Comparative Genomic Analysis of Asian Cultivated Rice and Its Wild Progenitor (Oryza rufipogon) Has Revealed Evolutionary Innovation of the Pentatricopeptide Repeat Gene Family through Gene Duplication
Abstract
:1. Introduction
2. Results
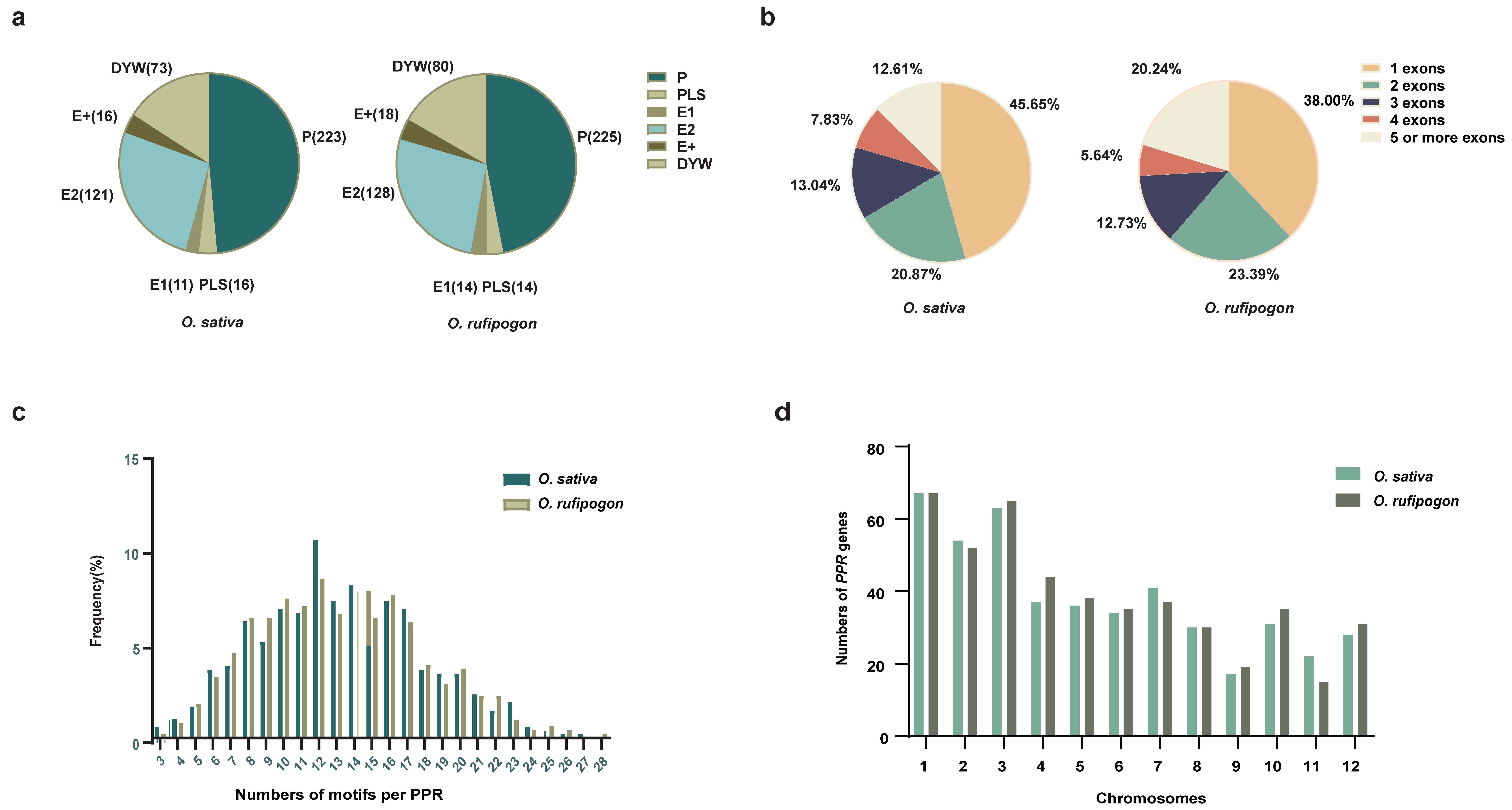
2.1. Identification and Classification of PPR Genes in the O. sativa and O. rufipogon Genomes
2.2. Phylogenetic Analysis of PPR Genes in the O. sativa and O. rufipogon Genomes
2.3. Comparative Genomic Analysis of PPR Gene Families between O. sativa and O. rufipogon
2.4. Expression Patterns of PPR Genes in O. sativa and O. rufipogon
2.5. Segmental Duplication Promotes the Structural Diversity of PPR Genes through Incomplete Gene Duplications
3. Discussion
4. Materials and Methods
4.1. Identification of PPR Genes
4.2. Chromosomal Location and Gene Structure and Cis-Acting Element Analysis
4.3. Phylogenetic Analysis and Gene Ontology (GO) Enrichment Analysis
4.4. Analysis of PPR Gene Expression and Classification of Expression Patterns of Sister Duplicates
4.5. Identification of Shared and Species-Specific PPR Genes in O. sativa and O. rufipogon and Ks Analysis
4.6. Identification of Gene Duplications and Segmental Duplications
4.7. Identification of Domain and Visualization of Segmental Duplication
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Small, I.D.; Peeters, N. The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000, 25, 45–47. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Small, I. Pentatricopeptide Repeat Proteins in Plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Manna, S. An overview of pentatricopeptide repeat proteins and their applications. Biochimie 2015, 113, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, B.; Royan, S.; Schallenberg-Rüdinger, M.; Lenz, H.; Castleden, I.R.; Mcdowell, R.; Vacher, M.A.; Tonti-Filippini, J.; Bond, C.S.; Knoop, V.; et al. The Expansion and Diversification of Pentatricopeptide Repeat RNA-Editing Factors in Plants. Mol. Plant. 2020, 13, 215–230. [Google Scholar] [CrossRef]
- Xing, H.; Fu, X.; Yang, C.; Tang, X.; Guo, L.; Li, C.; Xu, C.; Luo, K. Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci. Rep. 2018, 8, 2817. [Google Scholar] [CrossRef]
- Ma, X.; Wang, D.; Xue, G.; Zheng, X.; Lu, Y.; Shi, J.; Hao, Z.; Chen, J. Characterization of the Liriodendron chinense Pentatricopeptide Repeat (PPR) Gene Family and Its Role in Osmotic Stress Response. Genes 2023, 14, 1125. [Google Scholar] [CrossRef]
- Lurin, C.; Andreés, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyeère, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-Wide Analysis of Arabidopsis Pentatricopeptide Repeat Proteins Reveals Their Essential Role in Organelle Biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef]
- Cheng, S.; Gutmann, B.; Zhong, X.; Ye, Y.; Fisher, M.F.; Bai, F.; Castleden, I.; Song, Y.; Song, B.; Huang, J.; et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016, 85, 532–547. [Google Scholar] [CrossRef]
- Coquille, S.; Filipovska, A.; Chia, T.; Rajappa, L.; Lingford, J.P.; Razif, M.F.M.; Thore, S.; Rackham, O. An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 2014, 5, 5729. [Google Scholar] [CrossRef]
- Barkan, A.; Rojas, M.; Fujii, S.; Yap, A.; Chong, Y.S.; Bond, C.S.; Small, I. A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins. PLoS Genet. 2012, 8, e1002910. [Google Scholar] [CrossRef]
- Yagi, Y.; Hayashi, S.; Kobayashi, K.; Hirayama, T.; Nakamura, T. Elucidation of the RNA Recognition Code for Pentatricopeptide Repeat Proteins Involved in Organelle RNA Editing in Plants. PLoS ONE 2013, 8, e57286. [Google Scholar] [CrossRef]
- Takenaka, M.; Zehrmann, A.; Brennicke, A.; Graichen, K. Improved Computational Target Site Prediction for Pentatricopeptide Repeat RNA Editing Factors. PLoS ONE 2013, 8, e65343. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, M.; Tasaki, E.; Sugita, C.; Sugita, M. A PPR-DYW protein is required for splicing of a group II intron of cox1 pre-mRNA in Physcomitrella patens. Plant J. 2012, 70, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Dong, J.; Lu, J.; Li, J.; Jiang, D.; Yu, H.; Ye, S.; Bu, W.; Liu, Z.; Zhou, H.; et al. A cytosolic pentatricopeptide repeat protein is essential for tapetal plastid development by regulating OsGLK1 transcript levels in rice. New Phytol. 2022, 234, 1678–1695. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Zhang, Q.; Qin, X.; Xu, Y.; Ni, C.; Huang, J.; Zhu, L.; Zhong, F.; Liu, W.; Yao, G.; et al. Rice PPS1 encodes a DYW motif-containing pentatricopeptide repeat protein required for five consecutive RNA-editing sites of nad3 in mitochondria. New Phytol. 2018, 220, 878–892. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Huang, Z.; Ma, B.; Yang, Y.; Xiu, Z.; Wang, L.; Tan, B. Maize PPR-E proteins mediate RNA C-to-U editing in mitochondria by recruiting the trans deaminase PCW1. Plant Cell 2023, 35, 529–551. [Google Scholar] [CrossRef]
- An, H.; Ke, X.; Li, L.; Liu, Y.; Yuan, S.; Wang, Q.; Hou, X.; Zhao, J. ALBINO EMBRYO AND SEEDLING is required for RNA splicing and chloroplast homeostasis in Arabidopsis. Plant Physiol. 2023, 193, 483–501. [Google Scholar] [CrossRef]
- Meierhoff, K.; Felder, S.; Nakamura, T.; Bechtold, N.; Schuster, G. HCF152, an Arabidopsis RNA Binding Pentatricopeptide Repeat Protein Involved in the Processing of Chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 2003, 15, 1480–1495. [Google Scholar] [CrossRef]
- Nishimura, Y.; Shikanai, T. Maturation and longevity of mRNA in chloroplasts: The functions of PPR proteins in RNA editing and the molecular mechanisms of RNA stability control in chloroplasts. Tanpakushitsu Kakusan Koso 2009, 54, 2098–2101. [Google Scholar]
- Macedo-Osorio, K.S.; Martínez-Antonio, A.; Badillo-Corona, J.A. Pas de Trois: An Overview of Penta-, Tetra-, and Octo-Tricopeptide Repeat Proteins From Chlamydomonas reinhardtii and Their Role in Chloroplast Gene Expression. Front. Plant Sci. 2021, 12, 775366. [Google Scholar] [CrossRef]
- Hammani, K.; Colas Des Francs-Small, C.; Takenaka, M.; Tanz, S.K.; Okuda, K.; Shikanai, T.; Brennicke, A.; Small, I. The Pentatricopeptide Repeat Protein OTP87 Is Essential for RNA Editing of nad7 and atp1 Transcripts in Arabidopsis Mitochondria. J. Biol. Chem. 2011, 286, 21361–21371. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Linneweber, C.; Williams-Carrier, R.; Barkan, A. RNA Immunoprecipitation and Microarray Analysis Show a Chloroplast Pentatricopeptide Repeat Protein to Be Associated with the 5′ Region of mRNAs Whose Translation It Activates. Plant Cell 2005, 17, 2791–2804. [Google Scholar] [CrossRef] [PubMed]
- Pfalz, J.; Bayraktar, O.A.; Prikryl, J.; Barkan, A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009, 28, 2042–2052. [Google Scholar] [CrossRef] [PubMed]
- Prikryl, J.; Rojas, M.; Schuster, G.; Barkan, A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. USA 2011, 108, 415–420. [Google Scholar] [CrossRef]
- Barkan, A. Expression of Plastid Genes: Organelle-Specific Elaborations on a Prokaryotic Scaffold. Plant Physiol. 2011, 155, 1520–1532. [Google Scholar] [CrossRef]
- Melonek, J.; Zhou, R.; Bayer, P.E.; Edwards, D.; Stein, N.; Small, I. High intraspecific diversity of Restorer-of-fertility-like genes in barley. Plant J. 2019, 97, 281–295. [Google Scholar] [CrossRef]
- Ma, Q.; Hedden, P.; Zhang, Q. Heterosis in rice seedlings: Its relationship to gibberellin content and expression of gibberellin metabolism and signaling genes. Plant Physiol. 2011, 156, 1905–1920. [Google Scholar] [CrossRef]
- Labroo, M.R.; Studer, A.J.; Rutkoski, J.E. Heterosis and Hybrid Crop Breeding: A Multidisciplinary Review. Front. Genet. 2021, 12, 643761. [Google Scholar] [CrossRef]
- Takatsuka, A.; Kazama, T.; Toriyama, K. Cytoplasmic Male Sterility-Associated Mitochondrial Gene orf312 Derived from Rice (Oryza sativa L.) Cultivar Tadukan. Rice 2021, 14, 46. [Google Scholar] [CrossRef]
- Chase, C.D. Cytoplasmic male sterility: A window to the world of plant mitochondria nuclear interactions. Trends Genet. 2007, 23, 81–90. [Google Scholar] [CrossRef]
- Brown, G.G.; Formanová, N.; Jin, H.; Wargachuk, R.; Dendy, C.; Patil, P.; Laforest, M.; Zhang, J.; Cheung, W.Y.; Landry, B.S. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 2003, 35, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y. Male Sterility and Fertility Restoration in Crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, K.; Huang, W.; Liu, G.; Gao, Y.; Wang, J.; Huang, Q.; Ji, Y.; Qin, X.; Wan, L.; et al. The Rice Pentatricopeptide Repeat Protein RF5 Restores Fertility in Hong-Lian Cytoplasmic Male-Sterile Lines via a Complex with the Glycine-Rich Protein GRP162. Plant Cell 2012, 24, 109–122. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, N.; Hattori, M.; Andres, C.; Iida, K.; Lurin, C.; Schmitz-Linneweber, C.; Sugita, M.; Small, I. On the Expansion of the Pentatricopeptide Repeat Gene Family in Plants. Mol. Biol. Evol. 2008, 25, 1120–1128. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Small, I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 2008, 13, 663–670. [Google Scholar] [CrossRef]
- Fujii, S.; Small, I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011, 191, 37–47. [Google Scholar] [CrossRef]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella Genome Reveals Evolutionary Insights into the Conquest of Land by Plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef]
- Rivals, E.; Bruyère, C.; Toffano-Nioche, C.; Lecharny, A. Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol. 2006, 141, 825–839. [Google Scholar] [CrossRef]
- Taylor, J.S.; Raes, J. Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 2004, 38, 615–643. [Google Scholar] [CrossRef]
- Muller, H.J. Bar duplication. Science 1936, 83, 528–530. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Wolf, U.; Atkin, N.B. Evolution from fish to mammals by gene duplication. Hereditas 1968, 59, 169–187. [Google Scholar] [CrossRef]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 2004, 20, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- De Bodt, S.; Maere, S.; Van de Peer, Y. Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 2005, 20, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Vollger, M.R.; Guitart, X.; Dishuck, P.C.; Mercuri, L.; Harvey, W.T.; Gershman, A.; Diekhans, M.; Sulovari, A.; Munson, K.M.; Lewis, A.P.; et al. Segmental duplications and their variation in a complete human genome. Science 2022, 376, eabj6965. [Google Scholar] [CrossRef]
- Bailey, J.A.; Gu, Z.; Clark, R.A.; Reinert, K.; Samonte, R.V.; Schwartz, S.; Adams, M.D.; Myers, E.W.; Li, P.W.; Eichler, E.E. Recent segmental duplications in the human genome. Science 2002, 297, 1003–1007. [Google Scholar] [CrossRef]
- Bailey, J.A.; Yavor, A.M.; Massa, H.F.; Trask, B.J.; Eichler, E.E. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001, 11, 1005–1017. [Google Scholar] [CrossRef]
- Bailey, J.A.; Eichler, E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006, 7, 552–564. [Google Scholar] [CrossRef]
- Li, K.; Jiang, W.; Hui, Y.; Kong, M.; Feng, L.; Gao, L.; Li, P.; Lu, S. Gapless indica rice genome reveals synergistic contributions of active transposable elements and segmental duplications to rice genome evolution. Mol. Plant. 2021, 14, 1745–1756. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, G.; Zhang, S.; Chen, S.; Wang, Y.; Wen, P.; Ma, X.; Shi, Y.; Qi, R.; Yang, Y.; et al. Genomes of the Banyan Tree and Pollinator Wasp Provide Insights into Fig-Wasp Coevolution. Cell 2020, 183, 875–889. [Google Scholar] [CrossRef] [PubMed]
- Giannuzzi, G.; D’Addabbo, P.; Gasparro, M.; Martinelli, M.; Carelli, F.N.; Antonacci, D.; Ventura, M. Analysis of high-identity segmental duplications in the grapevine genome. BMC Genom. 2011, 12, 436. [Google Scholar] [CrossRef] [PubMed]
- Antonacci, F.; Dennis, M.Y.; Huddleston, J.; Sudmant, P.H.; Steinberg, K.M.; Rosenfeld, J.A.; Miroballo, M.; Graves, T.A.; Vives, L.; Malig, M.; et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet. 2014, 46, 1293–1302. [Google Scholar] [CrossRef]
- Dennis, M.Y.; Eichler, E.E. Human adaptation and evolution by segmental duplication. Curr. Opin. Genet. Dev. 2016, 41, 44–52. [Google Scholar] [CrossRef]
- Dennis, M.Y.; Nuttle, X.; Sudmant, P.H.; Antonacci, F.; Graves, T.A.; Nefedov, M.; Rosenfeld, J.A.; Sajjadian, S.; Malig, M.; Kotkiewicz, H.; et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 2012, 149, 912–922. [Google Scholar] [CrossRef]
- Ciccarelli, F.D.; von Mering, C.; Suyama, M.; Harrington, E.D.; Izaurralde, E.; Bork, P. Complex genomic rearrangements lead to novel primate gene function. Genome Res. 2005, 15, 343–351. [Google Scholar] [CrossRef]
- Liu, S.; Melonek, J.; Boykin, L.M.; Small, I.; Howell, K.A. PPR-SMRs: Ancient proteins with enigmatic functions. RNA Biol. 2013, 10, 1501–1510. [Google Scholar] [CrossRef]
- Chateigner-Boutin, A.; Small, I. Plant RNA editing. RNA Biol. 2010, 7, 213–219. [Google Scholar] [CrossRef]
- Chen, G.; Zou, Y.; Hu, J.; Ding, Y. Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genom. 2018, 19, 720. [Google Scholar] [CrossRef]
- Subburaj, S.; Tu, L.; Lee, K.; Park, G.; Lee, H.; Chun, J.; Lim, Y.; Park, M.; Mcgregor, C.; Lee, G. A Genome-Wide Analysis of the Pentatricopeptide Repeat (PPR) Gene Family and PPR-Derived Markers for Flesh Color in Watermelon (Citrullus lanatus). Genes 2020, 11, 1125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, Y.; Meng, Y.; Xiao, Y.; Zhao, J.; Xiao, B.; An, C.; Gao, Y. PPR proteins in the tea plant (Camellia sinensis) and their potential roles in the leaf color changes. Sci. Hortic. 2022, 293, 110745. [Google Scholar] [CrossRef]
- Wei, K.; Han, P. Pentatricopeptide repeat proteins in maize. Mol. Breed. 2016, 36, 170. [Google Scholar] [CrossRef]
- Zhang, J.; Guo, Y.; Fang, Q.; Zhu, Y.; Zhang, Y.; Liu, X.; Lin, Y.; Barkan, A.; Zhou, F. The PPR-SMR Protein ATP4 Is Required for Editing the Chloroplast rps8 mRNA in Rice and Maize. Plant Physiol. 2020, 184, 2011–2021. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, H.; Shen, J.; Sun, F.; Wang, M.; Xu, C.; Tan, B. PPR-SMR1 is required for the splicing of multiple mitochondrial introns, interacts with Zm-mCSF1, and is essential for seed development in maize. J. Exp. Bot. 2019, 70, 5245–5258. [Google Scholar] [CrossRef]
- Lan, X.; Pritchard, J.K. Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. Science 2016, 352, 1009–1013. [Google Scholar] [CrossRef]
- Li, W.; Li, K.; Huang, Y.; Shi, C.; Hu, W.; Zhang, Y.; Zhang, Q.; Xia, E.; Hutang, G.; Zhu, X.; et al. SMRT sequencing of the Oryza rufipogon genome reveals the genomic basis of rice adaptation. Commun. Biol. 2020, 3, 167. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic. Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Gu, Z.; Hübschmann, D. Make Interactive Complex Heatmaps in R. Bioinformatics 2022, 38, 1460–1462. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.; Fraser, M.; Li, W.; Mcanulla, C.; Mcwilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and Collinearity in Plant Genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Gel, B.; Serra, E. karyoploteR: An R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 2017, 33, 3088–3090. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, L.-Y.; Lin, P.-F.; Xu, R.-J.; Kang, H.-Q.; Gao, L.-Z. Comparative Genomic Analysis of Asian Cultivated Rice and Its Wild Progenitor (Oryza rufipogon) Has Revealed Evolutionary Innovation of the Pentatricopeptide Repeat Gene Family through Gene Duplication. Int. J. Mol. Sci. 2023, 24, 16313. https://doi.org/10.3390/ijms242216313
Feng L-Y, Lin P-F, Xu R-J, Kang H-Q, Gao L-Z. Comparative Genomic Analysis of Asian Cultivated Rice and Its Wild Progenitor (Oryza rufipogon) Has Revealed Evolutionary Innovation of the Pentatricopeptide Repeat Gene Family through Gene Duplication. International Journal of Molecular Sciences. 2023; 24(22):16313. https://doi.org/10.3390/ijms242216313
Chicago/Turabian StyleFeng, Li-Ying, Pei-Fan Lin, Rong-Jing Xu, Hai-Qi Kang, and Li-Zhi Gao. 2023. "Comparative Genomic Analysis of Asian Cultivated Rice and Its Wild Progenitor (Oryza rufipogon) Has Revealed Evolutionary Innovation of the Pentatricopeptide Repeat Gene Family through Gene Duplication" International Journal of Molecular Sciences 24, no. 22: 16313. https://doi.org/10.3390/ijms242216313
APA StyleFeng, L.-Y., Lin, P.-F., Xu, R.-J., Kang, H.-Q., & Gao, L.-Z. (2023). Comparative Genomic Analysis of Asian Cultivated Rice and Its Wild Progenitor (Oryza rufipogon) Has Revealed Evolutionary Innovation of the Pentatricopeptide Repeat Gene Family through Gene Duplication. International Journal of Molecular Sciences, 24(22), 16313. https://doi.org/10.3390/ijms242216313





