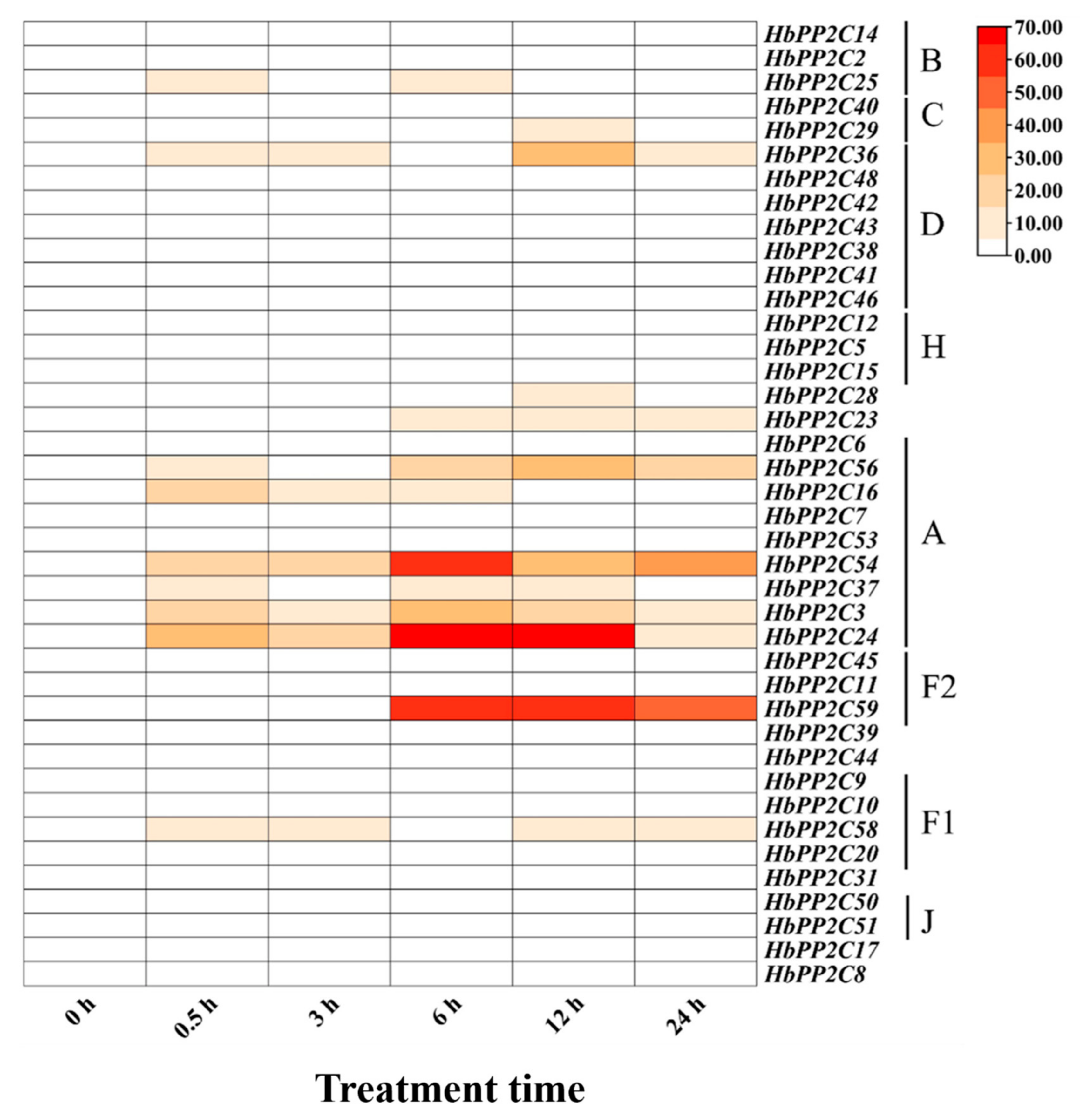
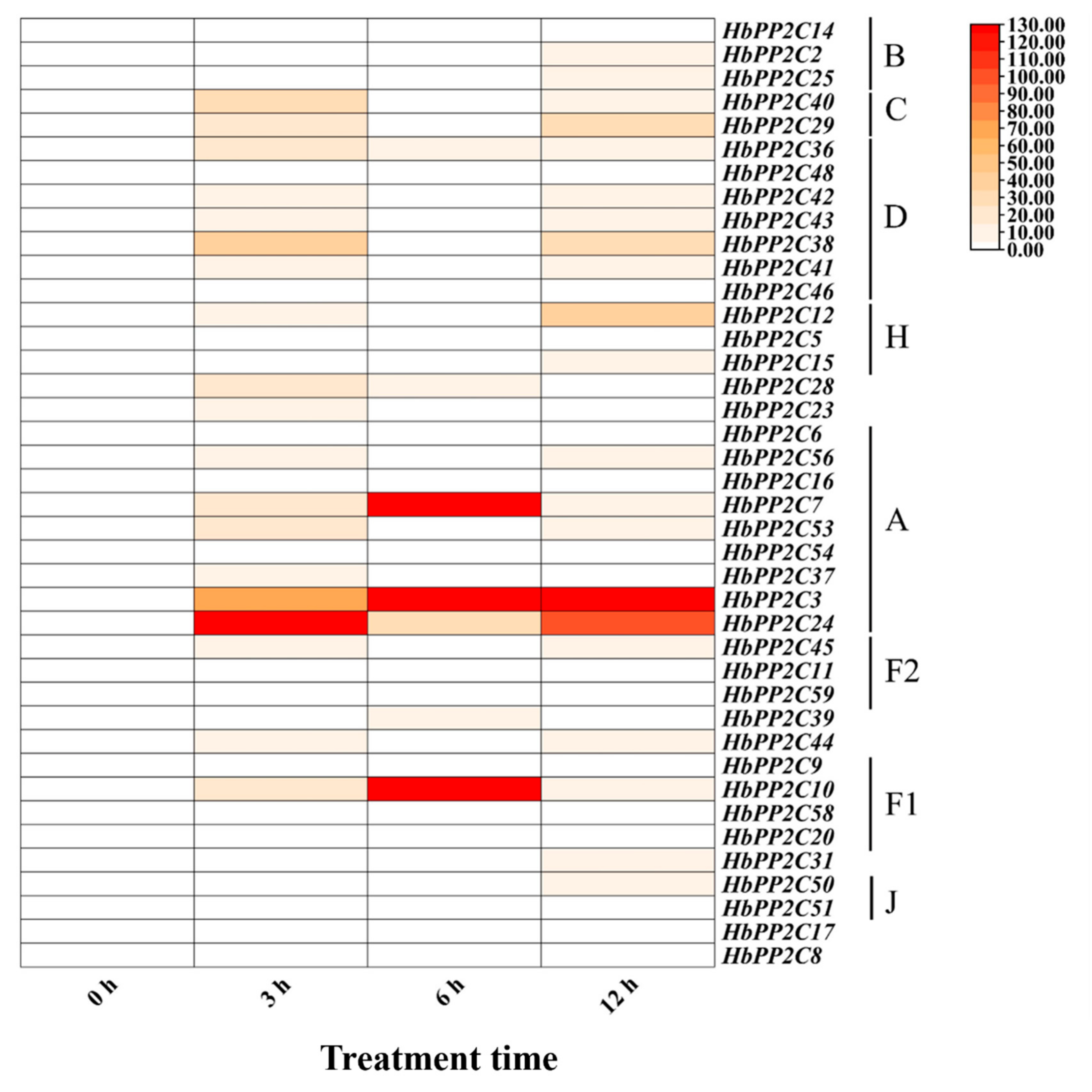
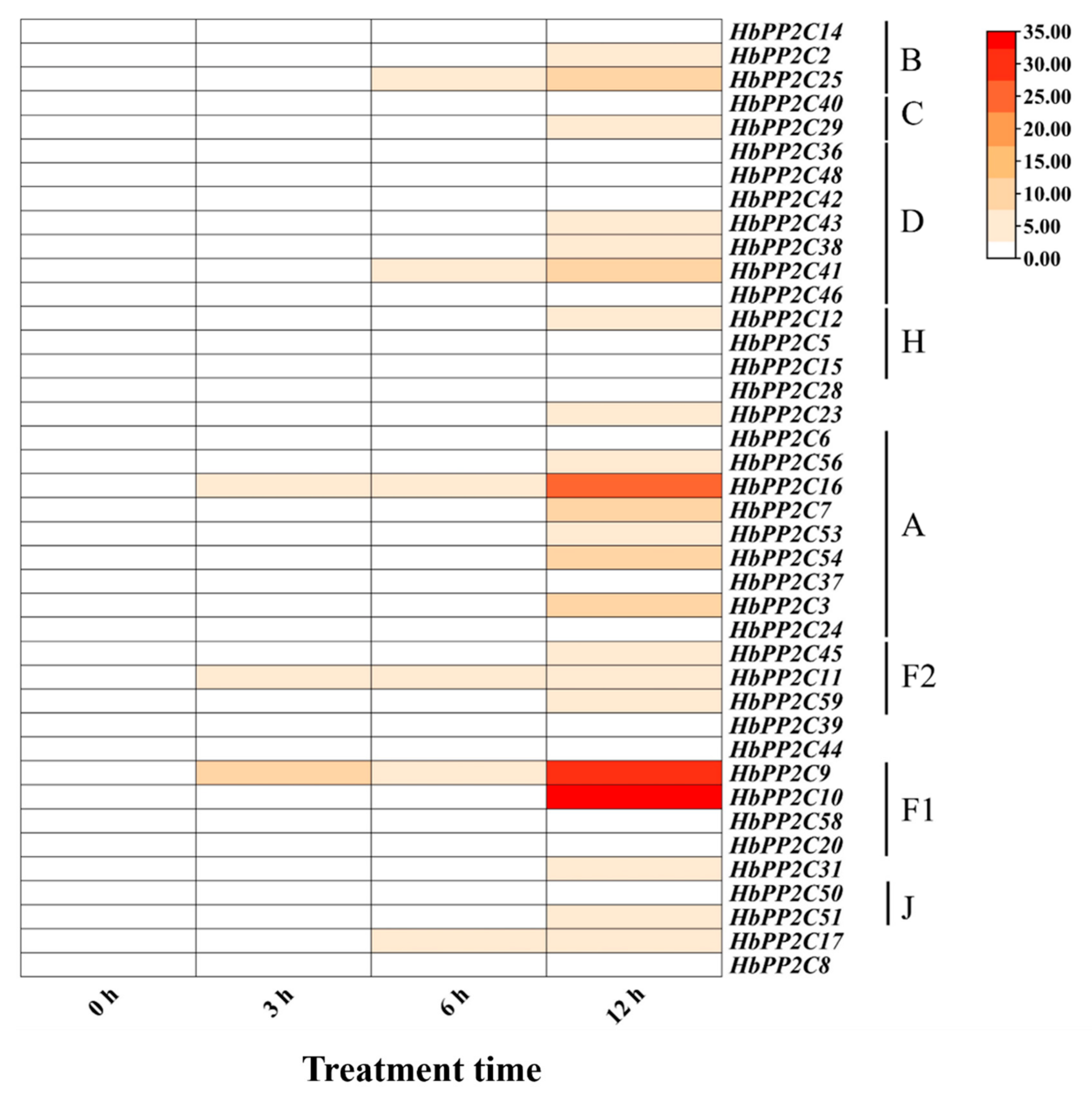
2.8. Expression of the HbPP2C Gene under Biotic Stress

3. Discussion
3.1. Detailed Characterization and Evolution of HbPP2Cs in Rubber Tree
3.2. HbPP2C Genes Are Specifically Expressed in Branches
3.3. HbPP2C Genes Play Important Roles in the ABA Pathway and Drought Stress Response
3.4. HbPP2C Genes Regulate the Temperature Stress Response in Rubber Tree
3.5. HbPP2Cs Play an Influential Role in the Response to Biological Adversity Stresses
4. Materials and Methods
4.1. Plant Material
4.2. Plant Treatment
4.3. Gene Sequence Acquisition
4.4. HbPP2C Structural and Functional Analyses
4.5. Sequence Alignment and Phylogenetic Analysis
4.6. Expression Analysis of HbPP2Cs Using qRT–PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.; Li, X.; He, Z.; Zhao, X.; Wang, Q.; Zhou, B.; Yu, D.; Huang, X.; Tang, D.; Guo, X.; et al. Molecular character of a phosphatase 2C (PP2C) gene relation to stress tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 2013, 40, 2633–2644. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liang, Y.; Zhang, R.; Zhang, B.; Song, X.; Liu, J.; Lu, M.; Qin, Z.; Li, D.; Li, S.; et al. Genome-wide identification of the PP2C gene family and analyses with their expression profiling in response to cold stress in wild sugarcane. Plants 2023, 12, 2418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, Z.; Luo, S.; Li, X.; Lyu, J.; Liu, Z.; Wan, Z.; Yu, J. Genome-wide identification and expression analysis of the cucumber PP2C gene family. BMC Genom. 2022, 23, 563. [Google Scholar] [CrossRef]
- Cohen, P.; Cohen, P.T. Protein phosphatases come of age. J. Biol. Chem. 1989, 264, 21435–21438. [Google Scholar] [CrossRef] [PubMed]
- Shazadee, H.; Khan, N.; Wang, J.; Wang, C.; Zeng, J.; Huang, Z.; Wang, X. Identification and expression profiling of protein phosphatases (PP2C) gene family in Gossypium hirsutum L. Int. J. Mol. Sci. 2019, 20, 1395. [Google Scholar] [CrossRef]
- Zheng, X.; Fei, S.; Wang, S.; He, Y.; Zhu, Z.; Liu, Y. Identification of the functional modules of SlPP2C.D—SlSAUR and their roles in abscisic acid-mediated inhibition of tomato hypocotyl elongation. Agronomy 2022, 12, 2542. [Google Scholar] [CrossRef]
- Née, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Yao, L.; Yu, L.; Qiao, Z.; Tang, M.; Wei, F.; Huang, X.; Zhou, Y. Transcriptome and proteome analyses reveal the potential mechanism of seed dormancy release in Amomum tsaoko during warm stratification. BMC Genom. 2023, 24, 99. [Google Scholar] [CrossRef]
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, J.R., 3rd; et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 2018, 9, 2132. [Google Scholar] [CrossRef]
- Giska, F.; Martin, G.B. PP2C phosphatase Pic1 negatively regulates the phosphorylation status of Pti1b kinase, a regulator of flagellin-triggered immunity in tomato. Biochem. J. 2019, 476, 1621–1635. [Google Scholar] [CrossRef]
- Sobol, G.; Chakraborty, J.; Martin, G.; Sessa, G. The emerging role of PP2C phosphatases in tomato immunity. Mol. Plant Microbe Interact. 2022, 35, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, H.; Kondo, S.; Tanaka, T.; Imamura, C.; Muramoto, N.; Hattori, E.; Ogawa, K.; Mitsukawa, N.; Ohto, C. Overexpression of a novel Arabidopsis PP2C isoform, AtPP2CF1, enhances plant biomass production by increasing inflorescence stem growth. J. Exp. Bot. 2014, 65, 5385–5400. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Zuo, T.; Liu, Y.; Tao, H.; Mo, Y.; Li, C.; Zhao, L.; Gao, J. Phylogenetic analysis of PP2C proteins and interactive proteins analyze of BjuPP2C52 in Brassica juncea. Plant Physiol. Biochem. 2022, 179, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yu, Y.; Ding, X.; Liu, B.; Duanmu, H.; Zhu, D.; Sun, X.; Cao, L.; Zaib Un, N.; Li, Q.; et al. Genome-wide analysis and expression profiling of PP2C clade D under saline and alkali stresses in wild soybean and Arabidopsis. Protoplasma 2018, 255, 643–654. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, K.; Niu, X.; Wang, Q.; Wan, Y.; Yang, F.; Li, G.; Wang, Y.; Wang, R. Genome-wide identification of PP2C genes and their expression profiling in response to drought and cold stresses in Medicago truncatula. Sci. Rep. 2018, 8, 12841. [Google Scholar] [CrossRef]
- He, Z.; Wu, J.; Sun, X.; Dai, M. The maize clade A PP2C phosphatases play critical roles in multiple abiotic stress responses. Int. J. Mol. Sci. 2019, 20, 3573. [Google Scholar] [CrossRef]
- Wu, X.T.; Xiong, Z.P.; Chen, K.X.; Zhao, G.R.; Feng, K.R.; Li, X.H.; Li, X.R.; Tian, Z.; Huo, F.L.; Wang, M.X.; et al. Genome-wide identification and transcriptional expression profiles of PP2C in the barley (Hordeum vulgare L.) Pan-Genome. Genes 2022, 13, 834. [Google Scholar] [CrossRef]
- Wei, L.; Zhang, J.; Wei, S.; Wang, C.; Deng, Y.; Hu, D.; Liu, H.; Gong, W.; Pan, Y.; Liao, W. Nitric oxide alleviates salt stress through protein S-nitrosylation and transcriptional regulation in tomato seedlings. Planta 2022, 256, 101. [Google Scholar] [CrossRef]
- Sisi, C.; Jieru, D.; Peidong, C.; Zhaolong, Z.; Yihang, W.; Shuwen, C.; Yan, T.; Tianyu, W.; Guiyan, Y. Transcriptome-wide identification of walnut PP2C family genes in response to external stimulus. BMC Genom. 2022, 23, 640. [Google Scholar] [CrossRef]
- Li, M.; Wu, T.; Wang, S.; Duan, T.; Huang, S.; Xie, Y. The modulation of sucrose nonfermenting 1-related protein kinase 2.6 state by persulfidation and phosphorylation: Insights from molecular dynamics simulations. Int. J. Mol. Sci. 2023, 24, 11512. [Google Scholar] [CrossRef]
- Bhalothia, P.; Sangwan, C.; Alok, A.; Mehrotra, S.; Mehrotra, R. PP2C-like Promoter and its deletion variants are induced by ABA but not by MeJA and SA in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 547. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Wang, D.; Zhang, S.; Ehlting, J.; Ni, F.; Jakab, S.; Zheng, C.; Zhong, Y. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genom. 2008, 9, 550. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Ke, H.; Hu, C.M.; Naseri, E.; Haider, M.S.; Ayaz, A.; Amjad Khan, W.; Wang, J.; Hou, X. Genome-wide identification, evolution, and transcriptional profiling of PP2C gene family in Brassica rapa. BioMed Res. Int. 2019, 2019, 2965035. [Google Scholar] [CrossRef]
- Haider, M.S.; Khan, N.; Pervaiz, T.; Zhongjie, L.; Nasim, M.; Jogaiah, S.; Mushtaq, N.; Jiu, S.; Jinggui, F. Genome-wide identification, evolution, and molecular characterization of the PP2C gene family in woodland strawberry. Gene 2019, 702, 27–35. [Google Scholar] [CrossRef]
- Cao, J.; Jiang, M.; Li, P.; Chu, Z. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon. BMC Genom. 2016, 17, 175. [Google Scholar] [CrossRef]
- Fan, K.; Yuan, S.; Chen, J.; Chen, Y.; Li, Z.; Lin, W.; Zhang, Y.; Liu, J.; Lin, W. Molecular evolution and lineage-specific expansion of the PP2C family in Zea mays. Planta 2019, 250, 1521–1538. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Luo, L.; Wan, Y.; Liu, F. Genome-wide characterization of the PP2C gene family in peanut (Arachis hypogaea L.) and the identification of candidate genes involved in salinity-stress response. Front. Plant Sci. 2023, 14, 1093913. [Google Scholar] [CrossRef]
- Xin, H.; Li, Q.; Wu, X.; Yin, B.; Li, J.; Zhu, J. The Arabidopsis thaliana integrin-like gene AT14A improves drought tolerance in Solanum lycopersicum. J. Plant Res. 2023, 136, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Naveed, Z.A.; Ali, G.S. Comparative transcriptome analysis between a resistant and a susceptible wild tomato accession in response to Phytophthora parasitica. Int. J. Mol. Sci. 2018, 19, 3735. [Google Scholar] [CrossRef]
- Wang, J.; Li, C.; Li, L.; Gao, L.; Hu, G.; Zhang, Y.; Reynolds, M.P.; Zhang, X.; Jia, J.; Mao, X.; et al. DIW1 encoding a clade I PP2C phosphatase negatively regulates drought tolerance by dephosphorylating TaSnRK1.1 in wheat. J. Integr. Plant Biol. 2023, 256, 101. [Google Scholar] [CrossRef]
- Fan, K.; Chen, Y.; Mao, Z.; Fang, Y.; Li, Z.; Lin, W.; Zhang, Y.; Liu, J.; Huang, J.; Lin, W. Pervasive duplication, biased molecular evolution and comprehensive functional analysis of the PP2C family in Glycine max. BMC Genom. 2020, 21, 465. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Shi, Y.; Wang, Y.; Liu, F.; Li, Z.; Qi, J.; Wang, Y.; Zhang, J.; Yang, S.; Wang, Y.; et al. The clade F PP2C phosphatase ZmPP84 negatively regulates drought tolerance by repressing stomatal closure in maize. New Phytol. 2022, 237, 1728–1744. [Google Scholar] [CrossRef]
- Soon, F.F.; Ng, L.M.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.H.; Suino-Powell, K.M.; He, Y.; Xu, Y.; Chalmers, M.J.; et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012, 335, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Tovar-Mendez, A.; Miernyk, J.A.; Hoyos, E.; Randall, D.D. A functional genomic analysis of Arabidopsis thaliana PP2C clade D. Protoplasma 2014, 251, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Han, J.; Li, L.; Zhang, Q.; Yang, G.; He, G. Wheat PP2C-a10 regulates seed germination and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 2020, 39, 635–651. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Zhang, D.; Liu, Q.; Liu, Y.; Qin, B.; Liang, X.; Wang, L.; Zhang, Y. Characterization of 33 HbbZIP gene family members and analysis of their expression profiles in rubber tree in response to ABA, glyphosate and powdery mildew treatment. Forests 2023, 14, 556. [Google Scholar] [CrossRef]
- Hu, X.; Liu, L.; Xiao, B.; Li, D.; Xing, X.; Kong, X.; Li, D. Enhanced tolerance to low temperature in tobacco by over-expression of a new maize protein phosphatase 2C, ZmPP2C2. J. Plant Physiol. 2010, 167, 1307–1315. [Google Scholar] [CrossRef]
- Urrea Castellanos, R.; Friedrich, T.; Petrovic, N.; Altmann, S.; Brzezinka, K.; Gorka, M.; Graf, A.; Baurle, I. FORGETTER2 protein phosphatase and phospholipase D modulate heat stress memory in Arabidopsis. Plant J. 2020, 104, 7–17. [Google Scholar] [CrossRef]
- Jeon, J.E.; Kim, J.G.; Fischer, C.R.; Mehta, N.; Dufour-Schroif, C.; Wemmer, K.; Mudgett, M.B.; Sattely, E. A pathogen-responsive gene cluster for highly modified fatty acids in tomato. Cell 2020, 180, 176–187.e119. [Google Scholar] [CrossRef] [PubMed]
- Birch, P.; Qiu, M.; Tian, M.; Yong, S.; Sun, Y.; Cao, J.; Li, Y.; Zhang, X.; Zhai, C.; Ye, W.; et al. Phase-specific transcriptional patterns of the oomycete pathogen Phytophthora sojae unravel genes essential for asexual development and pathogenic processes. PLoS Pathog. 2023, 19, e1011256. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Qin, B.; Zhang, D.; Liang, X.; Yang, Y.; Wang, L.; Wang, M.; Zhang, Y. Identification and Characterization of the HbPP2C Gene Family and Its Expression in Response to Biotic and Abiotic Stresses in Rubber Tree. Int. J. Mol. Sci. 2023, 24, 16061. https://doi.org/10.3390/ijms242216061
Liu Q, Qin B, Zhang D, Liang X, Yang Y, Wang L, Wang M, Zhang Y. Identification and Characterization of the HbPP2C Gene Family and Its Expression in Response to Biotic and Abiotic Stresses in Rubber Tree. International Journal of Molecular Sciences. 2023; 24(22):16061. https://doi.org/10.3390/ijms242216061
Chicago/Turabian StyleLiu, Qifeng, Bi Qin, Dong Zhang, Xiaoyu Liang, Ye Yang, Lifeng Wang, Meng Wang, and Yu Zhang. 2023. "Identification and Characterization of the HbPP2C Gene Family and Its Expression in Response to Biotic and Abiotic Stresses in Rubber Tree" International Journal of Molecular Sciences 24, no. 22: 16061. https://doi.org/10.3390/ijms242216061
APA StyleLiu, Q., Qin, B., Zhang, D., Liang, X., Yang, Y., Wang, L., Wang, M., & Zhang, Y. (2023). Identification and Characterization of the HbPP2C Gene Family and Its Expression in Response to Biotic and Abiotic Stresses in Rubber Tree. International Journal of Molecular Sciences, 24(22), 16061. https://doi.org/10.3390/ijms242216061





