Analysis of Growth Trajectories and Verification of Related SNPs in Populus deltoides
Abstract
:1. Introduction
2. Results
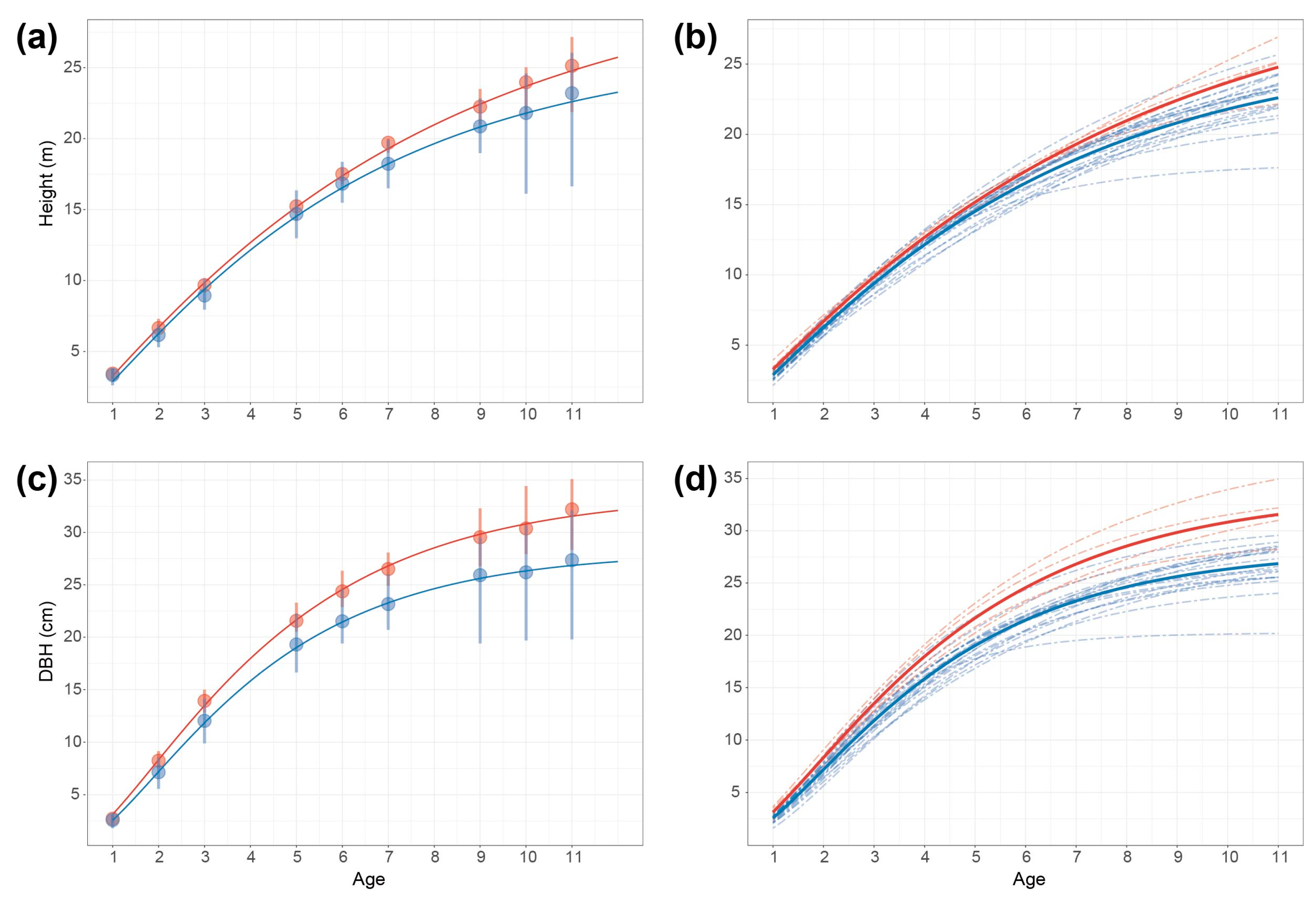
2.1. Phenotypic Variation of Growth Traits in 24 P. deltoides Clones
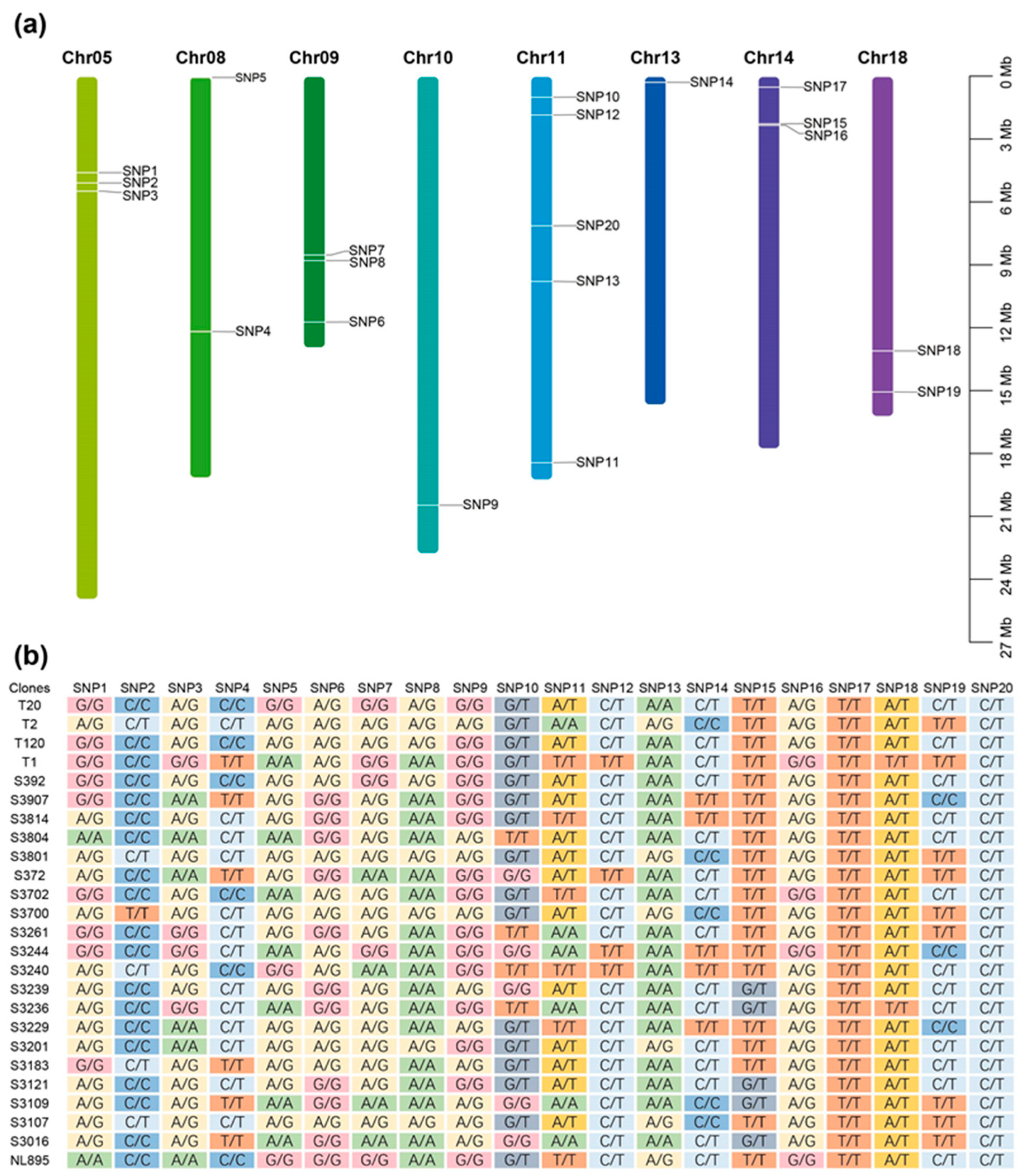
2.2. SNP Identification Using Sequencing Data
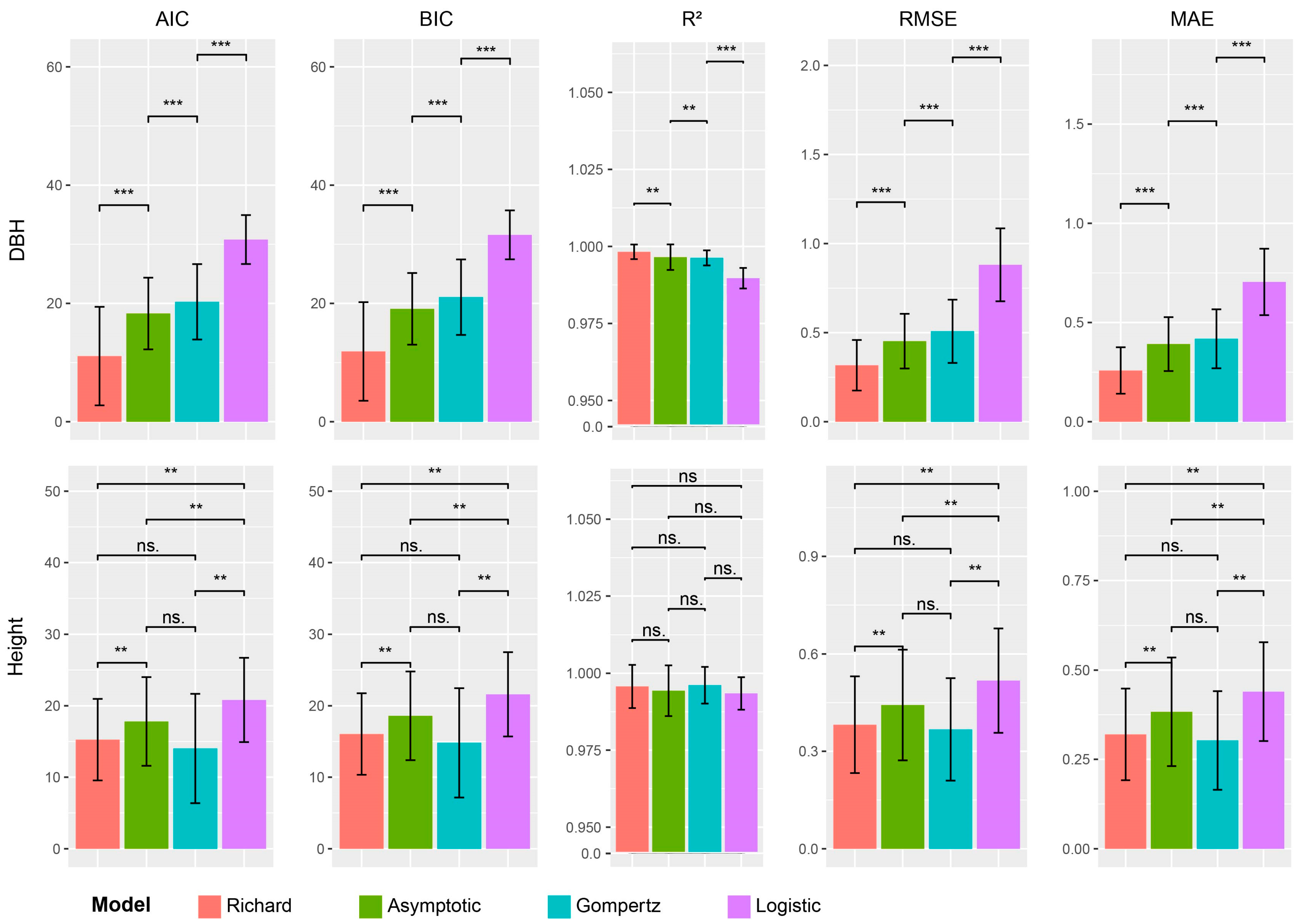
2.3. Selection of Growth Curve Models
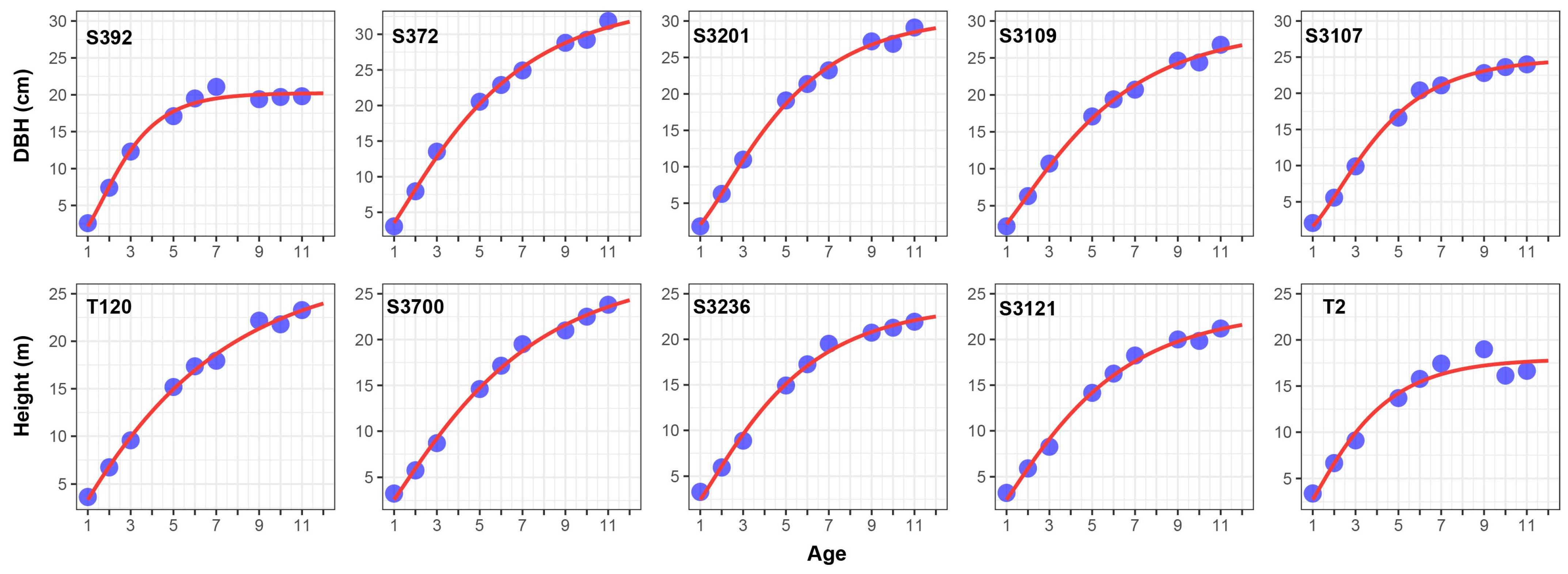
2.4. Estimation of Parameters in Richard Growth Model
2.5. Determination of Ontogenetic Stages
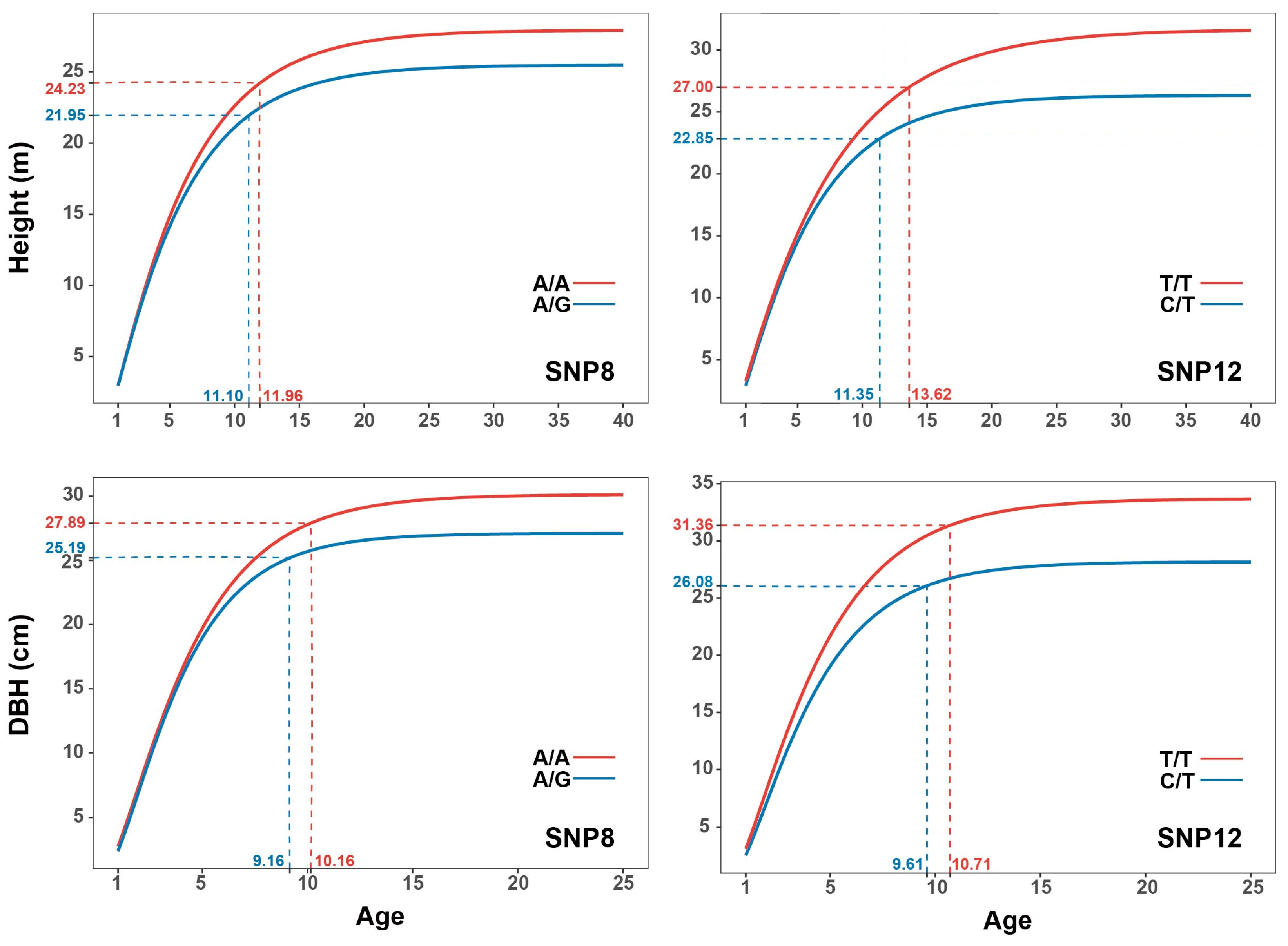
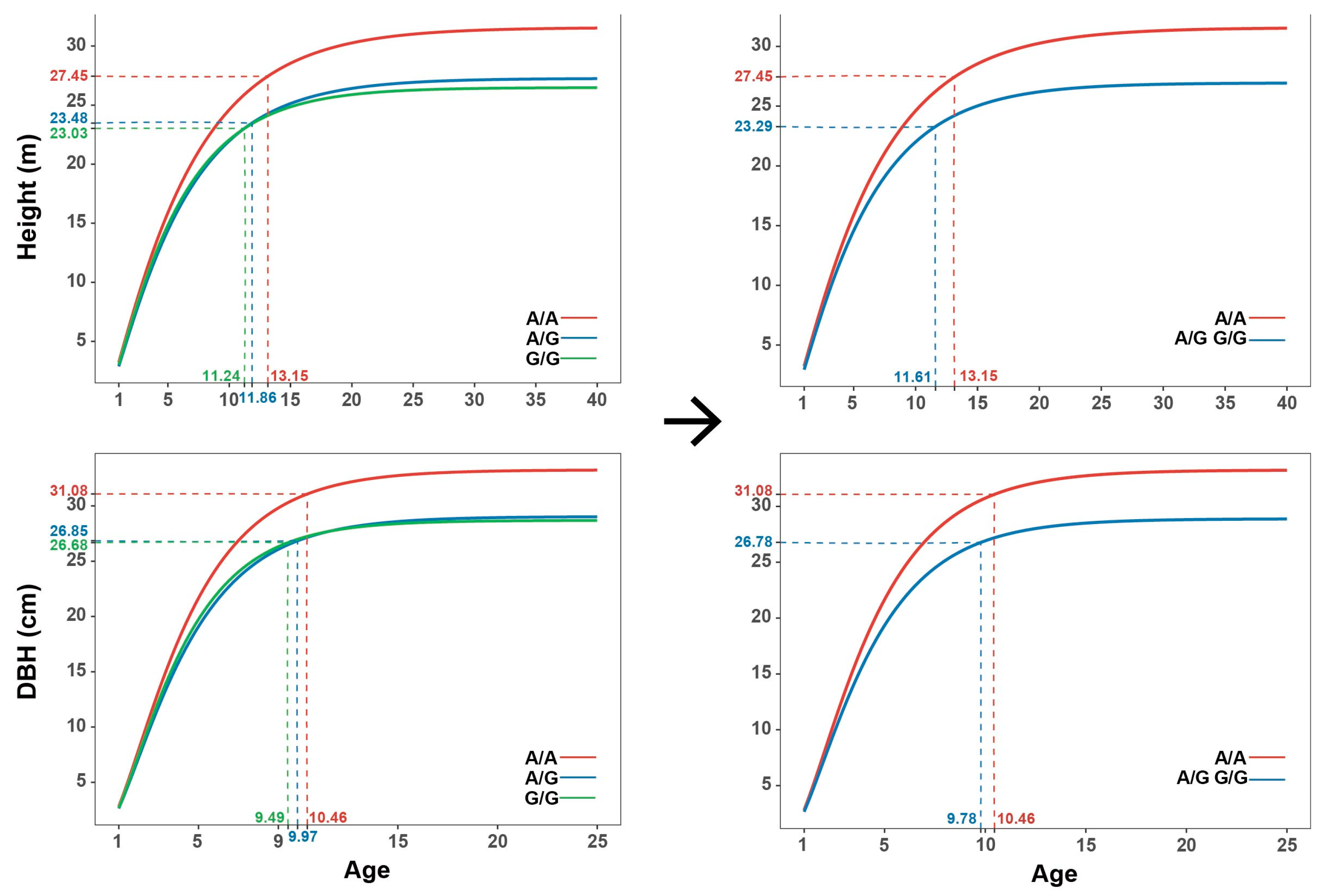
2.6. Effect Modes of Significant SNPs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Trait Measurements
4.2. DNA Extraction, Resequencing, and Genotyping
4.3. Model Evaluation
4.4. Hypothesis Testing
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, C.; Ding, C.; Zhang, J.; Li, B.; Chu, Y.; Su, X.; Huang, Q. Population Structure Analysis and Core Collection Construction of Populus deltoides. Sci. Silvae Sin. 2020, 56, 67–76. [Google Scholar]
- Stanton, B.J.B.; Neale, D.; Li, S. Populus Breeding: From the Classical to the Genomic Approach. In Genetics and Genomics of Populus; Jansson, S., Bhalerao, R., Groover, A., Eds.; Springer: New York, NY, USA, 2010; pp. 309–348. [Google Scholar]
- Li, Z.; Lathe, R.S.; Li, J.; He, H.; Bhalerao, R.P. Towards understanding the biological foundations of perenniality. Trends Plant Sci. 2022, 27, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Archontoulis, S.V.; Miguez, F.E. Nonlinear Regression Models and Applications in Agricultural Research. Agron. J. 2015, 107, 786–798. [Google Scholar] [CrossRef]
- Zhang, Q.; Su, Z.; Guo, Y.; Zhang, S.; Jiang, L.; Wu, R. Genome-wide association studies of callus differentiation for the desert tree, Populus euphratica. Tree Physiol. 2020, 40, 1762–1777. [Google Scholar] [CrossRef]
- Hao, S.; Zhao, T.; Xia, X.; Yin, W. Genome-wide comparison of two poplar genotypes with different growth rates. Plant Mol. Biol. 2011, 76, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yan, J.; Ma, X.; Teng, W.; Wu, W.; Dai, J.; Dhillon, B.S.; Melchinger, A.E.; Li, J. Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theor. Appl. Genet. 2009, 120, 333–340. [Google Scholar] [CrossRef]
- Wei, D.; Cui, K.; Ye, G.; Pan, J.; Xiang, J.; Huang, J.; Nie, L. QTL mapping for nitrogen-use efficiency and nitrogen-deficiency tolerance traits in rice. Plant Soil 2012, 359, 281–295. [Google Scholar] [CrossRef]
- Xia, W.; Xiao, Z.; Cao, P.; Zhang, Y.; Du, K.; Wang, N. Construction of a high-density genetic map and its application for leaf shape QTL mapping in poplar. Planta 2018, 248, 1173–1185. [Google Scholar] [CrossRef]
- Chen, X.; Xiong, C.; Lou, Y.; Xu, H.; Cheng, Q.; Sun, S.; Xiao, F. High-Density Genetic Map and QTL Analysis in Cunninghamia lanceolate: Insights into Growth and Wood-Color Traits. Forests 2023, 14, 1591. [Google Scholar] [CrossRef]
- Li, Y.; Wang, D.; Li, Z.; Wei, J.; Jin, C.; Liu, M. A Molecular Genetic Linkage Map of Eucommia ulmoides and Quantitative Trait Loci (QTL) Analysis for Growth Traits. Int. J. Mol. Sci. 2014, 15, 2053–2074. [Google Scholar] [CrossRef]
- Du, Q.; Yang, X.; Xie, J.; Quan, M.; Xiao, L.; Lu, W.; Tian, J.; Gong, C.; Chen, J.; Li, B.; et al. Time-specific and pleiotropic quantitative trait loci coordinately modulate stem growth in Populus. Plant Biotechnol. J. 2019, 17, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Yang, G.; Yang, Y.; Yin, T. Time-sequential detection of quantitative trait loci and candidate genes underlying the dynamic growth of Salix suchowensis. Tree Physiol. 2022, 42, 877–890. [Google Scholar] [CrossRef] [PubMed]
- Francisco, F.R.; Aono, A.H.; da Silva, C.C.; Gonçalves, P.S.; Junior, E.J.S.; Le Guen, V.; Fritsche-Neto, R.; Souza, L.M.; de Souza, A.P. Unravelling Rubber Tree Growth by Integrating GWAS and Biological Network-Based Approaches. Front. Plant Sci. 2021, 12, 768589. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Dong, X.; Liu, Q.; Wang, R.; Li, Y.; Huang, X.; Zhao, Y. SLAF-Seq Technology-Based Genome-Wide Association and Population Structure Analyses of Ancient Camellia sinensis (L.) Kuntze in Sandu County, China. Forests 2022, 13, 1885. [Google Scholar] [CrossRef]
- Bond, B.J. Age-related changes in photosynthesis of woody plants. Trends Plant Sci. 2000, 5, 349–353. [Google Scholar] [CrossRef]
- Tang, M.; Bai, X.; Niu, L.-J.; Chai, X.; Chen, M.-S.; Xu, Z.-F. miR172 Regulates both Vegetative and Reproductive Development in the Perennial Woody Plant Jatropha curcas. Plant Cell Physiol. 2018, 59, 2549–2563. [Google Scholar] [CrossRef]
- Ahsan, M.U.; Hayward, A.; Irihimovitch, V.; Fletcher, S.; Tanurdzic, M.; Pocock, A.; Beveridge, C.A.; Mitter, N. Juvenility and Vegetative Phase Transition in Tropical/Subtropical Tree Crops. Front. Plant Sci. 2019, 10, 729. [Google Scholar] [CrossRef]
- Xu, M.; Jiang, L.; Zhu, S.; Zhou, C.; Ye, M.; Mao, K.; Sun, L.; Su, X.; Pan, H.; Zhang, S.; et al. A computational framework for mapping the timing of vegetative phase change. New Phytol. 2016, 211, 750–760. [Google Scholar] [CrossRef]
- Wu, R.; Lin, M. Functional mapping—How to map and study the genetic architecture of dynamic complex traits. Nat. Rev. Genet. 2006, 7, 229–237. [Google Scholar] [CrossRef]
- Xiao, L.; Fang, Y.; Zhang, H.; Quan, M.; Zhou, J.; Li, P.; Wang, D.; Ji, L.; Ingvarsson, P.K.; Wu, H.X.; et al. Natural variation in the prolyl 4-hydroxylase gene PtoP4H9 contributes to perennial stem growth in Populus. Plant Cell 2023, 35, 4046–4065. [Google Scholar] [CrossRef]
- Xia, H.; Hao, Z.; Shen, Y.; Tu, Z.; Yang, L.; Zong, Y.; Li, H. Genome-wide association study of multiyear dynamic growth traits in hybrid Liriodendron identifies robust genetic loci associated with growth trajectories. Plant J. 2023, 115, 1544–1563. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wei, X.; Cong, J.; Zou, J.; Wan, L.; Xu, S. Structural insights into mechanism and specificity of the plant protein O-fucosyltransferase SPINDLY. Nat. Commun. 2022, 13, 7424. [Google Scholar] [CrossRef] [PubMed]
- Xing, D.; Wang, Y.; Xu, R.; Ye, X.; Yang, D.; Li, Q.Q. The regulatory role of Pcf11-similar-4 (PCFS4) in Arabidopsis development by genome-wide physical interactions with target loci. BMC Genom. 2013, 14, 598. [Google Scholar] [CrossRef]
- Plomion, C.; Durel, C.E.; O’Malley, D.M. Genetic dissection of height in maritime pine seedlings raised under accelerated growth conditions. Theor. Appl. Genet. 1996, 93, 849–858. [Google Scholar] [CrossRef]
- Sperandio, G.; Acampora, A.; Del Giudice, A.; Civitarese, V. Models for the Evaluation of Productivity and Costs of Mechanized Felling on Poplar Short Rotation Coppice in Italy. Forests 2021, 12, 954. [Google Scholar] [CrossRef]
- Niemczyk, M. The effects of cultivar and rotation length (5 vs. 10 years) on biomass production and sustainability of poplar (Populus spp.) bioenergy plantation. GCB Bioenergy 2021, 13, 999–1014. [Google Scholar] [CrossRef]
- Emebiri, L.C.; Devey, M.E.; Matheson, A.C.; Slee, M.U. Age-related changes in the expression of QTLs for growth in radiata pine seedlings. Theor. Appl. Genet. 1998, 97, 1053–1061. [Google Scholar] [CrossRef]
- Bac-Molenaar, J.A.; Granier, C.; Keurentjes, J.J.B.; Vreugdenhil, D. Genome-wide association mapping of time-dependent growth responses to moderate drought stress in Arabidopsis. Plant Cell Environ. 2015, 39, 88–102. [Google Scholar] [CrossRef]
- Knoch, D.; Abbadi, A.; Grandke, F.; Meyer, R.C.; Samans, B.; Werner, C.R.; Snowdon, R.J.; Altmann, T. Strong temporal dynamics of QTL action on plant growth progression revealed through high-throughput phenotyping in canola. Plant Biotechnol. J. 2019, 18, 68–82. [Google Scholar] [CrossRef]
- Li, K.; Wang, J.; Kuang, L.; Tian, Z.; Wang, X.; Dun, X.; Tu, J.; Wang, H. Genome-wide association study and transcriptome analysis reveal key genes affecting root growth dynamics in rapeseed. Biotechnol. Biofuels 2021, 14, 178. [Google Scholar] [CrossRef]
- Kushwaha, H.R.; Singh, A.K.; Sopory, S.K.; Singla-Pareek, S.L.; Pareek, A. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genom. 2009, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Pribat, A.; Sormani, R.; Rousseau-Gueutin, M.; Julkowska, M.M.; Testerink, C.; Joubès, J.; Castroviejo, M.; Laguerre, M.; Meyer, C.; Germain, V.; et al. A novel class of PTEN protein in Arabidopsis displays unusual phosphoinositide phosphatase activity and efficiently binds phosphatidic acid. Biochem. J. 2011, 441, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Zhang, X.; Zhang, L.; Ye, S.; Zeng, L.; Liu, J.; Li, Q.; He, Z. CURVED CHIMERIC PALEA 1 encoding an EMF1-like protein maintains epigenetic repression of OsMADS58 in rice palea development. Plant J. 2015, 82, 12–24. [Google Scholar] [CrossRef]
- Xing, D.; Zhao, H.; Xu, R.; Li, Q.Q. Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J. Cell Mol. Biol. 2008, 54, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Ma, N.; Sun, F.; Ye, M.; Wu, R. Functional mapping of gravitropism and phototropism for a desert tree, Populus euphratica. Front. Biosci. Landmark 2021, 26, 988–1000. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Align-ment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Bates, D.M.; Watts, D.G. Nonlinear Regression Analysis and Its Applications; Wiley: New York, NY, USA, 1988. [Google Scholar]
- Sakamoto, Y.; Ishiguro, M.; Kitagawa, G. Akaike Information Criterion Statistics. D. Reidel 1986, 81, 26853. [Google Scholar]
- Crainiceanu, C.M.; Ruppert, D. Likelihood ratio tests for goodness-of-fit of a nonlinear regression model. J. Multivar. Anal. 2004, 91, 35–52. [Google Scholar] [CrossRef]
- Dean, D.C., 3rd; O’Muircheartaigh, J.; Dirks, H.; Waskiewicz, N.; Lehman, K.; Walker, L.; Han, M.; Deoni, S.C. Modeling healthy male white matter and myelin development: 3 through 60 months of age. Neuroimage 2014, 84, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.; Vilhjálmsson, B.J.; Segura, V.; Platt, A.; Long, Q.; Nordborg, M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat. Genet. 2012, 44, 1066–1071. [Google Scholar] [CrossRef] [PubMed]

| SNP | Alleles | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Product Size (bp) |
|---|---|---|---|---|
| SNP1 | A/G | TGGTGAACAGTCAAAGTAACCT | GCATTAAAACTGAGAATGGACA | 364 |
| SNP2 | C/T | GGCCACAAAATGAATTTCAGAAG | GCTATCACAGTTGCAAATTGTCA | 384 |
| SNP3 | A/G | GTTTATTGATTGGGTGTTGTTTG | GATGAAATGCGCTAAAAATCC | 376 |
| SNP4 | C/T | CAACCAAAACTGCTCCACATA | GCAATCAAACTCATCCAATACA | 362 |
| SNP5 | A/G | AGTCTACACGAGCCCTTCATTT | ATGGAAGCAAGATGGCTAGACT | 371 |
| SNP6 | A/G | GAAGCACTTGAGGAGCCTTGA | CACCGTGCTTGTCCTGTTTCT | 359 |
| SNP7 | A/G | GTCCCTGGATTTCAATCTCA | TACGAATCCCATATGTACCTCC | 354 |
| SNP8 | A/G | TGATATTATCAGCTGTGTTGGGG | TCCTGGTTCTGAAGTTAGCTTCT | 364 |
| SNP9 | A/G | ATCAACAGGGTGCATAAGAGAA | TTTTATGCTTTTGCTGCCTTC | 341 |
| SNP10 | G/T | ACTGCTTTAACCAGCTTGTATCCT | CTCAACCCAAATCCACACAATA | 380 |
| SNP11 | A/T | TTAGGCTGAGAGAACCTCGTG | TTCCTCGTATACCGCTCTCTA | 370 |
| SNP12 | C/T | TTGTGCACTGTCTTGGAAGGT | CCTTTAGCGGTCAGCAATGTT | 388 |
| SNP13 | A/G | TACAATGTTCACAAACCTTCC | GAAAAGTTTTCTTTAACATGCTG | 346 |
| SNP14 | C/T | GGAATTCAAGAACCTGCAATCA | TTCAATTGCTAGAACTCTGGGTC | 353 |
| SNP15 | G/T | CCAAGTATATCTGACCTAGTTTGC | TAAGAAGCTATGCTGCTCAGTT | 348 |
| SNP16 | A/G | GAAAGAAGCGTAAGGGCACTG | ACACGATTGGATGAGGCAAG | 358 |
| SNP17 | C/T | TATGCGGTGTCTCAGACTCTCA | CCGGTATTCTGCTTTGTTTATCC | 366 |
| SNP18 | A/T | CGCGAGTTGAAATTGAAGAGTC | CATTTTCCCAACAAGCATCGT | 362 |
| SNP19 | C/T | ATGCATGGTGGCTAGCTAAC | GCCTAGGAAGCTGAACTTGTG | 358 |
| SNP20 | C/T | TTTGGACTTGATGACTGCAAAC | AAGCTGAATGCATAGCCTGAA | 364 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wang, Z.; Zhu, S.; Pan, H.; Ding, C.; Xu, M. Analysis of Growth Trajectories and Verification of Related SNPs in Populus deltoides. Int. J. Mol. Sci. 2023, 24, 16192. https://doi.org/10.3390/ijms242216192
Wang Y, Wang Z, Zhu S, Pan H, Ding C, Xu M. Analysis of Growth Trajectories and Verification of Related SNPs in Populus deltoides. International Journal of Molecular Sciences. 2023; 24(22):16192. https://doi.org/10.3390/ijms242216192
Chicago/Turabian StyleWang, Yaolin, Zesen Wang, Sheng Zhu, Huixin Pan, Changjun Ding, and Meng Xu. 2023. "Analysis of Growth Trajectories and Verification of Related SNPs in Populus deltoides" International Journal of Molecular Sciences 24, no. 22: 16192. https://doi.org/10.3390/ijms242216192
APA StyleWang, Y., Wang, Z., Zhu, S., Pan, H., Ding, C., & Xu, M. (2023). Analysis of Growth Trajectories and Verification of Related SNPs in Populus deltoides. International Journal of Molecular Sciences, 24(22), 16192. https://doi.org/10.3390/ijms242216192







