Linear Dichroism Measurements for the Study of Protein-DNA Interactions
Abstract
1. Introduction
2. Principles and Measurements
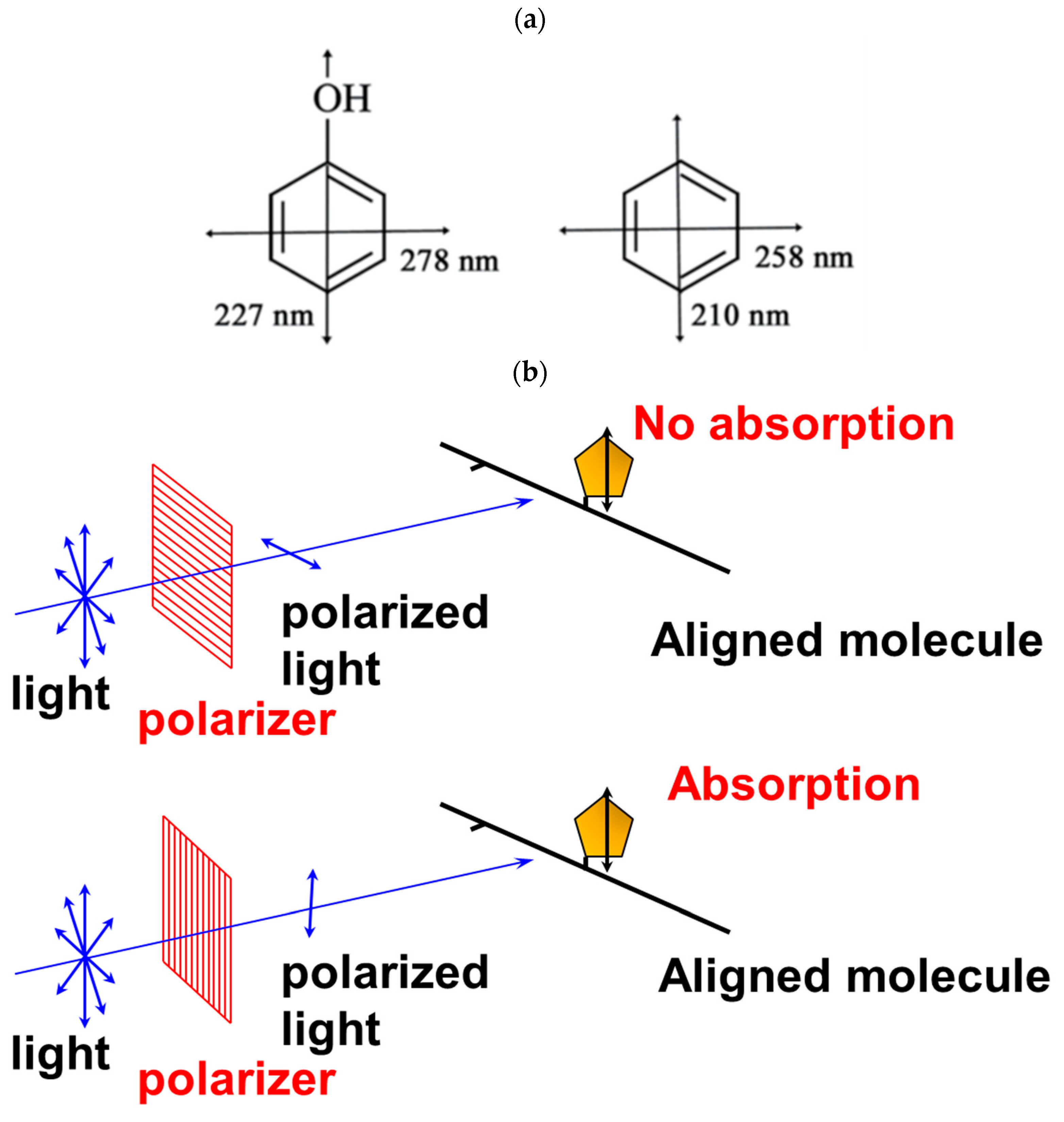
2.1. Principle of Linear Dichroism
2.2. Utility and Advantages of LD Measurements
Detection of Changes in Shape and Flexibility of DNA
2.3. Binding Mode (Intercalation or Groove Binding)
2.4. Advantages of LD Measurements
2.5. Difficulties in the Analysis of LD Signals
3. Measurements
4. Methods of Sample Alignment
4.1. Couette Cells
4.2. Flow Cells
4.3. Application of Electric or Magnetic Field
4.4. Deformation of Sample-Loaded Film or Gel
5. Examples of LD Applications
5.1. Determination of the Binding Mode of Ligands (Intercalation or Groove Binding)
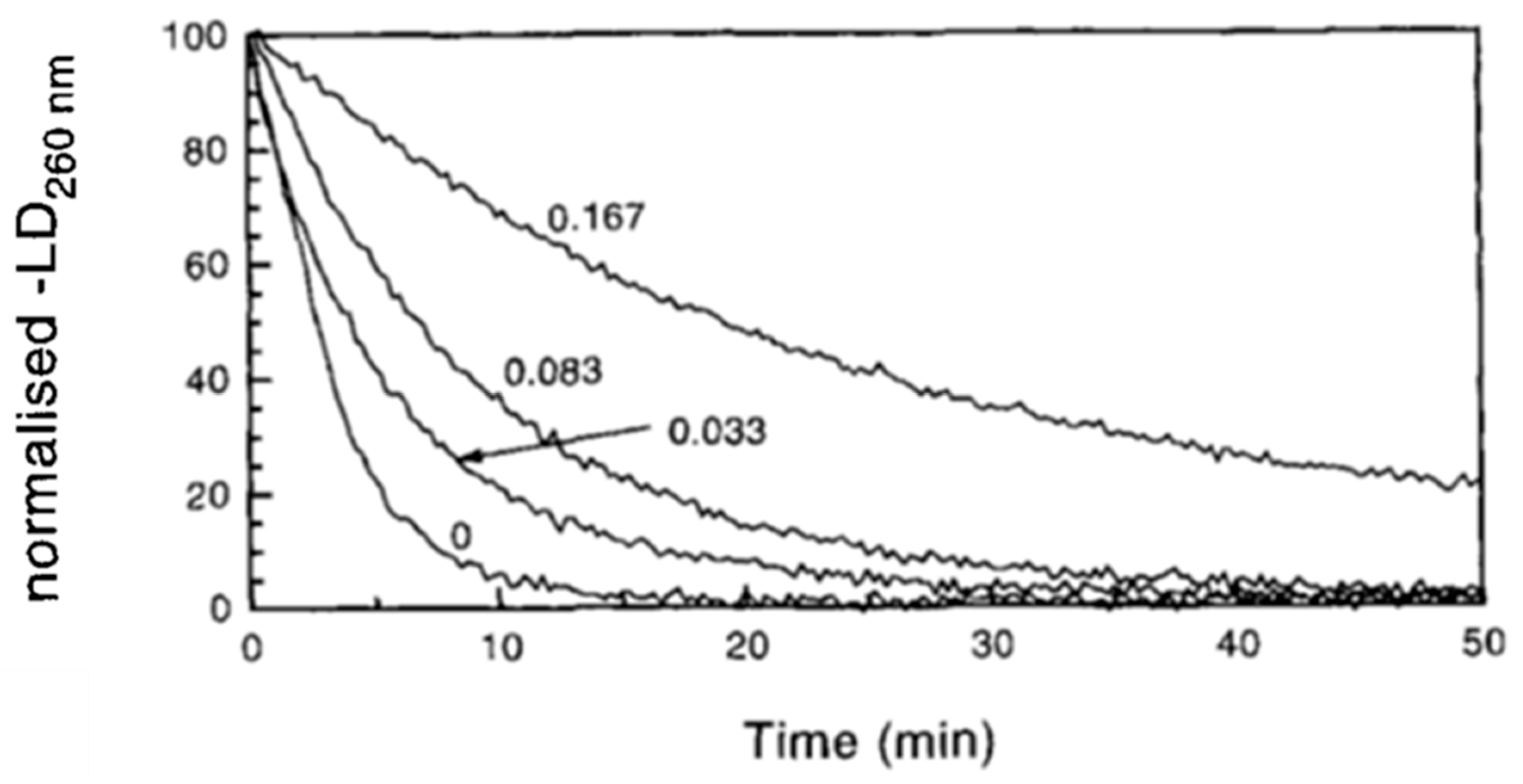
5.2. Kinetic Analysis of DNA Digestion by Endonucleases
5.3. DNA Bending by UvrB DNA Repair Protein
5.4. DNA Bending by CRP Transcription Activator
5.5. Structure of Single-Stranded DNA in Complexes with RecA and Rad51 Recombinases
5.6. Stiffening of Single-Stranded DNA by RecA and Rad51 Recombinases
5.7. Immobilization of Nucleobases of Single-Stranded DNA in the Presence of Strand Exchange Activating Agents
5.8. Perpendicular Orientation of DNA Bases in RecA and Rad51 Filaments
- (a)
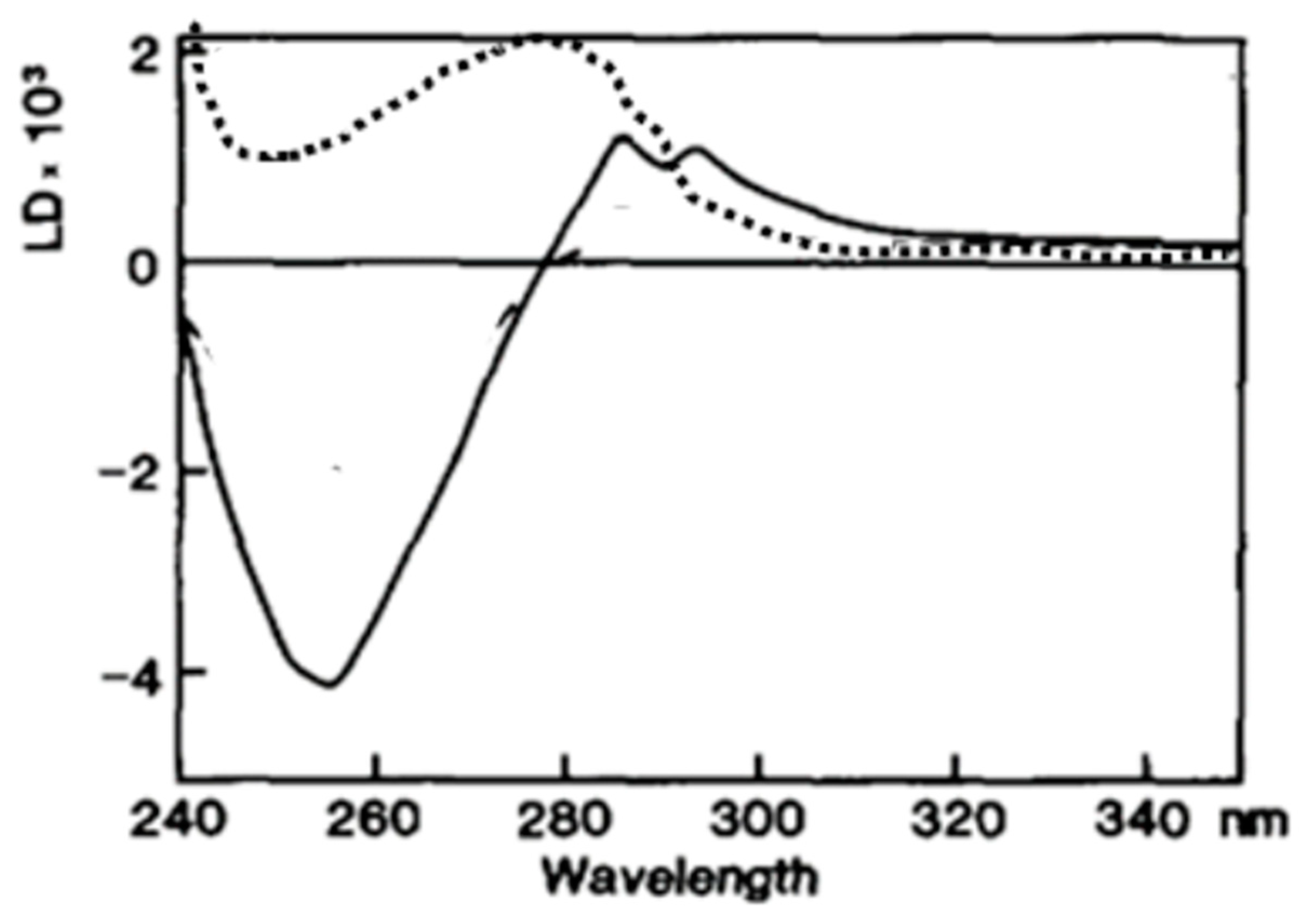
- A DNA analog (poly(dεA)) which absorbs UV light above 310 nm at a wavelength at where no other reaction constituents absorb light, was used [4,55] to unambiguously determine the orientation of poly(dεA) bases. Indeed, as expected, a negative LD signal was observed above 310 nm in the presence of ATPγS, supporting the perpendicular orientation of DNA bases in the recombinase DNA/complex (Figure 6) [9,55].
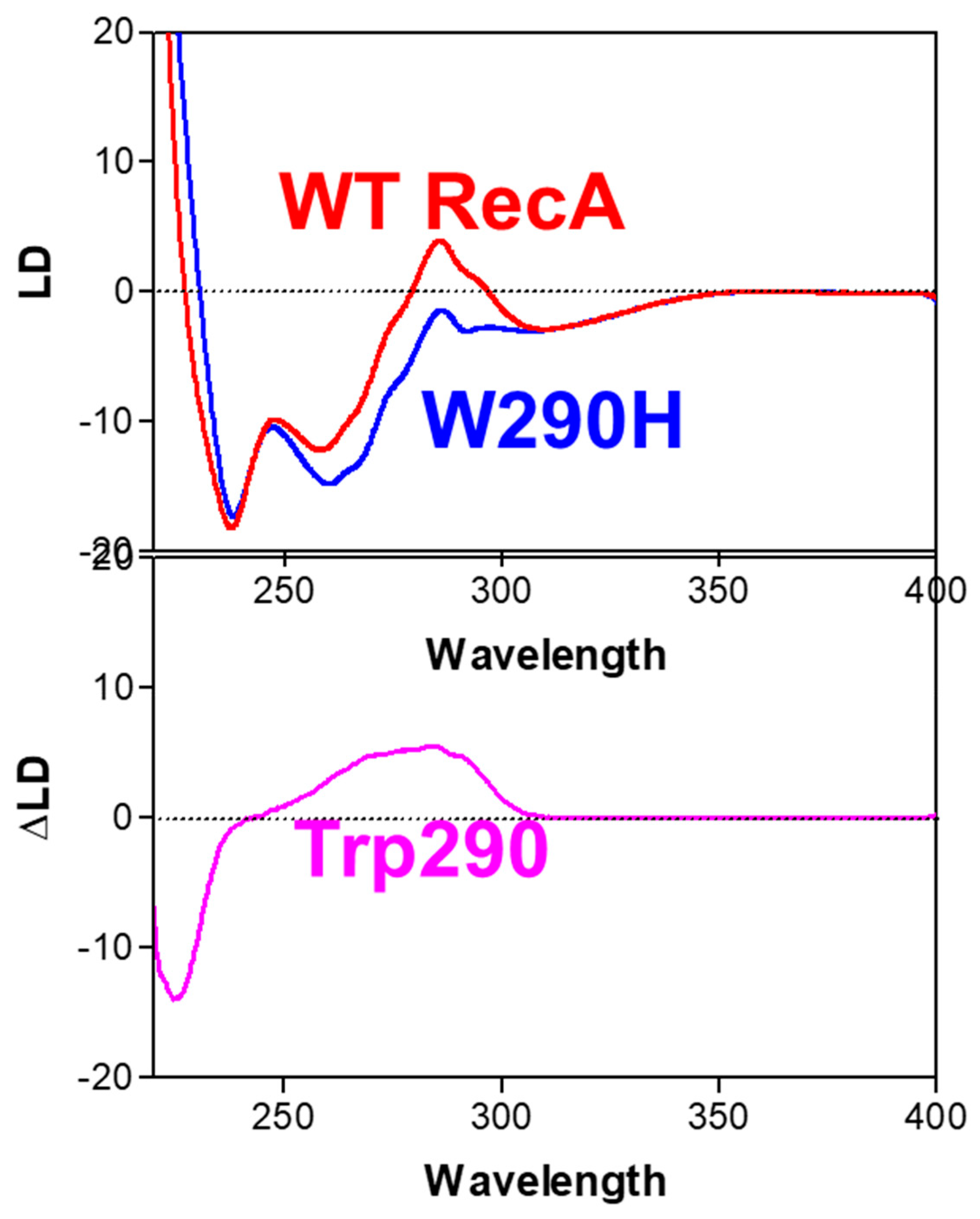
- (b)
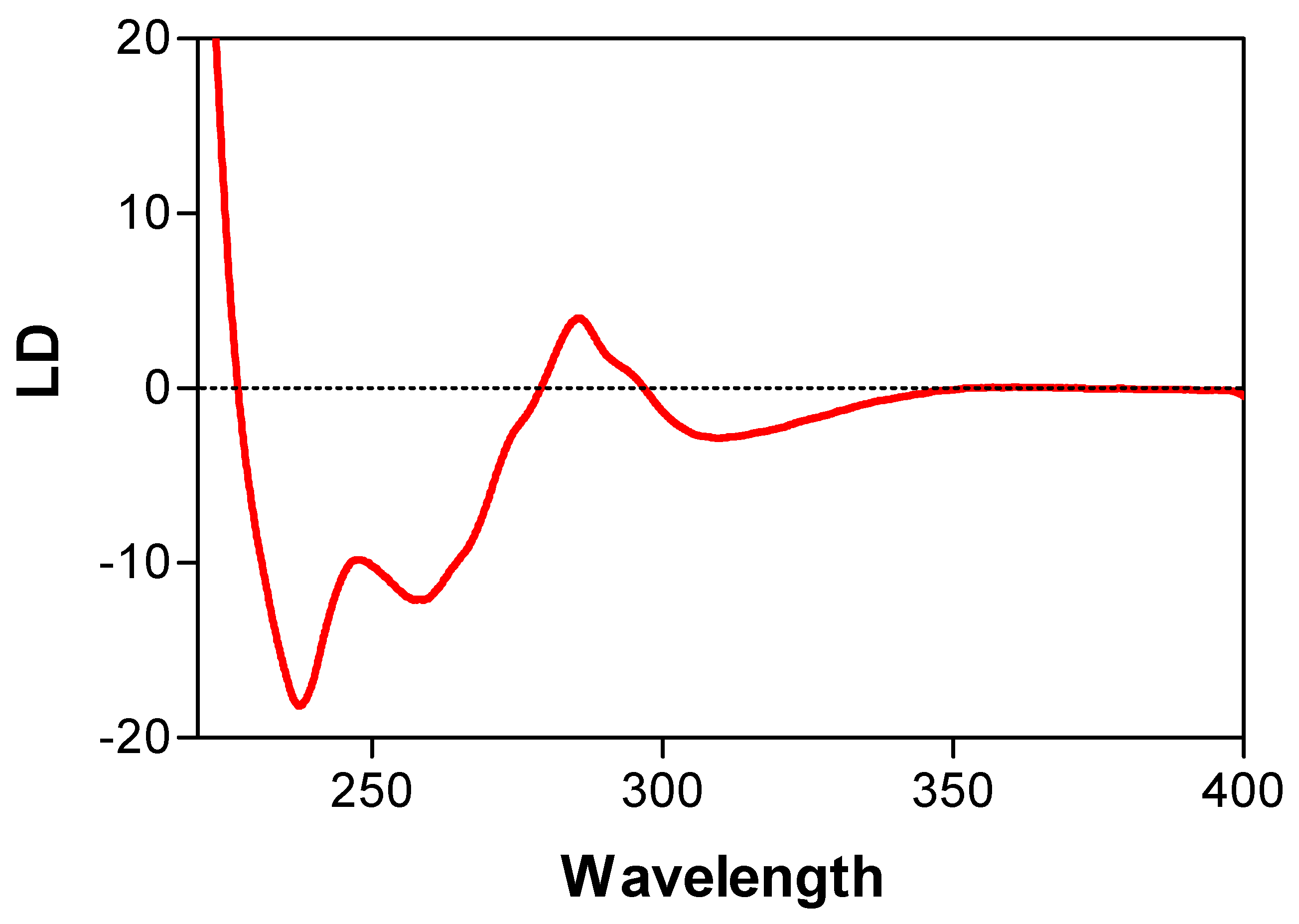
- A genetically engineered RecA protein was employed, in which one of its two Trp residues, the primary UV-absorbing residues, was replaced with a structurally similar residue (Tyr or His) with weaker UV-absorbance [56,57]. The poly(dεA)-engineered RecA complex also produced a negative LD signal above 310 nm, that was identical to that of the complex formed with wild-type RecA (Figure 7). However, as expected, there was a decrease in the positive band around 280 nm (Figure 7), indicating that the positive signal was produced by the Trp residues of RecA and not by the DNA bases, thus confirming the perpendicular orientation of the DNA bases in the active complex. This conclusion was later confirmed by crystallographic and cryo-EM analyses [58,59].
5.9. Building a Molecular Model of RecA/DNA and Rad51/DNA Complexes
6. Conclusions and Future Directions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Norden, B.; Kubista, M.; Kurucsev, T. Linear dichroism spectroscopy of nucleic acids. Q. Rev. Biophys. 1992, 25, 51–170. [Google Scholar] [CrossRef]
- Rodgers, A. How to study DNA and proteins by linear dichroism spectroscopy. Sci. Prog. 2008, 91, 377–396. [Google Scholar] [CrossRef] [PubMed]
- Bloemendal, M.; van Grondelle, R. Linear-dichroism spectroscopy for the study of structural properties of proteins. Mol. Biol. Rep. 1993, 18, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Norden, B.; Elvingson, C.; Kubista, M.; Sjöberg, B.; Ryberg, H.; Ryberg, M.; Mortensen, K.; Takahashi, M. Structure of RecA-DNA complexes studied by combination of linear dichroism and small angle neutron scattering measurements on flow-oriented samples. J. Mol. Biol. 1992, 226, 1175–1191. [Google Scholar] [CrossRef] [PubMed]
- Tuite, E.; Sehlstedt, U.; Hagmar, P.; Norden, B.; Takahashi, M. Effects of minor and major groove binding drugs and intercalators on the DNA association of minor-groove binding proteins RecA and DNaseI detected by flow linear dichroism. Eur. J. Biochem. 1997, 243, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Hicks, M.; Rodger, A.; Thomas, C.; Batt, S.; Dafforn, T. Restriction enzyme kinetics monitored by U.V. linear dichroism. Biochemistry 2006, 45, 8912–8917. [Google Scholar] [CrossRef]
- Porschke, D.; Hillen, W.; Takahashi, M. The change of DNA structure by specific binding of the cAMP receptor protein from rotation diffusion and dichroism measurement. EMBO J. 1984, 3, 2873–2878. [Google Scholar] [CrossRef]
- Takahashi, M.; Bertrand, E.; Fuchs, R.P.P.; Norden, B. Structure of UvrABC excinuclease-DNA complexes studied by flow linear dichroism: DNA curved by UvrB and UvrC. FEBS Lett. 1992, 314, 10–12. [Google Scholar] [CrossRef]
- Takahashi, M.; Kubista, M.; Nordén, B. Linear dichroism study of RecA-DNA complexes: Structural evidence and stoichiometries. J. Biol. Chem. 1987, 262, 8109–8111. [Google Scholar] [CrossRef]
- Maeshima, K.; Maraboeuf, F.; Morimatsu, K.; Horii, T.; Takahashi, M. Nucleotide dependent structural and kinetic changes in Xenopus Rad51.1-DNA complex stimulating the strand exchange reaction: Destacking of DNA bases and restriction of their local motion. J. Mol. Biol. 1998, 284, 689–697. [Google Scholar] [CrossRef]
- Hagmar, P.; Dahlman, K.; Takahashi, M.; Carlstedt-Duke, J.; Gustafsson, J.A.; Norden, B. Unspecific DNA binding of the DNA binding domain of the glucocorticoid receptor studied with flow linear dichroism. FEBS Lett. 1989, 253, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Nordén, B.; Kurucsev, T. Analysing DNA complexes by circular and linear dichroism. J. Mol. Recognit. 1994, 7, 141–155. [Google Scholar] [CrossRef]
- Colson, P.; Bailly, C.; Houssier, C. Electric linear dichroism as a new tool to study sequence preference in drug binding to DNA. Biophys. Chem. 1996, 58, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, P.; Nordén, B. DNA Binding Geometries of Ruthenium(II) Complexes with 1,10-Phenanthroline and 2,2′-Bipyridine Ligands Studied with Linear Dichroism Spectroscopy. J. Phys. Chem. B 1998, 102, 9583–9594. [Google Scholar] [CrossRef]
- Tuite, E.M.; Nordén, B. Linear and circular dichroism characterization of thionine binding mode with DNA polynucleotides. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 189, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Nordén, B. Linear dichroism studies of nucleic acids. II. Calculation of reduced dichroism curves of A-and B-form DNA. Biopolymers 1982, 21, 2433–2452. [Google Scholar] [CrossRef] [PubMed]
- Fornander, L.H.; Renodon-Corniere, A.; Kuwabara, N.; Ito, K.; Tsutsui, Y.; Shimizu, T.; Iwasaki, H.; Norden, B.; Takahashi, M. Swi5-Sfr1 protein stimulates Rad51-mediated DNA strand exchange reaction through organization of DNA bases in the presynaptic filament. Nucleic Acids Res. 2014, 42, 2358–2365. [Google Scholar] [CrossRef]
- Fornander, L.H.; Frykholm, K.; Reymer, A.; Renodon-Cornière, A.; Takahashi, M.; Nordén, B. Ca2+ improves organization of single-stranded DNA bases in human Rad51 filament, explaining stimulatory effect on gene recombination. Nucleic Acids Res. 2011, 40, 4904–4913. [Google Scholar] [CrossRef]
- Marrington, R.; Dafforn, T.R.; Halsall, D.J.; Rodger, A. Micro-volume Couette flow sample orientation for absorbance and fluorescence linear dichroism. Biophys. J. 2004, 87, 2002–2012. [Google Scholar] [CrossRef][Green Version]
- Morimatsu, K.; Takahashi, M. Structural analysis of RecA protein-DNA complexes by fluorescence-detected linear dichroism: Absence of structural change of filament for pairing of complementary DNA strands. Anal. Biochem. 2006, 358, 192–198. [Google Scholar] [CrossRef]
- Wemyss, A.M.; Chmel, N.P.; Lobo, D.P.; Sutherland, J.A.; Dafforn, T.R.; Rodger, A. Fluorescence detected linear dichroism spectroscopy: A selective and sensitive probe for fluorophores in flow-oriented systems. Chirality 2018, 30, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Phelps, C.; Lee, W.; Jose, D.; Marcus, A.H. Single-molecule FRET and linear dichroism studies of DNA breathing and helicase binding at replication fork junctions. Proc. Natl. Acad. Sci. USA 2013, 110, 17320–17325. [Google Scholar] [CrossRef]
- Adachi, R.; Yamaguchi, K.; Yagi, H.; Sakurai, K.; Naiki, H.; Goto, Y. Flow-induced alignment of amyloid protofilaments revealed by Linear Dichroism. J. Biol. Chem. 2007, 282, 8978–8983. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, S.; Hillen, W.; Morgeneyer, B.; Wells, R.D.; Pörschke, D. Orientation relaxation of DNA restriction fragments and the internal mobility of the double helix. Biophys. Chem. 1982, 15, 263–270. [Google Scholar] [CrossRef]
- Diekmann, S.; Pörschke, D. Electro-optical analysis of ‘curved’ DNA fragments. Biophys Chem. 1987, 26, 207–216. [Google Scholar] [CrossRef]
- Porschke, D. Electric dichroism and bending amplitudes of DNA fragments according to a simple orientation function for weakly bent rods. Biopolymers 1989, 28, 1383–1396. [Google Scholar] [CrossRef]
- Jonsson, M.; Jacobsson, U.; Takahashi, M.; Norden, B. Orientation of large DNA during free solution electrophoresis studied by linear dichroism. J. Chem. Soc. Faraday Trans. 1993, 89, 2791–2798. [Google Scholar] [CrossRef]
- Choi, E.; Lee, S.-H.; Kang, K.-T. DNA Alignment-Induced Linear Dichroism via Magnetic Field Assisted Electrospray: Implications for Optical Applications. ACS Appl. Nano Mater. 2023, 6, 7150–7155. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Nordén, B. Linear Dichroism Studies of Nucleic Acid Bases In Stretched Poly(Vlnyl Alcohol) Molecular Orientation and Electronic Transition Moment Directions. J. Phys. Chem. 1982, 86, 1378–1386. [Google Scholar] [CrossRef]
- Razmkhah, K.; Chmel, N.; Gibson, M.; Rodger, A. Oxidized polyethylene films for orienting polar molecules for linear dichroism spectroscopy. Analyst 2014, 139, 1372–1382. [Google Scholar] [CrossRef]
- Van Amerongen, H.; Vasmel, H.; Van Grondelle, R. Linear dichroism of chlorosomes from Chloroflexus aurantiacus in compressed gels and electric fields. Biophys. J. 1998, 54, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lee, G.J.; Jang, K.J.; Cho, T.S.; Kim, S.K. Real-time detection of Fe·EDTA/H2O2 -induced DNA cleavage by linear dichroism. Nucleic Acids Res. 2008, 36, e85. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sancar, A.; Tang, M.S. Nucleotide excision repair. Photochem. Photobiol. 1993, 57, 905–921. [Google Scholar] [CrossRef]
- Hsu, D.S.; Takahashi, M.; Delagoutte, E.; Bertrand-Burggraf, E.; Wang, Y.H.; Norden, B.; Fuchs, R.P.P.; Griffith, J.; Sancar, A. Flow linear dichroism and electron microscopic analysis of protein-DNA complexes of a mutant UvrB protein which binds to but cannot kink DNA. J. Mol. Biol. 1994, 241, 645–650. [Google Scholar] [CrossRef]
- Verhoeven, E.E.; Wyman, C.; Moolenaar, G.F.; Hoeijmakers, J.H.; Goosen, N. Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J. 2001, 20, 601–611. [Google Scholar] [CrossRef]
- Pastan, I.; Adhya, S. Cyclic adenosine 5’-monophosphate in Escherichia coli. Bacteriol. Rev. 1996, 40, 527–551. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Constantinidou, C.; Hobman, J.L.; Minchin, S.D. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004, 232, 5874–5893. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.M.; Crothers, D.M. The locus of sequence-directed and protein-induced DNA bending. Nature 1989, 308, 509–513. [Google Scholar] [CrossRef]
- Kolb, A.; Spassky, A.; Chapon, C.; Blazy, B.; Buc, H. On the different binding affinities of CRP at the lac, gal and malT promoter regions. Nucleic Acids Res. 1993, 11, 7833–7852. [Google Scholar] [CrossRef]
- Schultz, S.C.; Shields, G.C.; Steitz, T.A. Crystal structure of a CAP-DNA complex: The DNA is bent by 90 degrees. Science 1991, 253, 1001–1007. [Google Scholar] [CrossRef]
- Meyer-Almes, F.J.; Porschke, D. The cyclic AMP receptor promoter DNA complex: A comparison of crystal and solution structure by quantitative molecular electrooptics. J. Mol. Biol. 1997, 269, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.L.; Nikolov, D.B.; Burley, S.K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 1993, 365, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Parekh, B.S.; Hatfield, G.W. Transcriptional activation by protein-induced DNA bending: Evidence for a DNA structural transmission model. Proc. Natl. Acad. Sci. USA 1996, 93, 1173–1177. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Heyer, W.-D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.; Klein, H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 739–750. [Google Scholar] [CrossRef]
- Tsuzuki, T.; Fujii, Y.; Sakumi, K.; Tominaga, Y.; Nakao, K.; Sekiguchi, M.; Matsushiro, A.; Yoshimura, Y. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 1996, 93, 6236–6240. [Google Scholar] [CrossRef]
- Wang, Z.; Jia, R.; Wang, L.; Yang, Q.; Hu, X.; Fu, Q.; Zhang, X.; Li, W.; Ren, Y. The Emerging Roles of Rad51 in Cancer and Its Potential as a Therapeutic Target. Front. Oncol. 2022, 12, 935593. [Google Scholar] [CrossRef]
- Ward, A.; Khanna, K.K.; Wiegmans, A.P. Targeting homologous recombination, new pre-clinical and clinical therapeutic combinations inhibiting RAD51. Cancer Treat. Rev. 2014, 41, 35–45. [Google Scholar] [CrossRef]
- Tsang, E.S.; Munster, P.N. Targeting RAD51-mediated homologous recombination as a treatment for advanced solid and hematologic malignancies: Opportunities and challenges ahead. Onco Targets Ther. 2022, 15, 1509–1518. [Google Scholar] [CrossRef]
- Howard-Flanders, P.; West, S.C.; Stasiak, A. Role of RecA protein spiral filaments in genetic recombination. Nature 1984, 309, 215–219. [Google Scholar] [CrossRef]
- Shibata, T.; DasGupta, C.; Cunningham, R.P.; Radding, C.M. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc. Natl. Acad. Sci. USA 1979, 76, 1638–1642. [Google Scholar] [CrossRef] [PubMed]
- McEntee, K.; Weinstock, G.M.; Lehman, I.R. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1979, 76, 2615–2619. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Murayama, Y.; Takahashi, M.; Iwasaki, H. Two three-strand intermediates are processed during Rad51-driven DNA strand exchange. Nat. Struct. Mol. Biol. 2018, 25, 29–36. [Google Scholar] [CrossRef]
- Bugreev, D.V.; Mazin, A.V. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. USA 2004, 101, 9988–9993. [Google Scholar] [CrossRef] [PubMed]
- Haruta, N.; Kurokawa, Y.; Murayama, Y.; Akamatsu, Y.; Unzai, S.; Tsutsui, Y.; Iwasaki, H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat. Struct. Mol. Biol. 2006, 13, 823–830. [Google Scholar] [CrossRef]
- Hagmar, P.; Norden, B.; Baty, D.; Chartier, M.; Takahashi, M. Structure of DNA-RecA complexes studied by residue differential linear dichroism and fluorescence spectroscopy for a genetically engineered RecA protein. J. Mol. Biol. 1992, 226, 1193–1205. [Google Scholar] [CrossRef]
- Morimatsu, K.; Takahashi, M.; Norden, B. Arrangement of RecA protein in its active filament determined by polarised-light spectroscopy. Proc. Natl. Acad. Sci. USA 2002, 99, 11688–11693. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, H.; Pavletich, N.P. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 2008, 453, 489–494. [Google Scholar] [CrossRef]
- Yang, H.; Pavletich, N.P. Insights into homology search from cryo-EM structures of RecA DNA recombination intermediates. Curr. Opin. Genet. Dev. 2021, 71, 188–194. [Google Scholar] [CrossRef]
- Reymer, A.; Frykholm, K.; Morimatsu, K.; Takahashi, M.; Norden, B. Structure of human Rad51 protein filament from molecular modeling and site-specific linear dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2009, 106, 13248–13253. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, L.; Xu, Y.; Zhao, W.; Sung, P.; Wang, W. Cryo-EM structures of human recombinase RAD51 filaments in the catalysis of DNA strand exchange. Nat. Struct. Mol. Biol. 2017, 24, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Story, R.M.; Weber, I.T.; Steitz, T.A. The structure of the E. coli recA protein monomer and polymer. Nature 1991, 355, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, L.; Yu, D.S.; Lo, T.; Anand, S.; Lee, M.; Blundell, T.L.; Venkitaraman, A.R. Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature 2002, 420, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Dafforn, T.R.; Rajendra, J.; Halsall, D.J.; Serpell, L.C.; Rodger, A. Protein Fiber Linear Dichroism for Structure Determination and Kinetics in a Low-Volume, Low-Wavelength Couette Flow Cell. Biophys. J. 2003, 86, 404–410. [Google Scholar] [CrossRef]
- Hicks, M.R.; Dennison, S.R.; Olamoyesan, A.; Rodger, A. Flow Linear Dichroism of Protein-Membrane Systems. Methods Mol. Biol. 2021, 2263, 449–463. [Google Scholar]
- Norden, B.; Lindblom, G.; Jonas, I. Linear dichroism spectroscopy as a tool for studying molecular orientation in model membrane systems. J. Phys. Chem. 1977, 81, 2086–2093. [Google Scholar] [CrossRef]
- Samorì, B.; Lenaz, G.; Battino, M.; Marconi, G.; Domini, I. On coenzyme Q orientation in membranes: A linear dichroism study of ubiquinones in a model bilayer. J. Membarin Biol. 1992, 128, 193–203. [Google Scholar] [CrossRef]
- Rodger, A.; Rajendra, J.; Marrington, R.; Ardhammar, M.; Nordén, B.; Hirst, J.; Gilbert, A.T.B.; Dafforn, T.R.; Halsall, D.J.; Woolhead, C.A.; et al. Flow oriented linear dichroism to probe protein orientation in membrane environments. Phys. Chem. Chem. Phys. 2002, 4, 4051–4057. [Google Scholar] [CrossRef]
- Matsuo, K.; Maki, Y.; Namatame, H.; Taniguchi, M.; Gekko, K. Conformation of membrane-bound proteins revealed by vacuum-ultraviolet circular-dichroism and linear-dichroism spectroscopy. Proteins Struct. Funct. Bioinform. 2016, 84, 349–359. [Google Scholar] [CrossRef]
- Kampmann, M.; Atkinson, C.E.; Mattheyses, A.L.; Simon, S.M. Mapping the orientation of nuclear pore proteins in living cells with polarized fluorescence microscopy. Nat. Struct. Mol. Biol. 2011, 18, 643–649. [Google Scholar] [CrossRef]
- Benninger, R.K.P.; Önfelt, B.; Neil, M.A.A.; Davis, D.M.; French, P.M.W. Fluorescence Imaging of Two-Photon Linear Dichroism: Cholesterol Depletion Disrupts Molecular Orientation in Cell Membranes. Biophys. J. 2005, 88, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Zhanghao, K.; Chen, X.; Liu, W.; Li, M.; Liu, Y.; Wang, Y.; Luo, S.; Wang, X.; Shan, C.; Xie, H.; et al. Super-resolution imaging of fluorescent dipoles via polarized structured illumination microscopy. Nat. Commun. 2019, 10, 4694. [Google Scholar] [CrossRef] [PubMed]
- Bondar, A.; Rybakova, O.; Melcr, J.; Dohnálek, J.; Khoroshyy, P.; Ticháček, O.; Timr, S.; Miclea, P.; Sakhi, A.; Marková, V.; et al. Quantitative linear dichroism imaging of molecular processes in living cells made simple by open software tools. Commun. Biol. 2021, 4, 189. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, M.; Norden, B. Linear Dichroism Measurements for the Study of Protein-DNA Interactions. Int. J. Mol. Sci. 2023, 24, 16092. https://doi.org/10.3390/ijms242216092
Takahashi M, Norden B. Linear Dichroism Measurements for the Study of Protein-DNA Interactions. International Journal of Molecular Sciences. 2023; 24(22):16092. https://doi.org/10.3390/ijms242216092
Chicago/Turabian StyleTakahashi, Masayuki, and Bengt Norden. 2023. "Linear Dichroism Measurements for the Study of Protein-DNA Interactions" International Journal of Molecular Sciences 24, no. 22: 16092. https://doi.org/10.3390/ijms242216092
APA StyleTakahashi, M., & Norden, B. (2023). Linear Dichroism Measurements for the Study of Protein-DNA Interactions. International Journal of Molecular Sciences, 24(22), 16092. https://doi.org/10.3390/ijms242216092






