MiR-138-5p Upregulation during Neuronal Maturation Parallels with an Increase in Neuronal Survival
Abstract
:1. Introduction
2. Results
2.1. Opposite Expression of miR-138-5p and Caspase 3 during Hippocampal Neuronal Maturation
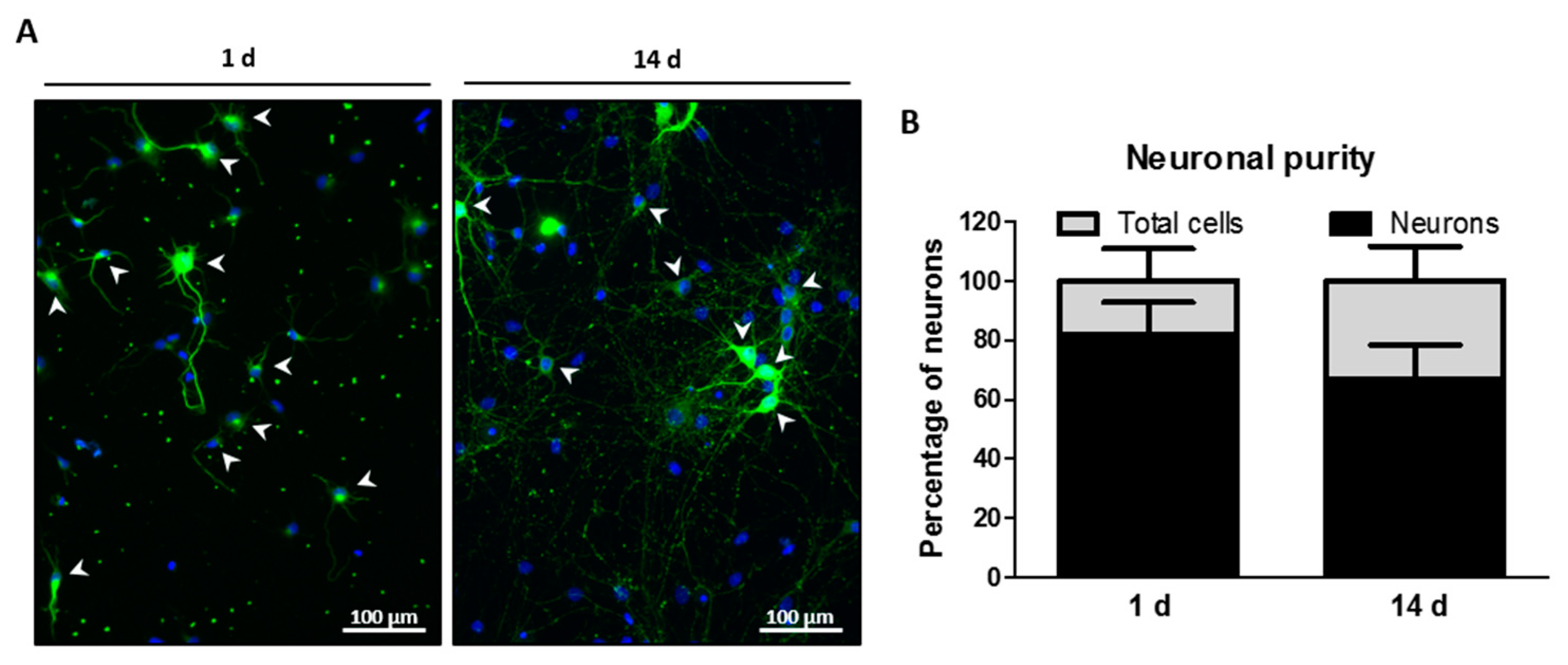
2.2. Neuronal Purity of Hippocampal Cell Cultures
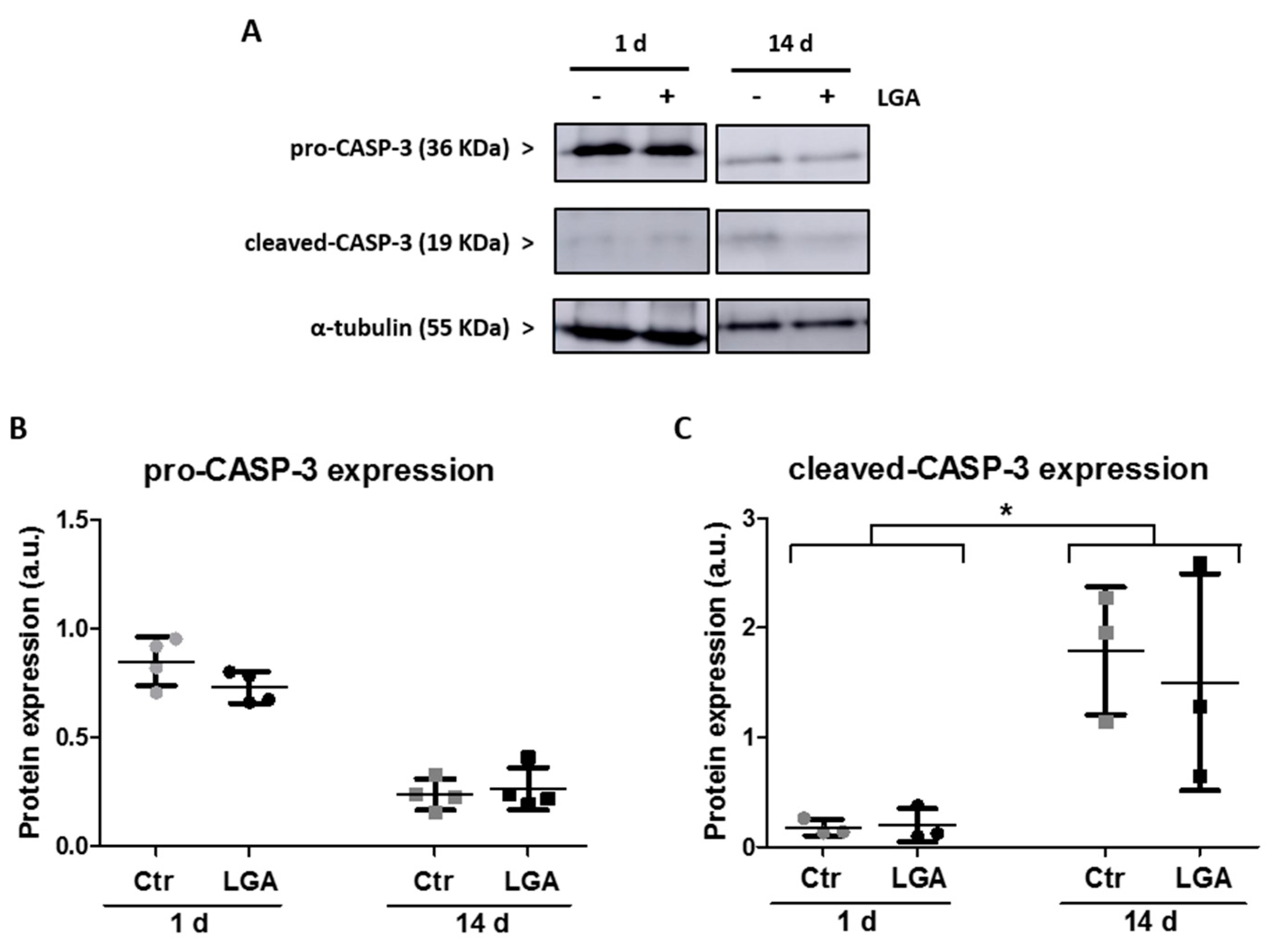
2.3. Neuronal Maturation Attenuates Caspase-Dependent Apoptosis
2.4. Neuronal Maturation Reduces Caspase Activity
2.5. Cell Maturation Promotes Neuronal Survival
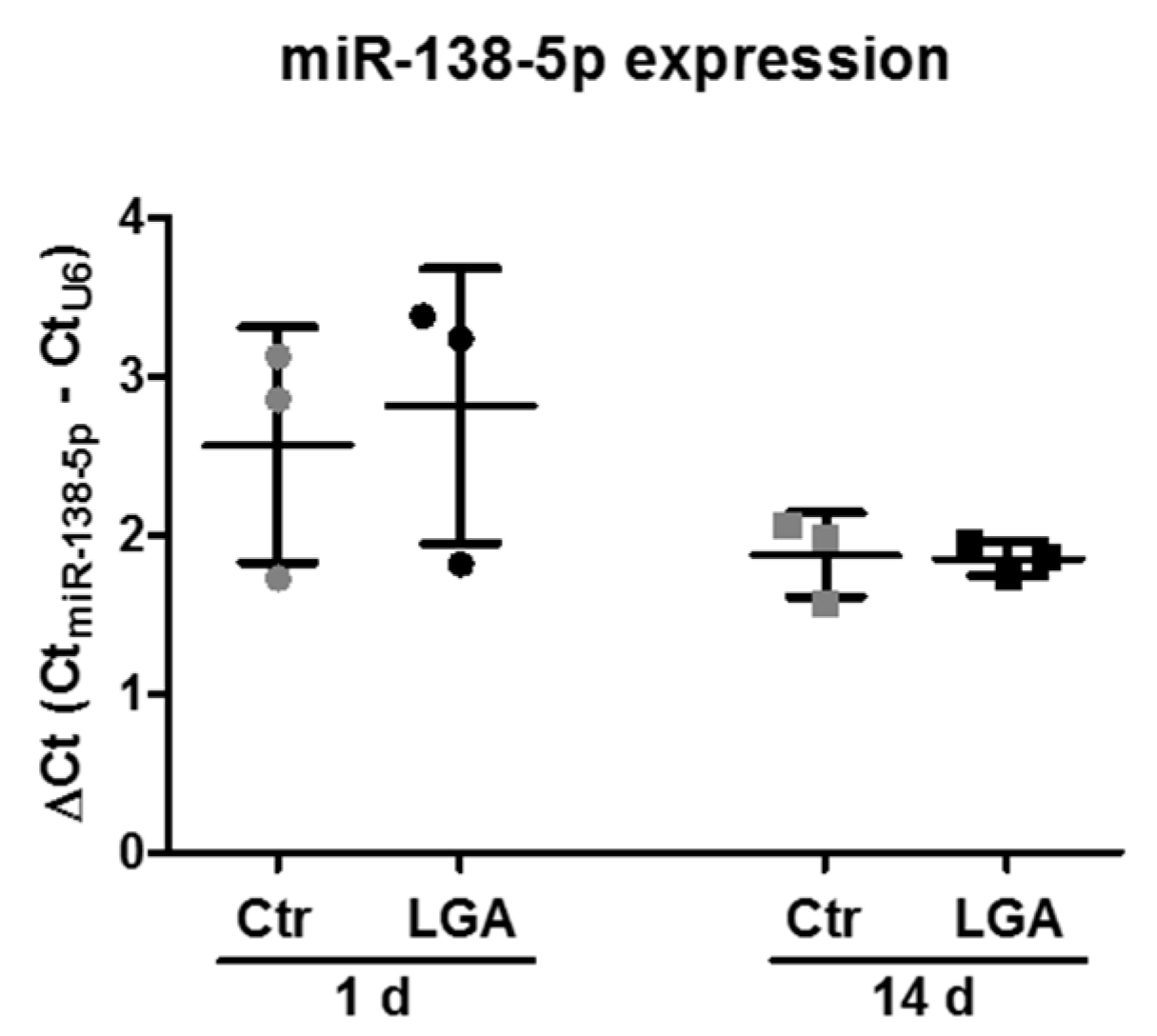
2.6. Neuronal Maturation Does Not Change the Increase in miR-138-5p Expression in Apoptotic-Stimulated Hippocampal Neurons
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Immunofluorescence Assay
4.3. RT-qPCR Analysis
4.4. Immunoblot Assay
4.5. Measurement of Caspase-3/7 Activity
4.6. Calcein/Propidium Iodide Assay
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aimone, J.B.; Wiles, J.; Gage, F.H. Potential role for adult neurogenesis in the encoding of time in new memories. Nat. Neurosci. 2006, 9, 723–727. [Google Scholar] [CrossRef] [PubMed]
- Ming, G.L.; Song, H. Adult neurogenesis in the mammalian brain: Significant answers and significant questions. Neuron 2011, 70, 687–702. [Google Scholar] [CrossRef] [PubMed]
- Bradke, F.; Di Giovanni, S.; Fawcett, J. Neuronal Maturation: Challenges and Opportunities in a Nascent Field. Trends Neurosci. 2020, 43, 360–362. [Google Scholar] [CrossRef] [PubMed]
- Piatti, V.C.; Espósito, M.S.; Schinder, A.F. The timing of neuronal development in adult hippocampal neurogenesis. Neuroscientist 2006, 12, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Piatti, V.C.; Davies-Sala, M.G.; Espósito, M.S.; Mongiat, L.A.; Trinchero, M.F.; Schinder, A.F. The timing for neuronal maturation in the adult hippocampus is modulated by local network activity. J. Neurosci. 2011, 31, 7715–7728. [Google Scholar] [CrossRef] [PubMed]
- Tooley, U.A.; Bassett, D.S.; Mackey, A.P. Environmental influences on the pace of brain development. Nat. Rev. Neurosci. 2021, 22, 372–384. [Google Scholar] [CrossRef]
- Crutcher, K.A. The role of growth factors in neuronal development and plasticity. CRC Crit. Rev. Clin. Neurobiol. 1986, 2, 297–333. [Google Scholar]
- Landreth, G.E. Classes of Growth Factors Acting in the Nervous System. In Basic Neurochemistry: Molecular, Cellular and Medical Aspects, 6th ed.; Siegel, G.J., Agranoff, B.W., Albers, R.W., Fisher, S.K., Uhler, M.D., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 1999. Available online: https://www.ncbi.nlm.nih.gov/books/NBK28185/ (accessed on 2 May 2023).
- Aloe, L.; Rocco, M.L.; Balzamino, B.O.; Micera, A. Nerve Growth Factor: A Focus on Neuroscience and Therapy. Curr. Neuropharmacol. 2015, 13, 294–303. [Google Scholar] [CrossRef]
- He, Z.; Yu, Q. Identification and characterization of functional modules reflecting transcriptome transition during human neuron maturation. BMC Genom. 2018, 19, 262. [Google Scholar] [CrossRef]
- Patel, T.; Hammelman, J.; Closser, M.; Gifford, D.K.; Wichterle, H. General and cell-type-specific aspects of the motor neuron maturation transcriptional program. BioRxiv 2021. [Google Scholar] [CrossRef]
- Kosik, K.S. MicroRNAs and cellular phenotypy. Cell 2010, 143, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Fiore, R.; Siegel, G.; Schratt, G. MicroRNA function in neuronal development, plasticity and disease. Biochim. Biophys. Acta 2008, 1779, 471–478. [Google Scholar] [CrossRef]
- Im, H.I.; Kenny, P.J. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012, 35, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Napoli, D.; Pizzorusso, T. Chapter 12—miRNA in Neuronal Networks Maturation and Plasticity. In Essentials of Noncoding RNA in Neuroscience; De Pietri-Tonelli, D., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 211–224. ISBN 9780128044025. [Google Scholar] [CrossRef]
- Rajman, M.; Schratt, G. MicroRNAs in neural development: From master regulators to fine-tuners. Development 2017, 144, 2310–2322. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.H.T.; Xu, B.; Blenkiron, C.; Fraser, M. Emerging Roles of miRNAs in Brain Development and Perinatal Brain Injury. Front. Physiol. 2019, 10, 227. [Google Scholar] [CrossRef]
- Oppenheim, R.W. Cell death during development of the nervous system. Annu. Rev. Neurosci. 1991, 14, 453–501. [Google Scholar] [CrossRef]
- Pfisterer, U.; Khodosevich, K. Neuronal survival in the brain: Neuron type-specific mechanisms. Cell Death Dis. 2017, 8, e2643. [Google Scholar] [CrossRef] [PubMed]
- Fricker, M.; Tolkovsky, A.M.; Borutaite, V.; Coleman, M.; Brown, G.C. Neuronal Cell Death. Physiol. Rev. 2018, 98, 813–880. [Google Scholar] [CrossRef]
- Kuan, C.Y.; Roth, K.A.; Flavell, R.A.; Rakic, P. Mechanisms of programmed cell death in the developing brain. Trends Neurosci. 2000, 23, 291–297. [Google Scholar] [CrossRef]
- Kole, A.J.; Annis, R.P.; Deshmukh, M. Mature neurons: Equipped for survival. Cell Death Dis. 2013, 4, e689. [Google Scholar] [CrossRef]
- Hollville, E.; Romero, S.E.; Deshmukh, M. Apoptotic cell death regulation in neurons. FEBS J. 2019, 286, 3276–3298. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tomooka, Y.; Noda, M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 1992, 185, 1155–1161. [Google Scholar] [CrossRef]
- Park, J.K.; Doseff, A.I.; Schmittgen, T.D. MicroRNAs Targeting Caspase-3 and -7 in PANC-1 Cells. Int. J. Mol. Sci. 2018, 19, 1206. [Google Scholar] [CrossRef] [PubMed]
- Maza, R.M.; Barreda-Manso, M.A.; Reigada, D.; Silván, Á.; Muñoz-Galdeano, T.; Soto, A.; Del Águila, Á.; Nieto-Díaz, M. MicroRNA-138-5p Targets Pro-Apoptotic Factors and Favors Neural Cell Survival: Analysis in the Injured Spinal Cord. Biomedicines 2022, 10, 1559. [Google Scholar] [CrossRef]
- Obernosterer, G.; Leuschner, P.J.; Alenius, M.; Martinez, J. Post-transcriptional regulation of microRNA expression. RNA 2006, 12, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Tatro, E.T.; Risbrough, V.; Soontornniyomkij, B.; Young, J.; Shumaker-Armstrong, S.; Jeste, D.V.; Achim, C.L. Short-term recognition memory correlates with regional CNS expression of microRNA-138 in mice. Am. J. Geriatr. Psychiatry 2013, 21, 461–473. [Google Scholar] [CrossRef]
- Daswani, R.; Gilardi, C.; Soutschek, M.; Nanda, P.M.; Weiss, K.; Bicker, S.; Fiore, R.; Dieterich, C.; Germain, P.L.; Winterer, J.; et al. MicroRNA-138 controls hippocampal interneuron function and short-term memory in mice. eLife 2022, 11, e74056. [Google Scholar] [CrossRef]
- Weiss, K.; Treiber, T.; Meister, G.; Schratt, G. The nuclear matrix protein Matr3 regulates processing of the synaptic microRNA-138-5p. Neurobiol. Learn. Mem. 2019, 159, 36–45. [Google Scholar] [CrossRef]
- Buss, R.R.; Sun, W.; Oppenheim, R.W. Adaptive roles of programmed cell death during nervous system development. Annu. Rev. Neurosci. 2006, 29, 1–35. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Miura, M. Programmed cell death in neurodevelopment. Dev. Cell 2015, 32, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.M.; Gillet, G.; Popgeorgiev, N. Caspases in the Developing Central Nervous System: Apoptosis and Beyond. Front. Cell Dev. Biol. 2021, 9, 702404. [Google Scholar] [CrossRef] [PubMed]
- Moujalled, D.; Strasser, A.; Liddell, J.R. Molecular mechanisms of cell death in neurological diseases. Cell Death Differ. 2021, 28, 2029–2044. [Google Scholar] [CrossRef] [PubMed]
- Belmonte-Mateos, C.; Pujades, C. From Cell States to Cell Fates: How Cell Proliferation and Neuronal Differentiation Are Coordinated During Embryonic Development. Front. Neurosci. 2022, 15, 781160. [Google Scholar] [CrossRef] [PubMed]
- De Bilbao, F.; Guarin, E.; Nef, P.; Vallet, P.; Giannakopoulos, P.; Dubois-Dauphin, M. Postnatal distribution of cpp32/caspase 3 mRNA in the mouse central nervous system: An in situ hybridization study. J. Comp. Neurol. 1999, 409, 339–357. [Google Scholar] [CrossRef]
- Zhang, X.; Szabo, E.; Michalak, M.; Opas, M. Endoplasmic reticulum stress during the embryonic development of the central nervous system in the mouse. Int. J. Dev. Neurosci. 2007, 25, 455–463. [Google Scholar] [CrossRef]
- Sousa, E.; Flames, N. Transcriptional regulation of neuronal identity. Eur. J. Neurosci. 2022, 55, 645–660. [Google Scholar] [CrossRef]
- Yakovlev, A.; Khafizova, M.; Abdullaev, Z.; Loukinov, D.; Kondratyev, A. Epigenetic regulation of caspase-3 gene expression in rat brain development. Gene 2010, 450, 103–108. [Google Scholar] [CrossRef]
- Kole, A.J.; Swahari, V.; Hammond, S.M.; Deshmukh, M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011, 25, 125–130. [Google Scholar] [CrossRef]
- Swahari, V.; Nakamura, A.; Hollville, E.; Stroud, H.; Simon, J.M.; Ptacek, T.S.; Beck, M.V.; Flowers, C.; Guo, J.; Plestant, C.; et al. MicroRNA-29 is an essential regulator of brain maturation through regulation of CH methylation. Cell Rep. 2021, 35, 108946. [Google Scholar] [CrossRef] [PubMed]
- Kisliouk, T.; Meiri, N. MiR-138 promotes the migration of cultured chicken embryonic hypothalamic cells by targeting reelin. Neuroscience 2013, 238, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.M.; Wang, R.Y.; Saijilafu-Jiao, Z.X.; Zhang, B.Y.; Zhou, F.Q. MicroRNA-138 and SIRT1 form a mutual negative feedback loop to regulate mammalian axon regeneration. Genes Dev. 2013, 27, 1473–1483. [Google Scholar] [CrossRef]
- Bicker, S.; Lackinger, M.; Weiß, K.; Schratt, G. MicroRNA-132, -134, and -138: A microRNA troika rules in neuronal dendrites. Cell. Mol. Life Sci. 2014, 71, 3987–4005. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, G.; Li, S.; Shi, Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009, 16, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.P.; Oksuz, I.; Svaren, J.; Awatramani, R. Egr2-dependent microRNA-138 is dispensable for peripheral nerve myelination. Sci. Rep. 2018, 8, 3817. [Google Scholar] [CrossRef]
- Zhang, W.; Thevapriya, S.; Kim, P.J.; Yu, W.P.; Je, H.S.; Tan, E.K.; Zeng, L. Amyloid precursor protein regulates neurogenesis by antagonizing miR-574-5p in the developing cerebral cortex. Nat. Commun. 2014, 5, 3330. [Google Scholar] [CrossRef]
- Nowak, J.S.; Choudhury, N.R.; de Lima Alves, F.; Rappsilber, J.; Michlewski, G. Lin28a regulates neuronal differentiation and controls miR-9 production. Nat. Commun. 2014, 5, 3687. [Google Scholar] [CrossRef]
- Gulyaeva, L.F.; Kushlinskiy, N.E. Regulatory mechanisms of microRNA expression. J. Transl. Med. 2016, 14, 143. [Google Scholar] [CrossRef]
- Dugas, J.C.; Cuellar, T.L.; Scholze, A.; Ason, B.; Ibrahim, A.; Emery, B.; Zamanian, J.L.; Foo, L.C.; McManus, M.T.; Barres, B.A. Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron 2010, 65, 597–611. [Google Scholar] [CrossRef]
- Li, H.; Mao, S.; Wang, H.; Zen, K.; Zhang, C.; Li, L. MicroRNA-29a modulates axon branching by targeting doublecortin in primary neurons. Protein Cell 2014, 5, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Nampoothiri, S.S.; Rajanikant, G.K. Decoding the ubiquitous role of microRNAs in neurogenesis. Mol. Neurobiol. 2017, 54, 2003–2011. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhang, M.; Li, H.; Frank, J.A.; Dai, L.; Liu, H.; Chen, G. MicroRNA-29b regulates ethanol-induced neuronal apoptosis in the developing cerebellum through SP1/RAX/PKR cascade. J. Biol. Chem. 2014, 289, 10201–10210. [Google Scholar] [CrossRef] [PubMed]
- Bittigau, P.; Sifringer, M.; Pohl, D.; Stadthaus, D.; Ishimaru, M.; Shimizu, H.; Ikeda, M.; Lang, D.; Speer, A.; Olney, J.W.; et al. Apoptotic neurodegeneration following trauma is markedly enhanced in the immature brain. Ann. Neurol. 1999, 45, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Pohl, D.; Bittigau, P.; Ishimaru, M.J.; Stadthaus, D.; Hübner, C.; Olney, J.W.; Turski, L.; Ikonomidou, C. N-Methyl-D-aspartate antagonists and apoptotic cell death triggered by head trauma in developing rat brain. Proc. Natl. Acad. Sci. USA 1999, 96, 2508–2513. [Google Scholar] [CrossRef]
- Hu, B.R.; Liu, C.L.; Ouyang, Y.; Blomgren, K.; Siesjö, B.K. Involvement of caspase-3 in cell death after hypoxia-ischemia declines during brain maturation. J. Cereb. Blood Flow Metab. 2000, 20, 1294–1300. [Google Scholar] [CrossRef]
- Yakovlev, A.G.; Ota, K.; Wang, G.; Movsesyan, V.; Bao, W.L.; Yoshihara, K.; Faden, A.I. Differential expression of apoptotic protease-activating factor-1 and caspase-3 genes and susceptibility to apoptosis during brain development and after traumatic brain injury. J. Neurosci. 2001, 21, 7439–7446. [Google Scholar] [CrossRef]
- Ertürk, A.; Wang, Y.; Sheng, M. Local pruning of dendrites and spines by caspase-3-dependent and proteasome-limited mechanisms. J. Neurosci. 2014, 34, 1672–1688. [Google Scholar] [CrossRef]
- Fadó, R.; Moubarak, R.S.; Miñano-Molina, A.J.; Barneda-Zahonero, B.; Valero, J.; Saura, C.A.; Moran, J.; Comella, J.X.; Rodríguez-Álvarez, J. X-linked inhibitor of apoptosis protein negatively regulates neuronal differentiation through interaction with cRAF and Trk. Sci. Rep. 2013, 3, 2397. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Headrick, T.C. Statistical Simulation: Power Method Polynomials and Other Transformations; Chapman & Hall/CRC: Boca Raton, FL, USA, 2010; 166p. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barreda-Manso, M.A.; Soto, A.; Muñoz-Galdeano, T.; Reigada, D.; Nieto-Díaz, M.; Maza, R.M. MiR-138-5p Upregulation during Neuronal Maturation Parallels with an Increase in Neuronal Survival. Int. J. Mol. Sci. 2023, 24, 16509. https://doi.org/10.3390/ijms242216509
Barreda-Manso MA, Soto A, Muñoz-Galdeano T, Reigada D, Nieto-Díaz M, Maza RM. MiR-138-5p Upregulation during Neuronal Maturation Parallels with an Increase in Neuronal Survival. International Journal of Molecular Sciences. 2023; 24(22):16509. https://doi.org/10.3390/ijms242216509
Chicago/Turabian StyleBarreda-Manso, María Asunción, Altea Soto, Teresa Muñoz-Galdeano, David Reigada, Manuel Nieto-Díaz, and Rodrigo M. Maza. 2023. "MiR-138-5p Upregulation during Neuronal Maturation Parallels with an Increase in Neuronal Survival" International Journal of Molecular Sciences 24, no. 22: 16509. https://doi.org/10.3390/ijms242216509
APA StyleBarreda-Manso, M. A., Soto, A., Muñoz-Galdeano, T., Reigada, D., Nieto-Díaz, M., & Maza, R. M. (2023). MiR-138-5p Upregulation during Neuronal Maturation Parallels with an Increase in Neuronal Survival. International Journal of Molecular Sciences, 24(22), 16509. https://doi.org/10.3390/ijms242216509





