Unbiased Phosphoproteome Mining Reveals New Functional Sites of Metabolite-Derived PTMs Involved in MASLD Development
Abstract
1. Introduction
2. Results
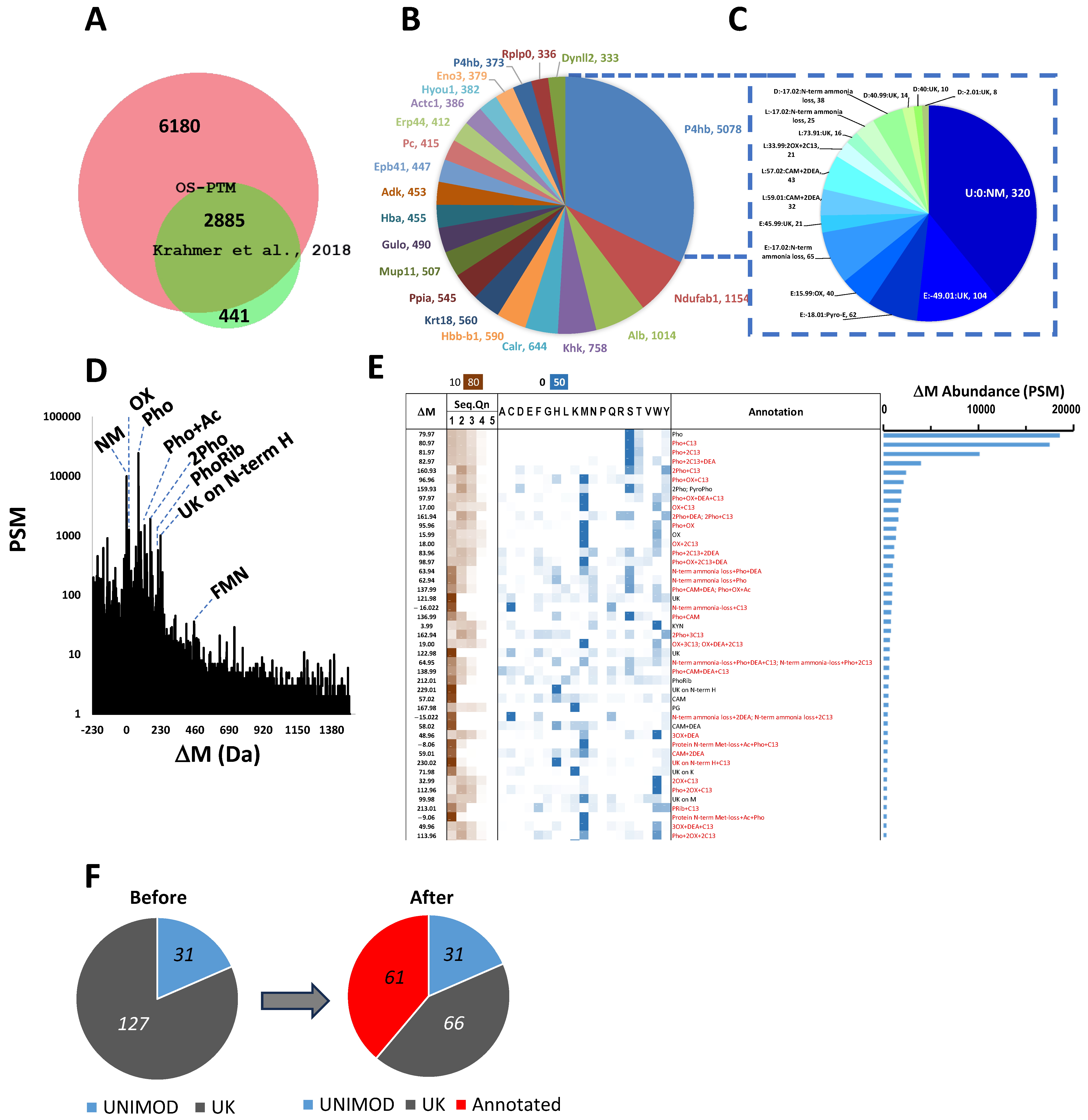
2.1. Unbiased Proteome-Wide OS PTM Analysis of Mouse Liver Phosphoproteome Reveals a Complex Picture
2.2. OS-PTM Uncovers Biologically Relevant Phosphate-Containing PTMs from Mouse Liver Phosphoproteome
2.2.1. ADP- and Phosphoribosylations
2.2.2. Glycerophosphorylations on Lys
2.2.3. Glycerylphosphorylethanolamine Modifications on Glu
2.2.4. Flavine Mononucleotide (FMN) on His
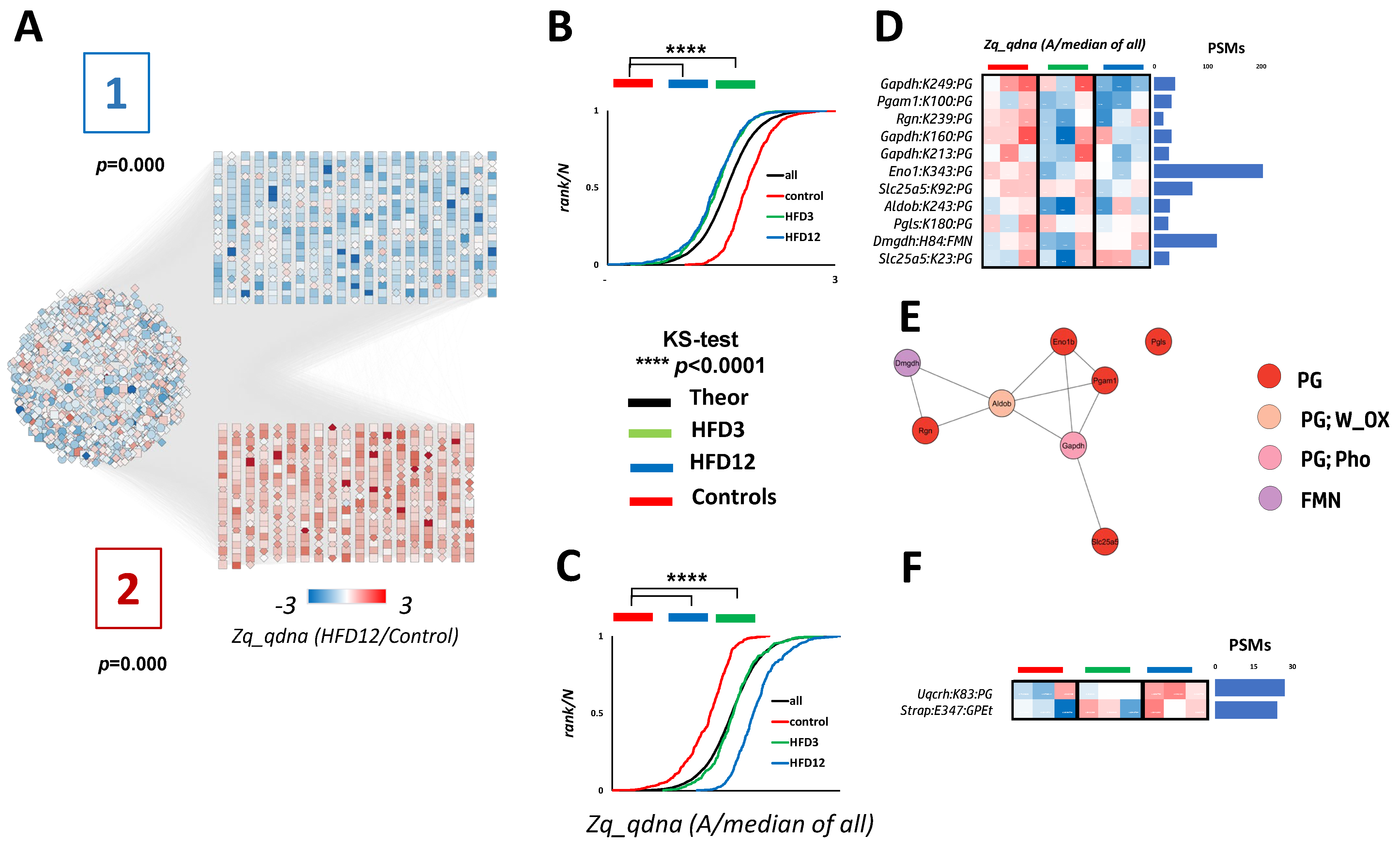
2.2.5. Some of the Newly Found Biologically Relevant PTMs Are Responsive to MASLD Development
2.2.6. Functional Implications of the Newly Found MASLD-Responsive PTMs in Their Respective Proteins
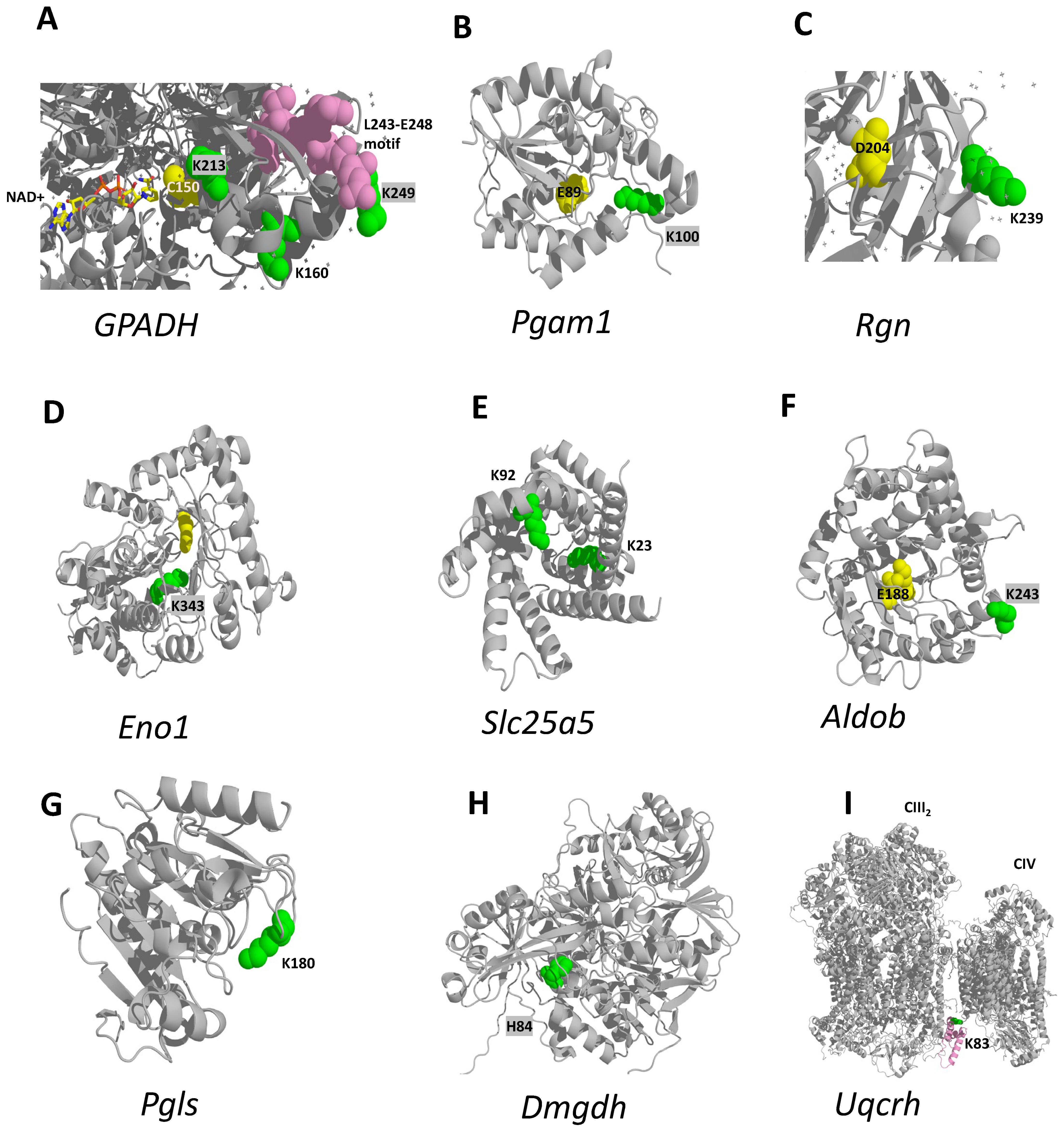
3. Discussion
4. Materials and Methods
4.1. MS Dataset
4.2. Data Analysis
4.3. Statistical Quantitative Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, S.; Xu, W.; Jiang, W.; Yu, W.; Lin, Y.; Zhang, T.; Yao, J.; Zhou, L.; Zeng, Y.; Li, H.; et al. Regulation of cellular metabolism by protein lysine acetylation. Science 2010, 327, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Moellering, R.E.; Cravatt, B.F. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science 2013, 341, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Prescott, N.A.; Maksimovic, I.; David, Y. (De)Toxifying the Epigenetic Code. Chem. Res. Toxicol. 2019, 32, 796–807. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Meana, M.; Minguet, M.; Bou-Teen, D.; Miro-Casas, E.; Castans, C.; Castellano, J.; Bonzon-Kulichenko, E.; Igual, A.; Rodriguez-Lecoq, R.; Vazquez, J.; et al. Ryanodine Receptor Glycation Favors Mitochondrial Damage in the Senescent Heart. Circulation 2019, 139, 949–964. [Google Scholar] [CrossRef]
- Herrero-Galan, E.; Martinez-Martin, I.; Sanchez-Gonzalez, C.; Vicente, N.; Bonzon-Kulichenko, E.; Calvo, E.; Suay-Corredera, C.; Pricolo, M.R.; Fernandez-Trasancos, A.; Velazquez-Carreras, D.; et al. Basal oxidation of conserved cysteines modulates cardiac titin stiffness and dynamics. Redox Biol. 2022, 52, 102306. [Google Scholar] [CrossRef]
- Rodriguez-Carrio, J.; Cerro-Pardo, I.; Lindholt, J.S.; Bonzon-Kulichenko, E.; Martinez-Lopez, D.; Roldan-Montero, R.; Escola-Gil, J.C.; Michel, J.B.; Blanco-Colio, L.M.; Vazquez, J.; et al. Malondialdehyde-modified HDL particles elicit a specific IgG response in abdominal aortic aneurysm. Free Radic. Biol. Med. 2021, 174, 171–181. [Google Scholar] [CrossRef]
- de la Fuente-Alonso, A.; Toral, M.; Alfayate, A.; Ruiz-Rodriguez, M.J.; Bonzon-Kulichenko, E.; Teixido-Tura, G.; Martinez-Martinez, S.; Mendez-Olivares, M.J.; Lopez-Maderuelo, D.; Gonzalez-Valdes, I.; et al. Aortic disease in Marfan syndrome is caused by overactivation of sGC-PRKG signaling by NO. Nat. Commun. 2021, 12, 2628. [Google Scholar] [CrossRef]
- Martinez-Lopez, D.; Camafeita, E.; Cedo, L.; Roldan-Montero, R.; Jorge, I.; Garcia-Marques, F.; Gomez-Serrano, M.; Bonzon-Kulichenko, E.; Blanco-Vaca, F.; Blanco-Colio, L.M.; et al. APOA1 oxidation is associated to dysfunctional high-density lipoproteins in human abdominal aortic aneurysm. eBioMedicine 2019, 43, 43–53. [Google Scholar] [CrossRef]
- Su, W.; Wu, S.; Yang, Y.; Guo, Y.; Zhang, H.; Su, J.; Chen, L.; Mao, Z.; Lan, R.; Cao, R.; et al. Phosphorylation of 17beta-hydroxysteroid dehydrogenase 13 at serine 33 attenuates nonalcoholic fatty liver disease in mice. Nat. Commun. 2022, 13, 6577. [Google Scholar] [CrossRef]
- Kalpage, H.A.; Wan, J.; Morse, P.T.; Zurek, M.P.; Turner, A.A.; Khobeir, A.; Yazdi, N.; Hakim, L.; Liu, J.; Vaishnav, A.; et al. Cytochrome c phosphorylation: Control of mitochondrial electron transport chain flux and apoptosis. Int. J. Biochem. Cell Biol. 2020, 121, 105704. [Google Scholar] [CrossRef]
- Friedrich, C.; Schallenberg, S.; Kirchner, M.; Ziehm, M.; Niquet, S.; Haji, M.; Beier, C.; Neudecker, J.; Klauschen, F.; Mertins, P. Comprehensive micro-scaled proteome and phosphoproteome characterization of archived retrospective cancer repositories. Nat. Commun. 2021, 12, 3576. [Google Scholar] [CrossRef] [PubMed]
- Fila, J.; Honys, D. Enrichment techniques employed in phosphoproteomics. Amino Acids 2012, 43, 1025–1047. [Google Scholar] [CrossRef] [PubMed]
- Kanshin, E.; Michnick, S.W.; Thibault, P. Displacement of N/Q-rich peptides on TiO2 beads enhances the depth and coverage of yeast phosphoproteome analyses. J. Proteome Res. 2013, 12, 2905–2913. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, F.; Ye, M.; Zou, H. Enrichment and separation techniques for large-scale proteomics analysis of the protein post-translational modifications. J. Chromatogr. A 2014, 1372C, 1–17. [Google Scholar] [CrossRef]
- Matic, I.; Ahel, I.; Hay, R.T. Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites. Nat. Methods 2012, 9, 771–772. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, J.; Liu, Z.; Wang, F. Identification of pyridoxal phosphate-modified proteins using mass spectrometry. Rapid Commun. Mass Spectrom. 2018, 32, 195–200. [Google Scholar] [CrossRef]
- Devabhaktuni, A.; Lin, S.; Zhang, L.; Swaminathan, K.; Gonzalez, C.G.; Olsson, N.; Pearlman, S.M.; Rawson, K.; Elias, J.E. TagGraph reveals vast protein modification landscapes from large tandem mass spectrometry datasets. Nat. Biotechnol. 2019, 37, 469–479. [Google Scholar] [CrossRef]
- Kong, A.T.; Leprevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef]
- Bagwan, N.; Bonzon-Kulichenko, E.; Calvo, E.; Lechuga-Vieco, A.V.; Michalakopoulos, S.; Trevisan-Herraz, M.; Ezkurdia, I.; Rodriguez, J.M.; Magni, R.; Latorre-Pellicer, A.; et al. Comprehensive Quantification of the Modified Proteome Reveals Oxidative Heart Damage in Mitochondrial Heteroplasmy. Cell Rep. 2018, 23, 3685–3697.e4. [Google Scholar] [CrossRef]
- Chick, J.M.; Kolippakkam, D.; Nusinow, D.P.; Zhai, B.; Rad, R.; Huttlin, E.L.; Gygi, S.P. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat. Biotechnol. 2015, 33, 743–749. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A multi-society Delphi consensus statement on new fatty liver disease nomenclature. Ann. Hepatol. 2023, 29, 101133. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.Y.; Zhai, Z.Z.; Li, Z.F.; Wang, L. High fat diet-triggered non-alcoholic fatty liver disease: A review of proposed mechanisms. Chem. Biol. Interact. 2020, 330, 109199. [Google Scholar] [CrossRef] [PubMed]
- Song, B.J.; Akbar, M.; Abdelmegeed, M.A.; Byun, K.; Lee, B.; Yoon, S.K.; Hardwick, J.P. Mitochondrial dysfunction and tissue injury by alcohol, high fat, nonalcoholic substances and pathological conditions through post-translational protein modifications. Redox Biol. 2014, 3, 109–123. [Google Scholar] [CrossRef]
- Krahmer, N.; Najafi, B.; Schueder, F.; Quagliarini, F.; Steger, M.; Seitz, S.; Kasper, R.; Salinas, F.; Cox, J.; Uhlenhaut, N.H.; et al. Organellar Proteomics and Phospho-Proteomics Reveal Subcellular Reorganization in Diet-Induced Hepatic Steatosis. Dev. Cell 2018, 47, 205–221.e7. [Google Scholar] [CrossRef] [PubMed]
- Boja, E.S.; Fales, H.M. Overalkylation of a protein digest with iodoacetamide. Anal. Chem. 2001, 73, 3576–3582. [Google Scholar] [CrossRef]
- Kuznetsova, K.G.; Levitsky, L.I.; Pyatnitskiy, M.A.; Ilina, I.Y.; Bubis, J.A.; Solovyeva, E.M.; Zgoda, V.G.; Gorshkov, M.V.; Moshkovskii, S.A. Cysteine alkylation methods in shotgun proteomics and their possible effects on methionine residues. J. Proteom. 2021, 231, 104022. [Google Scholar] [CrossRef]
- Crestfield, A.M.; Moore, S.; Stein, W.H. The preparation and enzymatic hydrolysis of reduced and S-carboxymethylated proteins. J. Biol. Chem. 1963, 238, 622–627. [Google Scholar] [CrossRef]
- Geoghegan, K.F.; Hoth, L.R.; Tan, D.H.; Borzilleri, K.A.; Withka, J.M.; Boyd, J.G. Cyclization of N-terminal S-carbamoylmethylcysteine causing loss of 17 Da from peptides and extra peaks in peptide maps. J. Proteome Res. 2002, 1, 181–187. [Google Scholar] [CrossRef]
- Simon, E.S.; Allison, J. Determination of pyridoxal-5′-phosphate (PLP)-bonding sites in proteins: A peptide mass fingerprinting approach based on diagnostic tandem mass spectral features of PLP-modified peptides. Rapid Commun. Mass Spectrom. 2009, 23, 3401–3408. [Google Scholar] [CrossRef]
- Vestling, M.M.; Kelly, M.A.; Fenselau, C. Optimization by mass spectrometry of a tryptophan-specific protein cleavage reaction. Rapid Commun. Mass Spectrom. 1994, 8, 786–790. [Google Scholar] [CrossRef]
- Zhang, Z.; Chow, S.Y.; De Guzman, R.; Joh, N.H.; Joubert, M.K.; Richardson, J.; Shah, B.; Wikstrom, M.; Zhou, Z.S.; Wypych, J. A Mass Spectrometric Characterization of Light-Induced Modifications in Therapeutic Proteins. J. Pharm. Sci. 2022, 111, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Polasky, D.A.; Geiszler, D.J.; Yu, F.; Li, K.; Teo, G.C.; Nesvizhskii, A.I. MSFragger-Labile: A Flexible Method to Improve Labile PTM Analysis in Proteomics. Mol. Cell Proteom. 2023, 22, 100538. [Google Scholar] [CrossRef] [PubMed]
- Hengel, S.M.; Goodlett, D.R. A Review of Tandem Mass Spectrometry Characterization of Adenosine Diphosphate-Ribosylated Peptides. Int. J. Mass Spectrom. 2012, 312, 114–121. [Google Scholar] [CrossRef]
- Moss, J.; Stanley, S.J. Amino acid-specific ADP-ribosylation. Identification of an arginine-dependent ADP-ribosyltransferase in rat liver. J. Biol. Chem. 1981, 256, 7830–7833. [Google Scholar] [CrossRef] [PubMed]
- Martello, R.; Leutert, M.; Jungmichel, S.; Bilan, V.; Larsen, S.C.; Young, C.; Hottiger, M.O.; Nielsen, M.L. Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat. Commun. 2016, 7, 12917. [Google Scholar] [CrossRef]
- Buch-Larsen, S.C.; Hendriks, I.A.; Lodge, J.M.; Rykaer, M.; Furtwangler, B.; Shishkova, E.; Westphall, M.S.; Coon, J.J.; Nielsen, M.L. Mapping Physiological ADP-Ribosylation Using Activated Ion Electron Transfer Dissociation. Cell Rep. 2020, 32, 108176. [Google Scholar] [CrossRef]
- Gehrig, P.M.; Nowak, K.; Panse, C.; Leutert, M.; Grossmann, J.; Schlapbach, R.; Hottiger, M.O. Gas-Phase Fragmentation of ADP-Ribosylated Peptides: Arginine-Specific Side-Chain Losses and Their Implication in Database Searches. J. Am. Soc. Mass Spectrom. 2021, 32, 157–168. [Google Scholar] [CrossRef]
- Thoden, J.B.; Wohlers, T.M.; Fridovich-Keil, J.L.; Holden, H.M. Human UDP-galactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J. Biol. Chem. 2001, 276, 15131–15136. [Google Scholar] [CrossRef]
- Fridovich-Keil, J.; Bean, L.; He, M.; Schroer, R. Epimerase Deficiency Galactosemia. In GeneReviews®; Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Park, J.; Chen, Y.; Tishkoff, D.X.; Peng, C.; Tan, M.; Dai, L.; Xie, Z.; Zhang, Y.; Zwaans, B.M.; Skinner, M.E.; et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 2013, 50, 919–930. [Google Scholar] [CrossRef]
- Rosenberry, T.L.; Krall, J.A.; Dever, T.E.; Haas, R.; Louvard, D.; Merrick, W.C. Biosynthetic incorporation of [3H]ethanolamine into protein synthesis elongation factor 1 alpha reveals a new post-translational protein modification. J. Biol. Chem. 1989, 264, 7096–7099. [Google Scholar] [CrossRef]
- Whiteheart, S.W.; Shenbagamurthi, P.; Chen, L.; Cotter, R.J.; Hart, G.W. Murine elongation factor 1 alpha (EF-1 alpha) is posttranslationally modified by novel amide-linked ethanolamine-phosphoglycerol moieties. Addition of ethanolamine-phosphoglycerol to specific glutamic acid residues on EF-1 alpha. J. Biol. Chem. 1989, 264, 14334–14341. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.R.; Kinzy, T.G. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 2005, 12, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Mittal, N.; Subramanian, G.; Butikofer, P.; Madhubala, R. Unique posttranslational modifications in eukaryotic translation factors and their roles in protozoan parasite viability and pathogenesis. Mol. Biochem. Parasitol. 2013, 187, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Signorell, A.; Jelk, J.; Rauch, M.; Butikofer, P. Phosphatidylethanolamine is the precursor of the ethanolamine phosphoglycerol moiety bound to eukaryotic elongation factor 1A. J. Biol. Chem. 2008, 283, 20320–20329. [Google Scholar] [CrossRef]
- Luka, Z.; Pakhomova, S.; Loukachevitch, L.V.; Newcomer, M.E.; Wagner, C. Folate in demethylation: The crystal structure of the rat dimethylglycine dehydrogenase complexed with tetrahydrofolate. Biochem. Biophys. Res. Commun. 2014, 449, 392–398. [Google Scholar] [CrossRef][Green Version]
- Cook, R.J.; Misono, K.S.; Wagner, C. The amino acid sequences of the flavin-peptides of dimethylglycine dehydrogenase and sarcosine dehydrogenase from rat liver mitochondria. J. Biol. Chem. 1985, 260, 12998–13002. [Google Scholar] [CrossRef]
- Ha, M.N.; Graham, F.L.; D’Souza, C.K.; Muller, W.J.; Igdoura, S.A.; Schellhorn, H.E. Functional rescue of vitamin C synthesis deficiency in human cells using adenoviral-based expression of murine l-gulono-gamma-lactone oxidase. Genomics 2004, 83, 482–492. [Google Scholar] [CrossRef]
- Herbrich, S.M.; Cole, R.N.; West, K.P., Jr.; Schulze, K.; Yager, J.D.; Groopman, J.D.; Christian, P.; Wu, L.; O’Meally, R.N.; May, D.H.; et al. Statistical inference from multiple iTRAQ experiments without using common reference standards. J. Proteome Res. 2013, 12, 594–604. [Google Scholar] [CrossRef]
- Jia, J.; Arif, A.; Terenzi, F.; Willard, B.; Plow, E.F.; Hazen, S.L.; Fox, P.L. Target-selective protein S-nitrosylation by sequence motif recognition. Cell 2014, 159, 623–634. [Google Scholar] [CrossRef]
- Jia, J.; Arif, A.; Willard, B.; Smith, J.D.; Stuehr, D.J.; Hazen, S.L.; Fox, P.L. Protection of extraribosomal RPL13a by GAPDH and dysregulation by S-nitrosylation. Mol. Cell 2012, 47, 656–663. [Google Scholar] [CrossRef]
- Sampath, P.; Mazumder, B.; Seshadri, V.; Gerber, C.A.; Chavatte, L.; Kinter, M.; Ting, S.M.; Dignam, J.D.; Kim, S.; Driscoll, D.M.; et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: Gene-specific silencing of translation. Cell 2004, 119, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.S.; Jia, J.; Yao, P.; Majumder, M.; Hatzoglou, M.; Fox, P.L. A stress-responsive RNA switch regulates VEGFA expression. Nature 2009, 457, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Weinert, B.T.; Scholz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F.; Pierri, C.L.; De Grassi, A.; Nunes-Nesi, A.; Fernie, A.R. Evolution, structure and function of mitochondrial carriers: A review with new insights. Plant J. 2011, 66, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Augustin, P.; Hromic, A.; Pavkov-Keller, T.; Gruber, K.; Macheroux, P. Structure and biochemical properties of recombinant human dimethylglycine dehydrogenase and comparison to the disease-related H109R variant. FEBS J. 2016, 283, 3587–3603. [Google Scholar] [CrossRef] [PubMed]
- Willie, A.; Edmondson, D.E.; Jorns, M.S. Sarcosine oxidase contains a novel covalently bound FMN. Biochemistry 1996, 35, 5292–5299. [Google Scholar] [CrossRef] [PubMed]
- Steenkamp, D.J. Identification of the prosthetic groups of dimethylamine dehydrogenase from Hyphomicrobium X. Biochem. Biophys. Res. Commun. 1979, 88, 244–250. [Google Scholar] [CrossRef]
- Steenkamp, D.J.; McIntire, W.; Kenney, W.C. Structure of the covalently bound coenzyme of trimethylamine dehydrogenase. Evidence for a 6-substituted flavin. J. Biol. Chem. 1978, 253, 2818–2824. [Google Scholar] [CrossRef]
- Vercellino, I.; Sazanov, L.A. Structure and assembly of the mammalian mitochondrial supercomplex CIII(2)CIV. Nature 2021, 598, 364–367. [Google Scholar] [CrossRef]
- Rardin, M.J.; Newman, J.C.; Held, J.M.; Cusack, M.P.; Sorensen, D.J.; Li, B.; Schilling, B.; Mooney, S.D.; Kahn, C.R.; Verdin, E.; et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc. Natl. Acad. Sci. USA 2013, 110, 6601–6606. [Google Scholar] [CrossRef] [PubMed]
- Chari, A.; Golas, M.M.; Klingenhager, M.; Neuenkirchen, N.; Sander, B.; Englbrecht, C.; Sickmann, A.; Stark, H.; Fischer, U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell 2008, 135, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Seong, H.A.; Jung, H.; Choi, H.S.; Kim, K.T.; Ha, H. Regulation of transforming growth factor-beta signaling and PDK1 kinase activity by physical interaction between PDK1 and serine-threonine kinase receptor-associated protein. J. Biol. Chem. 2005, 280, 42897–42908. [Google Scholar] [CrossRef] [PubMed]
- Duque, S.I.; Arnold, W.D.; Odermatt, P.; Li, X.; Porensky, P.N.; Schmelzer, L.; Meyer, K.; Kolb, S.J.; Schumperli, D.; Kaspar, B.K.; et al. A large animal model of spinal muscular atrophy and correction of phenotype. Ann. Neurol. 2015, 77, 399–414. [Google Scholar] [CrossRef]
- Donlin-Asp, P.G.; Bassell, G.J.; Rossoll, W. A role for the survival of motor neuron protein in mRNP assembly and transport. Curr. Opin. Neurobiol. 2016, 39, 53–61. [Google Scholar] [CrossRef]
- Huh, H.D.; Ra, E.A.; Lee, T.A.; Kang, S.; Park, A.; Lee, E.; Choi, J.L.; Jang, E.; Lee, J.E.; Lee, S.; et al. STRAP Acts as a Scaffolding Protein in Controlling the TLR2/4 Signaling Pathway. Sci. Rep. 2016, 6, 38849. [Google Scholar] [CrossRef]
- Horie, Y.; Suzuki, A.; Kataoka, E.; Sasaki, T.; Hamada, K.; Sasaki, J.; Mizuno, K.; Hasegawa, G.; Kishimoto, H.; Iizuka, M.; et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J. Clin. Investig. 2004, 113, 1774–1783. [Google Scholar] [CrossRef]
- Keramida, G.; Hunter, J.; Peters, A.M. Hepatic glucose utilization in hepatic steatosis and obesity. Biosci. Rep. 2016, 36, e00402. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moltó, E.; Pintado, C.; Louzada, R.A.; Bernal-Mizrachi, E.; Andrés, A.; Gallardo, N.; Bonzon-Kulichenko, E. Unbiased Phosphoproteome Mining Reveals New Functional Sites of Metabolite-Derived PTMs Involved in MASLD Development. Int. J. Mol. Sci. 2023, 24, 16172. https://doi.org/10.3390/ijms242216172
Moltó E, Pintado C, Louzada RA, Bernal-Mizrachi E, Andrés A, Gallardo N, Bonzon-Kulichenko E. Unbiased Phosphoproteome Mining Reveals New Functional Sites of Metabolite-Derived PTMs Involved in MASLD Development. International Journal of Molecular Sciences. 2023; 24(22):16172. https://doi.org/10.3390/ijms242216172
Chicago/Turabian StyleMoltó, Eduardo, Cristina Pintado, Ruy Andrade Louzada, Ernesto Bernal-Mizrachi, Antonio Andrés, Nilda Gallardo, and Elena Bonzon-Kulichenko. 2023. "Unbiased Phosphoproteome Mining Reveals New Functional Sites of Metabolite-Derived PTMs Involved in MASLD Development" International Journal of Molecular Sciences 24, no. 22: 16172. https://doi.org/10.3390/ijms242216172
APA StyleMoltó, E., Pintado, C., Louzada, R. A., Bernal-Mizrachi, E., Andrés, A., Gallardo, N., & Bonzon-Kulichenko, E. (2023). Unbiased Phosphoproteome Mining Reveals New Functional Sites of Metabolite-Derived PTMs Involved in MASLD Development. International Journal of Molecular Sciences, 24(22), 16172. https://doi.org/10.3390/ijms242216172





