Integrated Transcriptome and Metabolomics to Reveal the Mechanism of Adipose Mesenchymal Stem Cells in Treating Liver Fibrosis
Abstract
:1. Introduction
2. Results
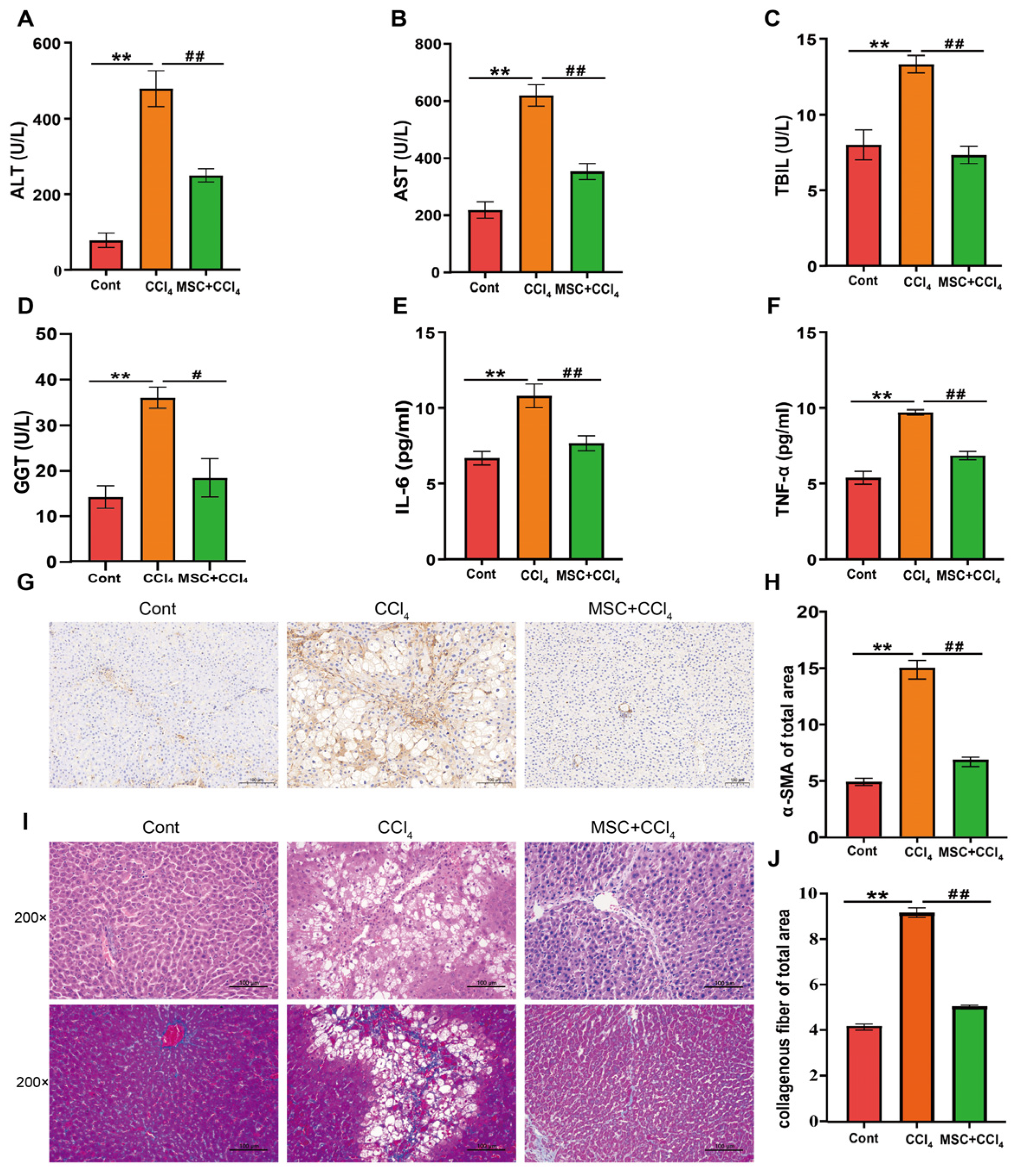
2.1. ADMSCs Alleviate Liver Injury and Fibrosis in Rats
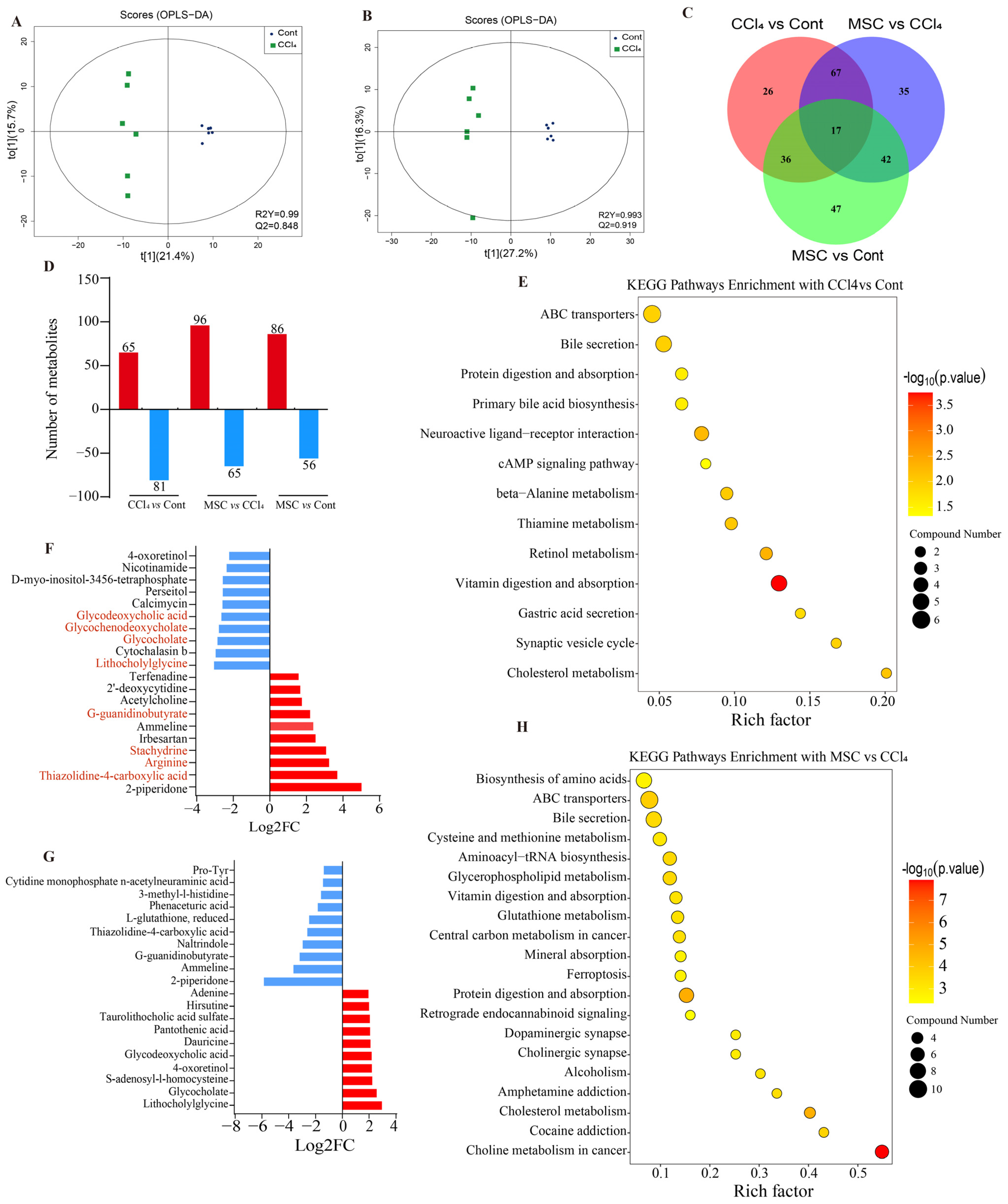
2.2. Changes in Liver Metabolic Profile after Adipose Mesenchymal Stem Cell Transplantation
2.3. Changes in Liver Transcriptome after Adipose Mesenchymal Stem Cell Transplantation
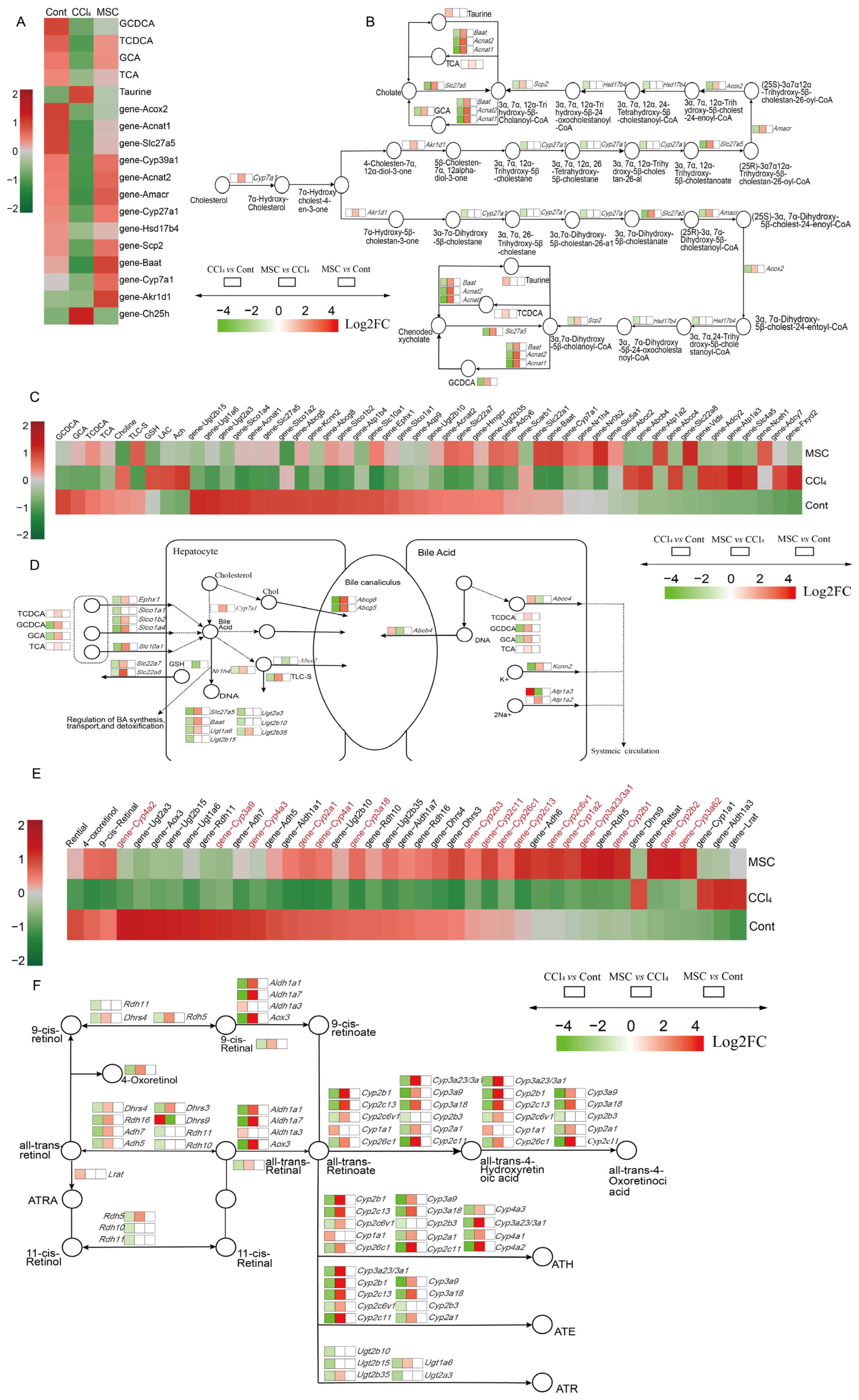
2.4. Alterations of Genes and Metabolites Related to Primary BA Biosynthesis and Bile Secretion
2.5. Alterations of Genes and Metabolites Related to Retinol Metabolism
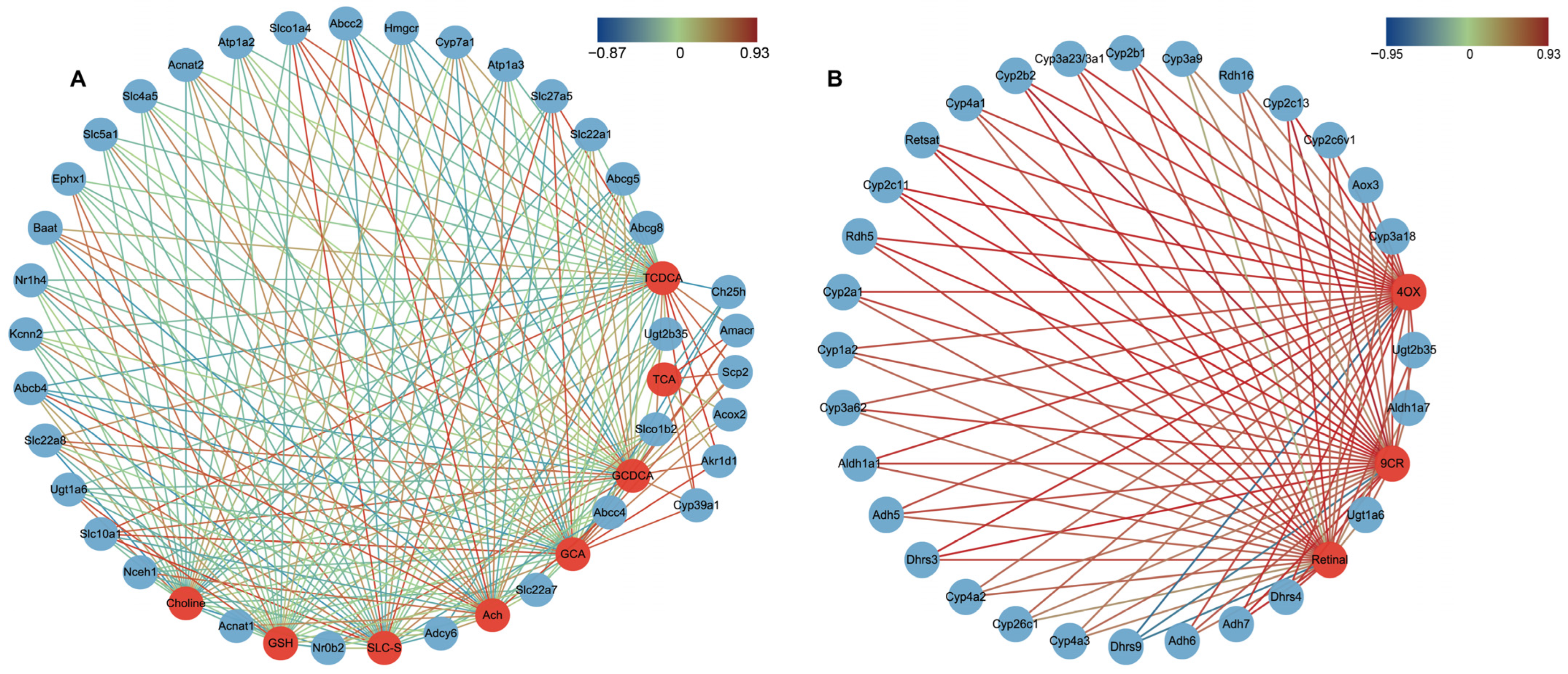
2.6. Correlations between DGEs and DMs Related to Primary BA Biosynthesis, Bile Secretion and Retinol Metabolism
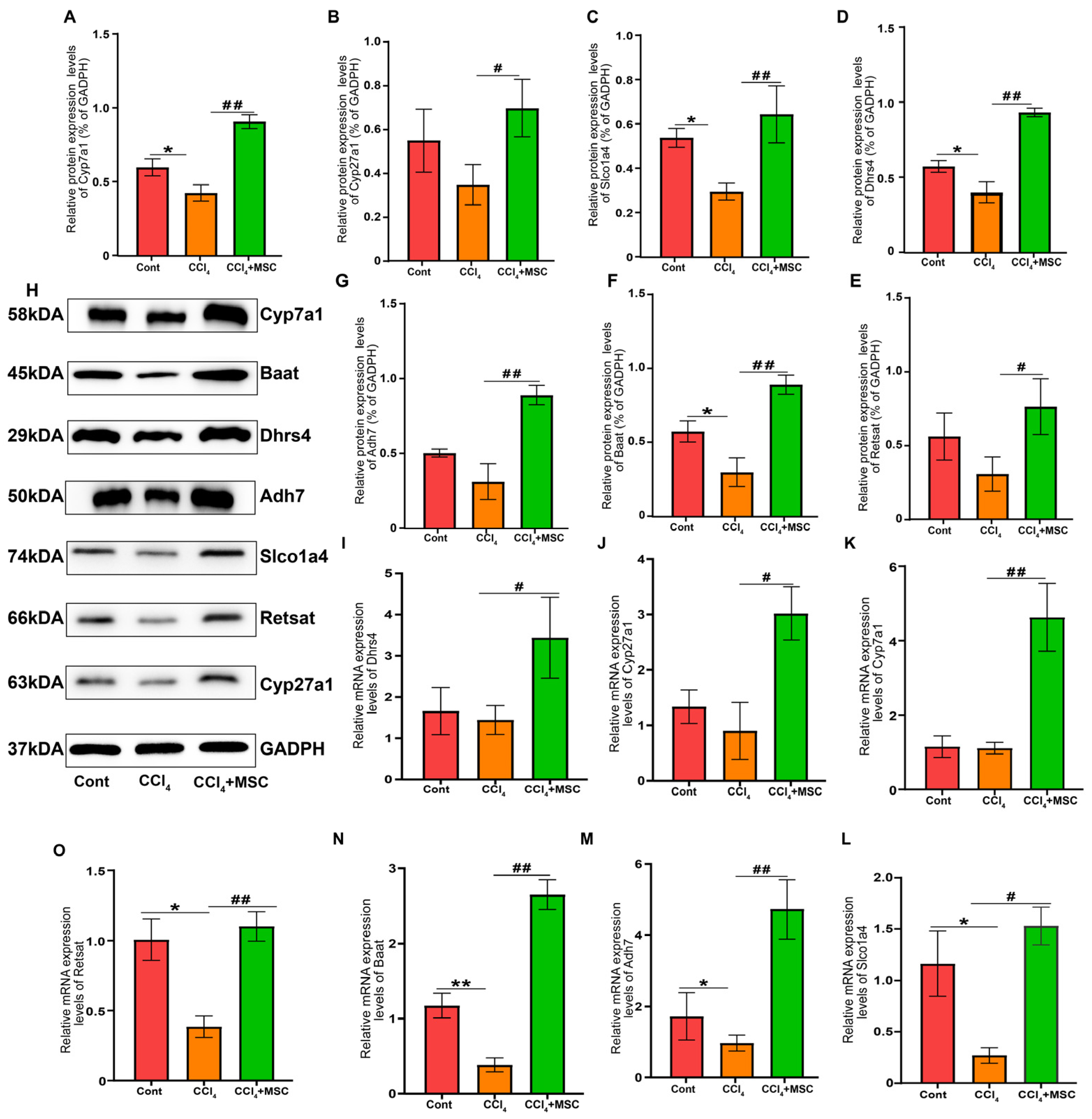
2.7. Candidate Gene Validation
3. Discussion
4. Materials and Methods
4.1. Animal Treatment and Sample Collection
4.2. Serum Biochemical and Inflammatory ELISA Factor Analysis
4.3. Histological and Immunohistochemistry Analysis
4.4. Transcriptomic Analysis
4.5. Metabolomic Analysis
4.6. Data Processing and Annotation
4.7. Statistical Analysis
4.8. Validation of DEGs by qPCR and Western Blot
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol. 2011, 6, 425–456. [Google Scholar] [CrossRef] [PubMed]
- Seki, E.; Brenner, D.A. Recent advancement of molecular mechanisms of liver fibrosis. J. Hepato-Biliary-Pancreat. Sci. 2015, 22, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Wang, K.; Aoyama, T.; Grivennikov, S.I.; Paik, Y.; Scholten, D.; Cong, M.; Iwaisako, K.; Liu, X.; Zhang, M.; et al. Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology 2012, 143, 765–776.e3. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.S.; Cao, Z.; Lis, R.; Nolan, D.J.; Guo, P.; Simons, M.; Penfold, M.E.; Shido, K.; Rabbany, S.Y.; Rafii, S. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature 2014, 505, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.N.; Zhang, G.B.; Lu, Y.Y.; Chen, Q.L.; Yang, L.; Wang, Z.T.; Liu, P.; Su, S.B. Huangqi decoction alleviates dimethylnitrosamine-induced liver fibrosis: An analysis of bile acids metabolic mechanism. J. Ethnopharmacol. 2016, 189, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Kirs, S.; Kliewers, A.; Mangelsdorfd, J. Roles of FGF19 in liver metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 139–144. [Google Scholar]
- Zollner, G.; Marschall, H.U.; Wagner, M.; Trauner, M. Role of nuclear receptors in the adaptive response to bile acids and cholestasis: Pathogenetic and therapeutic considerations. Mol. Pharm. 2006, 3, 231–251. [Google Scholar] [CrossRef]
- Hu, C.; Zhao, L.; Li, L. Current understanding of adipose-derived mesenchymal stem cell-based therapies in liver diseases. Stem Cell Res. Ther. 2019, 10, 199. [Google Scholar] [CrossRef]
- Yuan, Y.; Ni, S.; Zhuge, A.; Li, L.; Li, B. Adipose-Derived Mesenchymal Stem Cells Reprogram M1 Macrophage Metabolism via PHD2/HIF-1α Pathway in Colitis Mice. Front. Immunol. 2022, 13, 859806. [Google Scholar] [CrossRef]
- Zhai, L.; Shen, H.; Sheng, Y.; Guan, Q. ADMSC Exo-MicroRNA-22 improve neurological function and neuroinflammation in mice with Alzheimer’s disease. J. Cell. Mol. Med. 2021, 25, 7513–7523. [Google Scholar] [CrossRef]
- Freitag, J.; Bates, D.; Wickham, J.; Shah, K.; Huguenin, L.; Tenen, A.; Paterson, K.; Boyd, R. Adipose-derived mesenchymal stem cell therapy in the treatment of knee osteoarthritis: A randomized controlled trial. Regen. Med. 2019, 14, 213–230. [Google Scholar] [CrossRef] [PubMed]
- Natali, S.; Screpis, D.; Patania, E.; De Berardinis, L.; Benoni, A.; Piovan, G.; Iacono, V.; Magnan, B.; Gigante, A.P.; Zorzi, C. Efficacy and Long-Term Outcomes of Intra-Articular Autologous Micro-Fragmented Adipose Tissue in Individuals with Glenohumeral Osteoarthritis: A 36-Month Follow-Up Study. J. Pers. Med. 2023, 13, 1309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, S.; Zhou, Y.; Feng, F.; Wang, Q.; Zhu, X.; Ai, H.; Huang, X.; Zhang, X. Hepatocyte growgrthfactor gene-modified adipose-derived mesenchymal stem cells ameliorate radiation induced liver damage in a rat model. PLoS ONE 2014, 9, e114670. [Google Scholar]
- Almalkis, G.; Agrawald, K. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation 2016, 92, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Zhang, Q.; Li, H.; Bai, G.; Jiao, Z.; Wang, H. Adipose-derived stem cells alleviate liver apoptosis induced by ischemia-reperfusion and laparoscopic hepatectomy in swine. Sci. Rep. 2018, 8, 16878. [Google Scholar] [CrossRef]
- Ge, Y.; Zhang, Q.; Jiao, Z.; Li, H.; Bai, G.; Wang, H. Adipose-derived stem cells reduce liver oxidative stress and autophagy induced by ischemia-reperfusion and hepatectomy injury in swine. Life Sci. 2018, 214, 62–69. [Google Scholar] [CrossRef]
- Harn, H.J.; Lin, S.Z.; Hung, S.H.; Subeq, Y.M.; Li, Y.S.; Syu, W.S.; Ding, D.C.; Lee, R.P.; Hsieh, D.K.; Lin, P.C.; et al. Adipose-derived stem cells can abrogate chemical-induced liver fibrosis and facilitate recovery of liver function. Cell Transplant. 2012, 21, 2753–2764. [Google Scholar] [CrossRef]
- Okura, H.; Soeda, M.; Morita, M.; Fujita, M.; Naba, K.; Ito, C.; Ichinose, A.; Matsuyama, A. Therapeutic potential of human adipose tissue-derived multi-lineage progenitor cells in liver fibrosis. Biochem. Biophys. Res. Commun. 2015, 456, 860–865. [Google Scholar] [CrossRef]
- Yu, F.; Ji, S.; Su, L.; Wan, L.; Zhang, S.; Dai, C.; Wang, Y.; Fu, J.; Zhang, Q. Adipose-derived mesenchymal stem cells inhibit activation of hepatic stellate cells in vitro and ameliorate rat liver fibrosis in vivo. J. Formos. Med. Assoc. Taiwan Yi Zhi 2015, 114, 130–138. [Google Scholar] [CrossRef]
- Huang, K.C.; Chuang, M.H.; Lin, Z.S.; Lin, Y.-C.; Chen, C.-H.; Chang, C.-L.; Huang, P.-C.; Syu, W.-S.; Chiou, T.-W.; Hong, Z.-H.; et al. Transplantation with GXHPC1 for Liver Cirrhosis: Phase 1 Trial. Cell Transplant. 2019, 28 (Suppl. 1), 100s–111s. [Google Scholar] [CrossRef]
- Pannala, V.R.; Vinnakota, K.C.; Rawls, K.D.; Estes, S.K.; O’Brien, T.P.; Printz, R.L.; Papin, J.A.; Reifman, J.; Shiota, M.; Young, J.D.; et al. Mechanistic identification of biofluid metabolite changes as markers of acetaminophen-induced liver toxicity in rats. Toxicol. Appl. Pharmacol. 2019, 372, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, Z.; Wu, Y.; Jiang, H.; Zhou, F.; Xie, X.; Wang, R.; Hua, C. Untargeted metabonomics reveals intervention effects of chicory polysaccharide in a rat model of non-alcoholic fatty liver disease. Int. J. Biol. Macromol. 2019, 128, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.X.; Shi, D.H.; Chen, Y.X.; Cui, J.T.; Wang, Y.R.; Jiang, C.P.; Wu, J.H. The therapeutic effects of tectorigenin on chemically induced liver fibrosis in rats and an associated metabonomic investigation. Arch. Pharmacal Res. 2012, 35, 1479–1493. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Mo, J.; Zhao, Q.; Han, Q.; Kanerva, M.; Iwata, H.; Li, Q. De novo transcriptomic analysis predicts the effects of phenolic compounds in Ba River on the liver of female sharpbelly (Hemiculter lucidus). Environ. Pollut. 2020, 264, 114642. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Ji, X.; Wang, J.; Wu, H.; Han, J.; Zhang, H.; Xu, J.; Qian, M. Fungicide bromuconazole has the potential to induce hepatotoxicity at the physiological, metabolomic and transcriptomic levels in rats. Environ. Pollut. 2021, 280, 116940. [Google Scholar] [CrossRef]
- Chen, S.; Huang, Y.; Su, H.; Zhu, W.; Wei, Y.; Long, Y.; Shi, Y.; Wei, J. The Integrated Analysis of Transcriptomics and Metabolomics Unveils the Therapeutical Effect of Asiatic Acid on Alcoholic Hepatitis in Rats. Inflammation 2022, 45, 1780–1799. [Google Scholar] [CrossRef]
- Dong, S.; Chen, Q.L.; Song, Y.N.; Sun, Y.; Wei, B.; Li, X.Y.; Hu, Y.Y.; Liu, P.; Su, S.B. Mechanisms of CCl4-induced liver fibrosis with combined transcriptomic and proteomic analysis. J. Toxicol. Sci. 2016, 41, 561–572. [Google Scholar] [CrossRef]
- Weber, L.W.; Boll, M.; Stampfl, A. Hepatotoxicity and mechanism of action of haloalkanes: Carbon tetrachloride as a toxicological model. Crit. Rev. Toxicol. 2003, 33, 105–136. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, N.; Guo, Y.; Cui, S.; Ge, C.; He, Q.; Pan, X.; Wang, G.; Wang, H.; Hao, H. Combined obeticholic acid and apoptosis inhibitor treatment alleviates liver fibrosis. Acta Pharm. Sin. B 2019, 9, 526–536. [Google Scholar] [CrossRef]
- Wang, Y.; Lian, F.; Li, J.; Fan, W.; Xu, H.; Yang, X.; Liang, L.; Chen, W.; Yang, J. Adipose derived mesenchymal stem cells transplantation via portal vein improves microcirculation and ameliorates liver fibrosis induced by CCl4 in rats. J. Transl. Med. 2012, 10, 133. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, N.; Huang, Y.; Zhang, Q.; Fang, Y.; Fu, J.; Yuan, Y.; Chen, L.; Chen, X.; Xu, Z.; et al. Multiple Dimensions of using Mesenchymal Stem Cells for Treating Liver Diseases: From Bench to Beside. Stem Cell Rev. Rep. 2023, 19, 2192–2224. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Gahan, C.G.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [PubMed]
- La Frano, M.R.; Hernandez-Carretero, A.; Weber, N.; Borkowski, K.; Pedersen, T.L.; Osborn, O.; Newman, J.W. Diet-induced obesity and weight loss alter bile acid concentrations and bile acid-sensitive gene expression in insulin target tissues of C57BL/6J mice. Nutr. Res. 2017, 46, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.; Crawfordj, M. The pathology of cholestasis. Semin. Liver Dis. 2004, 24, 21–42. [Google Scholar]
- Jiao, N.A.; Baker, S.S.; Chapa-Rodriguez, A.; Liu, W.; Nugent, C.A.; Tsompana, M.; Mastrandrea, L.; Buck, M.J.; Baker, R.D.; Genco, R.J.; et al. Suppressed hepatic bile acid signalling despite elevated production of primary secondary bile acids in, NAFLD. Gut 2018, 67, 1881–1891. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, B.; Wu, Y.; Luo, X.; Chen, Z.; Sang, C.; Xiong, T. Modification effects of SanWei GanJiang Powder on liver and intestinal damage through reversing bile acid homeostasis. Biomed. Pharmacother. 2019, 116, 109044. [Google Scholar] [CrossRef]
- Wu, J.S.; Li, Y.F.; Li, Y.Y.; Dai, Y.; Li, W.K.; Zheng, M.; Shi, Z.C.; Shi, R.; Wang, T.M.; Ma, B.L.; et al. Huangqi Decoction Alleviates Alpha-Naphthylisothiocyanate Induced Intrahepatic Cholestasis by Reversing Disordered Bile Acid and Glutathione Homeostasis in Mice. Front. Pharmacol. 2017, 8, 938. [Google Scholar] [CrossRef]
- Xie, G.; Jiang, R.; Wang, X.; Liu, P.; Zhao, A.; Wu, Y.; Huang, F.; Liu, Z.; Rajani, C.; Zheng, X.; et al. Conjugated secondary 12α-hydroxylated bile acids promote liver fibrogenesis. EBioMedicine 2021, 66, 103290. [Google Scholar] [CrossRef]
- Schneider, K.M.; Candels, L.S.; Hov, J.R.; Myllys, M.; Hassan, R.; Schneider, C.V.; Wahlström, A.; Mohs, A.; Zühlke, S.; Liao, L.; et al. Gut microbiota depletion exacerbates cholestatic liver injury via loss of FXR signalling. Nat. Metab. 2021, 3, 1228–1241. [Google Scholar] [CrossRef]
- Cai, F.F.; Wu, R.; Song, Y.N.; Xiong, A.Z.; Chen, X.L.; Yang, M.D.; Yang, L.; Hu, Y.; Sun, M.Y.; Sun, S.B. Yinchenhao Decoction Alleviates Liver Fibrosis by Regulating Bile Acid Metabolism and TGF-β/Smad/ERK Signalling Pathway. Sci. Rep. 2018, 8, 15367. [Google Scholar] [CrossRef]
- Paridaens, A.; Raevens, S.; Colle, I.; Bogaerts, E.; Vandewynckel, Y.; Verhelst, X.; Hoorens, A.; van Grunsven, L.A.; Van Vlierberghe, H.; Geerts, A.; et al. Combination of tauroursodeoxycholic acid and N-acetylcysteine exceeds standard treatment for acetaminophen intoxication. Liver Int. 2017, 37, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Yang, J.; Hu, L.; Liao, B.; Qiao, L.; Shen, W.; Xie, F.; Zhu, G. Glycochenodeoxycholic acid induces stemness and chemoresistance via the STAT3 signaling pathway in hepatocellular carcinoma cells. Aging 2020, 12, 15546–15555. [Google Scholar] [CrossRef] [PubMed]
- Chiangj, Y.L.; Ferrellj, M. Up to date on cholesterol 7 alpha-hydroxylase (CYP7A1) in bile acid synthesis. Liver Res. 2020, 4, 47–63. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Mennone, A.; Boyer, J.L.; Cai, S.-Y. Combination of retinoic acid and ursodeoxycholic acid attenuates liver injury in bile duct-ligated rats and human hepatic cells. Hepatology 2011, 53, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Fader, K.A.; Nault, R.; Zhang, C.; Kumagai, K.; Harkema, J.R.; Zacharewski, T.R. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD)-elicited effects on bile acid homeostasis: Alterations in biosynthesis, enterohepatic circulation, and microbial metabolism. Sci. Rep. 2017, 7, 5921. [Google Scholar] [CrossRef] [PubMed]
- Doege, H.; Baillie, R.A.; Ortegon, A.M.; Tsang, B.; Wu, Q.; Punreddy, S.; Hirsch, D.; Watson, N.; Gimeno, R.E.; Stahl, A. Targeted deletion of FATP5 reveals multiple functions in liver metabolism: Alterations in hepatic lipid homeostasis. Gastroenterology 2006, 130, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.M.; Stahl, A. SLC27 fatty acid transport proteins. Mol. Asp. Med. 2013, 34, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.M.; Yu, M.; Ma, L.Y.; Zhang, H.W.; Zou, Z.M. Chaihu-Shu-Gan-San regulates phospholipids and bile acid metabolism against hepatic injury induced by chronic unpredictable stress in rat. J. Chromatogr. B 2017, 1064, 14–21. [Google Scholar] [CrossRef]
- Claytonp, P.T. Disorders of bile acid synthesis. J. Inherit. Metab. Dis. 2011, 34, 593–604. [Google Scholar] [CrossRef]
- Yu, X.H.; Qian, K.; Jiang, N.; Zheng, X.L.; Cayabyab, F.S.; Tang, C.K. ABCG5/ABCG8 in cholesterol excretion and atherosclerosis. Clin. Chim. Acta Int. J. Clin. Chem. 2014, 428, 82–88. [Google Scholar] [CrossRef]
- Hassan, A.S.; Bunick, D.; Lund, L.A.; Bottje, W.G. Glutathione and bile acid synthesis. Effect of GSH content of HepG2 cells on the activity and mRNA levels of cholesterol 7 alpha-hydroxylase. Biochem. Pharmacol. 1992, 44, 1475–1477. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, T.J.; Duester, G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 2015, 16, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Bono, M.R.; Tejon, G.; Flores-Santibañez, F.; Fernandez, D.; Rosemblatt, M.; Sauma, D. Retinoic Acid as a Modulator of T Cell Immunity. Nutrients 2016, 8, 349. [Google Scholar] [CrossRef] [PubMed]
- Podolak-Popinigis, J.; Górnikiewicz, B.; Ronowicz, A.; Sachadyn, P. Transcriptome profiling reveals distinctive traits of retinol metabolism and neonatal parallels in the MRL/MpJ mouse. BMC Genom. 2015, 16, 926. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Chen, L.; Wang, F.; Zhou, X. Potential role of retinoids in ovarian physiology and pathogenesis of polycystic ovary syndrome. Clin. Chim. Acta 2017, 469, 87–93. [Google Scholar] [CrossRef]
- Baybutt, R.C.; Hu, L.; Molteni, A. Vitamin A deficiency injures lung and liver parenchyma and impairs function of rat type II pneumocytes. J. Nutr. 2000, 130, 1159–1165. [Google Scholar]
- Chen, H.; Kan, Q.; Zhao, L.; Ye, G.; He, X.; Tang, H.; Shi, F.; Zou, Y.; Liang, X.; Song, X.; et al. Prophylactic effect of Tongxieyaofang polysaccharide on depressive behavior in adolescent male mice with chronic unpredictable stress through the microbiome-gut-brain axis. Biomed. Pharmacother. 2023, 161, 114525. [Google Scholar] [CrossRef]

| Gene | Primer Sequence (5-3′) | bp |
|---|---|---|
| Cyp7a1-Forward | CTGCCGGTACTAGACAGCAT | 20 |
| Cyp7a1-Reverse | TCCTCCTTAGCTGTGCGGAT | 20 |
| Cyp27a1 | GGAACAGGTCAAGACCGACC | 20 |
| Cyp27a1 | CTTGTTCAGCGCCTGGAG | 18 |
| Adh7 | TGGGCCAGTTGATAACCCAC | 20 |
| Adh7 | GGACAGTCCGAATGCTTTGC | 20 |
| Baat | CTGTCGAACTACGGTTTTGGC | 21 |
| Baat | GCTGTCAGCTTGGCCATTTT | 20 |
| Retsat | CATTCTGCCGAGCGTCTACT | 20 |
| Retsat | GCTGGGGGTTACTCCGTAAG | 20 |
| Slco1a4 | AGCTTCTTCATAAAAACAGCAGTAA | 25 |
| Slco1a4 | TGTTAATGCCAACAGAAACATCTTG | 25 |
| Dhrs4 | CTTGGCACCTGGACTCATCA | 20 |
| Dhrs4 | CTGGCTTGCCTAGCCTTCTA | 20 |
| GADPH | ACAGCAACAGGGTGGTGGAC | 20 |
| GADPH | TTTGAGGGTGCAGCGAACTT | 20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Wang, X.; Deng, H.; Huang, H.; Liu, Y.; Zhong, Z.; Shen, L.; Cao, S.; Ma, X.; Zhou, Z.; et al. Integrated Transcriptome and Metabolomics to Reveal the Mechanism of Adipose Mesenchymal Stem Cells in Treating Liver Fibrosis. Int. J. Mol. Sci. 2023, 24, 16086. https://doi.org/10.3390/ijms242216086
Liu H, Wang X, Deng H, Huang H, Liu Y, Zhong Z, Shen L, Cao S, Ma X, Zhou Z, et al. Integrated Transcriptome and Metabolomics to Reveal the Mechanism of Adipose Mesenchymal Stem Cells in Treating Liver Fibrosis. International Journal of Molecular Sciences. 2023; 24(22):16086. https://doi.org/10.3390/ijms242216086
Chicago/Turabian StyleLiu, Haifeng, Xinmiao Wang, Hongchuan Deng, Haocheng Huang, Yifan Liu, Zhijun Zhong, Liuhong Shen, Suizhong Cao, Xiaoping Ma, Ziyao Zhou, and et al. 2023. "Integrated Transcriptome and Metabolomics to Reveal the Mechanism of Adipose Mesenchymal Stem Cells in Treating Liver Fibrosis" International Journal of Molecular Sciences 24, no. 22: 16086. https://doi.org/10.3390/ijms242216086
APA StyleLiu, H., Wang, X., Deng, H., Huang, H., Liu, Y., Zhong, Z., Shen, L., Cao, S., Ma, X., Zhou, Z., Chen, D., & Peng, G. (2023). Integrated Transcriptome and Metabolomics to Reveal the Mechanism of Adipose Mesenchymal Stem Cells in Treating Liver Fibrosis. International Journal of Molecular Sciences, 24(22), 16086. https://doi.org/10.3390/ijms242216086








