Natural Inhibitors of Mammalian α-Amylases as Promising Drugs for the Treatment of Metabolic Diseases
Abstract
1. Introduction
2. General Aspects of Mammalian α-Amylases
3. Non-Proteinaceous α-Amylase Inhibitors
3.1. Pseudo-Oligosaccharides from Actinomycetes and Bacteria and Their Clinical Use
3.2. Secondary Metabolites from Plants and Prospects for Their Therapeutic Use
4. Proteinaceous α-Amylase Inhibitors
4.1. Inhibitors from Streptomyces
4.2. Inhibitors from Plants
4.3. Inhibitors from Sea Anemones
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [PubMed]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef] [PubMed]
- Temneanu, O.R.; Trandafir, L.M.; Purcarea, M.R. Type 2 diabetes mellitus in children and adolescents: A relatively new clinical problem within pediatric practice. J. Med. Life 2016, 9, 235–239. [Google Scholar]
- Alam, U.; Asghar, O.; Azmi, S. General aspects of diabetes mellitus. Handb. Clin. Neurol. 2014, 126, 211–222. [Google Scholar] [CrossRef]
- Pandey, A.; Chawla, S.; Guchhait, P. Type-2 diabetes: Current understanding and future perspectives. IUBMB Life 2015, 67, 506–513. [Google Scholar] [CrossRef]
- Jones, K.; Sim, L.; Mohan, S.; Kumarasamy, J.; Liu, H.; Avery, S.; Naim, H.Y.; Quezada-Calvillo, R.; Nichols, B.L.; Mario Pinto, B.; et al. Mapping the intestinal alpha-glucogenic enzyme specificities of starch digesting maltase-glucoamylase and sucrase-isomaltase. Bioorg. Med. Chem. 2011, 19, 3929–3934. [Google Scholar] [CrossRef]
- Ren, L.; Cao, X.; Geng, P.; Bai, F.; Bai, G. Study of the inhibition of two human maltase-glucoamylases catalytic domains by different α-glucosidase inhibitors. Carbohydr. Res. 2011, 346, 2688–2692. [Google Scholar] [CrossRef]
- Elferink, H.; Bruekers, J.P.J.; Veeneman, G.H.; Boltje, T.J. A comprehensive overview of substrate specificity of glycoside hydrolases and transporters in the small intestine: “A gut feeling”. Cell. Mol. Life Sci. 2020, 77, 4799–4826. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef]
- Da Lage, J.L.; Danchin, E.G.J.; Casane, D. Where do animal α-amylases come from? An interkingdom trip. FEBS Lett. 2007, 581, 3927–3935. [Google Scholar] [CrossRef]
- Janeček, Š. Sequence Similarities and Evolutionary Relationships of Microbial, Plant and Animal α-amylases. Eur. J. Biochem. 1994, 224, 519–524. [Google Scholar] [CrossRef]
- Janeček, Š. How many conserved sequence regions are there in the α-amylase family? Biol. Sect. Cell. Mol. Biol. 2002, 57, 29–41. [Google Scholar]
- D’Amico, S.; Gerday, C.; Feller, G. Structural similarities and evolutionary relationships in chloride-dependent α-amylases. Gene 2000, 253, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Brayer, G.D.; Sidhu, G.; Maurus, R.; Rydberg, E.H.; Braun, C.; Wang, Y.; Nguyen, N.T.; Overall, C.M.; Withers, S.G. Subsite mapping of the human pancreatic α-amylase active site through structural, kinetic, and mutagenesis techniques. Biochemistry 2000, 39, 4778–4791. [Google Scholar] [CrossRef]
- Rydberg, E.H.; Li, C.; Maurus, R.; Overall, C.M.; Brayer, G.D.; Withers, S.G. Mechanistic analyses of catalysis in human pancreatic α-amylase: Detailed kinetic and structural studies of mutants of three conserved carboxylic acids. Biochemistry 2002, 41, 4492–4502. [Google Scholar] [CrossRef] [PubMed]
- Buisson, G.; Duée, E.; Haser, R.; Payan, F. Three dimensional structure of porcine pancreatic alpha-amylase at 2.9 A resolution. Role of calcium in structure and activity. EMBO J. 1987, 6, 3909–3916. [Google Scholar] [CrossRef] [PubMed]
- Pasero, L.; Mazzéi-Pierron, Y.; Abadie, B.; Chicheportiche, Y.; Marchis-Mouren, G. Complete amino acid sequence and location of the five disulfide bridges in porcine pancreatic α-amylase. Biochim. Biophys. Acta (BBA)/Protein Struct. Mol. 1986, 869, 147–157. [Google Scholar] [CrossRef]
- Brayer, G.D.; Luo, Y.; Withers, S.G. The structure of human pancreatic alpha-amylase at 1.8 A resolution and comparisons with related enzymes. Protein Sci. 1995, 4, 1730–1742. [Google Scholar] [CrossRef]
- Gumucio, D.L.; Wiebauer, K.; Caldwell, R.M.; Samuelson, L.C.; Meisler, M.H. Concerted evolution of human amylase genes. Mol. Cell. Biol. 1988, 8, 1197–1205. [Google Scholar] [CrossRef]
- Sugino, H. Comparative genomic analysis of the mouse and rat amylase multigene family. FEBS Lett. 2007, 581, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Nishide, T.; Emi, M.; Nakamura, Y.; Matsubara, K. Corrected sequences of cDNAs for human salivary and pancreatic alpha-amylases [corrected]. Gene 1984, 28, 263–270. [Google Scholar] [PubMed]
- Wise, R.J.; Karn, R.C.; Larsen, S.H.; Hodes, M.E.; Gardell, S.J.; Rutter, W.J. A complementary DNA sequence that predicts a human pancreatic amylase primary structure consistent with the electrophoretic mobility of the common isozyme, Amy2 A. Mol. Biol. Med. 1984, 2, 307–322. [Google Scholar] [PubMed]
- Date, K.; Yamazaki, T.; Toyoda, Y.; Hoshi, K.; Ogawa, H. α-Amylase expressed in human small intestinal epithelial cells is essential for cell proliferation and differentiation. J. Cell. Biochem. 2020, 121, 1238–1249. [Google Scholar] [CrossRef]
- Yokouchi, H.; Horii, A.; Emi, M.; Tomita, N.; Doi, S.; Ogawa, M.; Mori, T.; Matsubara, K. Cloning and characterization of a third type of human α-amylase gene, AMY2B. Gene 1990, 90, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Seyama, K.; Nukiwa, T.; Takahashi, K.; Takahashi, H.; Kira, S. Amylase mRNA transcripts in normal tissues and neoplasms: The implication of different expressions of amylase isogenes. J. Cancer Res. Clin. Oncol. 1994, 120, 213–220. [Google Scholar] [CrossRef]
- Byman, E.; Schultz, N.; Fex, M.; Wennström, M. Brain alpha-amylase: A novel energy regulator important in Alzheimer disease? Brain Pathol. 2018, 28, 920–932. [Google Scholar] [CrossRef]
- Byman, E.; Schultz, N.; Blom, A.M.; Wennström, M. A Potential Role for α-Amylase in Amyloid-β-Induced Astrocytic Glycogenolysis and Activation. J. Alzheimer’s Dis. 2019, 68, 205–217. [Google Scholar] [CrossRef]
- Mahmud, T. The C7N aminocyclitol family of natural products. Nat. Prod. Rep. 2003, 20, 137–166. [Google Scholar] [CrossRef]
- Gilles, C.; Astier, J.P.; Marchis-Mouren, G.; Cambillau, C.; Payan, F. Crystal structure of pig pancreatic α-amylase isoenzyme II, in complex with the carbohydrate inhibitor acarbose. Eur. J. Biochem. 1996, 238, 561–569. [Google Scholar] [CrossRef]
- Yoon, S.H.; Robyt, J.F. Study of the inhibition of four alpha amylases by acarbose and its 4IV-α-maltohexaosyl and 4IV-α-maltododecaosyl analogues. Carbohydr. Res. 2003, 338, 1969–1980. [Google Scholar] [CrossRef] [PubMed]
- Singla, R.K.; Singh, R.; Dubey, A.K. Important Aspects of Post-Prandial Antidiabetic Drug, Acarbose. Curr. Top. Med. Chem. 2016, 16, 2625–2633. [Google Scholar] [CrossRef] [PubMed]
- Geng, P.; Bai, G. Two novel aminooligosaccharides isolated from the culture of Streptomyces coelicoflavus ZG0656 as potent inhibitors of α-amylase. Carbohydr. Res. 2008, 343, 470–476. [Google Scholar] [CrossRef]
- Geng, P.; Qiu, F.; Zhu, Y.; Bai, G. Four acarviosin-containing oligosaccharides identified from Streptomyces coelicoflavus ZG0656 are potent inhibitors of α-amylase. Carbohydr. Res. 2008, 343, 882–892. [Google Scholar] [CrossRef] [PubMed]
- McCranie, E.K.; Bachmann, B.O. Bioactive oligosaccharide natural products. Nat. Prod. Rep. 2014, 31, 1026–1042. [Google Scholar] [CrossRef]
- Qin, X.; Ren, L.; Yang, X.; Bai, F.; Wang, L.; Geng, P.; Bai, G.; Shen, Y. Structures of human pancreatic α-amylase in complex with acarviostatins: Implications for drug design against type II diabetes. J. Struct. Biol. 2011, 174, 196–202. [Google Scholar] [CrossRef]
- Hedrington, M.S.; Davis, S.N. Considerations when using alpha-glucosidase inhibitors in the treatment of type 2 diabetes. Expert Opin. Pharmacother. 2019, 20, 2229–2235. [Google Scholar] [CrossRef]
- Joshi, S.R.; Standl, E.; Tong, N.; Shah, P.; Kalra, S.; Rathod, R. Therapeutic potential of α-glucosidase inhibitors in type 2 diabetes mellitus: An evidence-based review. Expert Opin. Pharmacother. 2015, 16, 1959–1981. [Google Scholar] [CrossRef]
- Derosa, G.; Maffioli, P. Efficacy and Safety Profile Evaluation of Acarbose Alone and in Association with Other Antidiabetic Drugs: A Systematic Review. Clin. Ther. 2012, 34, 1221–1236. [Google Scholar] [CrossRef]
- Rosak, C.; Mertes, G. Critical evaluation of the role of acarbose in the treatment of diabetes: Patient considerations. Diabetes Metab. Syndr. Obes. Targets Ther. 2012, 5, 357–367. [Google Scholar] [CrossRef]
- Ríos, J.L.; Francini, F.; Schinella, G.R. Natural Products for the Treatment of Type 2 Diabetes Mellitus. Planta Med. 2015, 81, 975–994. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, Y.; Dai, Y.; Peng, J. Natural products for the treatment of type 2 diabetes mellitus: Pharmacology and mechanisms. Pharmacol. Res. 2018, 130, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Kashtoh, H.; Baek, K.H. Recent Updates on Phytoconstituent Alpha-Glucosidase Inhibitors: An Approach towards the Treatment of Type Two Diabetes. Plants 2022, 11, 2722. [Google Scholar] [CrossRef]
- Kashtoh, H.; Baek, K.H. New Insights into the Latest Advancement in α-Amylase Inhibitors of Plant Origin with Anti-Diabetic Effects. Plants 2023, 12, 2944. [Google Scholar] [CrossRef]
- Tundis, R.; Loizzo, M.R.; Menichini, F. Natural Products as alpha-Amylase and alpha-Glucosidase Inhibitors and their Hypoglycaemic Potential in the Treatment of Diabetes: An Update. Mini-Rev. Med. Chem. 2010, 10, 315–331. [Google Scholar] [CrossRef]
- Papoutsis, K.; Zhang, J.; Bowyer, M.C.; Brunton, N.; Gibney, E.R.; Lyng, J. Fruit, vegetables, and mushrooms for the preparation of extracts with α-amylase and α-glucosidase inhibition properties: A review. Food Chem. 2021, 338, 128119. [Google Scholar] [CrossRef] [PubMed]
- Shahwan, M.; Alhumaydhi, F.; Ashraf, G.M.; Hasan, P.M.Z.; Shamsi, A. Role of polyphenols in combating Type 2 Diabetes and insulin resistance. Int. J. Biol. Macromol. 2022, 206, 567–579. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, C.; Zhang, B.; Huang, Q. The inhibitory effects of flavonoids on α-amylase and α-glucosidase. Crit. Rev. Food Sci. Nutr. 2020, 60, 695–708. [Google Scholar] [CrossRef]
- Hossain, M.K.; Dayem, A.A.; Han, J.; Yin, Y.; Kim, K.; Saha, S.K.; Yang, G.M.; Choi, H.Y.; Cho, S.G. Molecular mechanisms of the anti-obesity and anti-diabetic properties of flavonoids. Int. J. Mol. Sci. 2016, 17, 569. [Google Scholar] [CrossRef]
- Vinayagam, R.; Xu, B. Antidiabetic properties of dietary flavonoids: A cellular mechanism review. Nutr. Metab. 2015, 12, 60. [Google Scholar] [CrossRef]
- Proença, C.; Ribeiro, D.; Freitas, M.; Fernandes, E. Flavonoids as potential agents in the management of type 2 diabetes through the modulation of α-amylase and α-glucosidase activity: A review. Crit. Rev. Food Sci. Nutr. 2020, 62, 3137–3207. [Google Scholar] [CrossRef] [PubMed]
- Proença, C.; Freitas, M.; Ribeiro, D.; Oliveira, E.F.T.; Sousa, J.L.C.; Tomé, S.M.; Ramos, M.J.; Silva, A.M.S.; Fernandes, P.A.; Fernandes, E. α-Glucosidase inhibition by flavonoids: An in vitro and in silico structure–activity relationship study. J. Enzym. Inhib. Med. Chem. 2017, 32, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Ni, X.; Kai, G.; Chen, X. A review on structure-activity relationship of dietary polyphenols inhibiting α-amylase. Crit. Rev. Food Sci. Nutr. 2013, 53, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Proença, C.; Freitas, M.; Ribeiro, D.; Tomé, S.M.; Oliveira, E.F.T.; Viegas, M.F.; Araújo, A.N.; Ramos, M.J.; Silva, A.M.S.; Fernandes, P.A.; et al. Evaluation of a flavonoids library for inhibition of pancreatic α-amylase towards a structure–activity relationship. J. Enzym. Inhib. Med. Chem. 2019, 34, 577–588. [Google Scholar] [CrossRef]
- Rasouli, H.; Hosseini-Ghazvini, S.M.B.; Adibi, H.; Khodarahmi, R. Differential α-amylase/α-glucosidase inhibitory activities of plant-derived phenolic compounds: A virtual screening perspective for the treatment of obesity and diabetes. Food Funct. 2017, 8, 1942–1954. [Google Scholar] [CrossRef]
- Koleckar, V.; Kubikova, K.; Rehakova, Z.; Kuca, K.; Jun, D.; Jahodar, L.; Opletal, L. Condensed and Hydrolysable Tannins as Antioxidants Influencing the Health. Mini-Rev. Med. Chem. 2008, 8, 436–447. [Google Scholar] [CrossRef]
- Lin, D.; Xiao, M.; Zhao, J.; Li, Z.; Xing, B.; Li, X.; Kong, M.; Li, L.; Zhang, Q.; Liu, Y.; et al. An overview of plant phenolic compounds and their importance in human nutrition and management of type 2 diabetes. Molecules 2016, 21, 1374. [Google Scholar] [CrossRef] [PubMed]
- de Sales, P.M.; de Souza, P.M.; Simeoni, L.A.; Magalhães, P.d.O.; Silveira, D. α-Amylase inhibitors: A review of raw material and isolated compounds from plant source. J. Pharm. Pharm. Sci. 2012, 15, 141–183. [Google Scholar] [CrossRef]
- Kandra, L.; Gyémánt, G.; Zajácz, Á.; Batta, G. Inhibitory effects of tannin on human salivary α-amylase. Biochem. Biophys. Res. Commun. 2004, 319, 1265–1271. [Google Scholar] [CrossRef] [PubMed]
- McDougall, G.J.; Shpiro, F.; Dobson, P.; Smith, P.; Blake, A.; Stewart, D. Different polyphenolic components of soft fruits inhibit α-amylase and α-glycosidase. J. Agric. Food Chem. 2005, 53, 2760–2766. [Google Scholar] [CrossRef]
- Etxeberria, U.; De La Garza, A.L.; Campin, J.; Martnez, J.A.; Milagro, F.I. Antidiabetic effects of natural plant extracts via inhibition of carbohydrate hydrolysis enzymes with emphasis on pancreatic alpha amylase. Expert Opin. Ther. Targets 2012, 16, 269–297. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Huang, W.; Xu, Z.; Cao, M.; Hu, Q.; Pan, C.; Guo, M.; Wei, J.F.; Yuan, H. Analysis of patents issued in China for antihyperglycemic therapies for type 2 diabetes mellitus. Front. Pharmacol. 2019, 10, 586. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, S.; Wang, S. Can highly cited herbs in ancient Traditional Chinese medicine formulas and modern publications predict therapeutic targets for diabetes mellitus? J. Ethnopharmacol. 2018, 213, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Zhao, Y.; Zhang, Y. Traditional Chinese medicines in treatment of patients with type 2 diabetes mellitus. Evid.-Based Complement. Altern. Med. 2011, 2011, 726723. [Google Scholar] [CrossRef]
- Jia, W.; Gaoz, W.; Tang, L. Antidiabetic Herbal Drugs Officially Approved in China. Phyther. Res. 2003, 17, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.S.; Lv, J.H.; Tao, J.; Li, G.M.; Li, H.D.; Ma, N. Antihyperglycemic effects of total flavonoids from Polygonatum odoratum in STZ and alloxan-induced diabetic rats. J. Ethnopharmacol. 2009, 124, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Alarcon-Aguilara, F.J.; Roman-Ramos, R.; Perez-Gutierrez, S.; Aguilar-Contreras, A.; Contreras-Weber, C.C.; Flores-Saenz, J.L. Study of the anti-hyperglycemic effect of plants used as antidiabetics. J. Ethnopharmacol. 1998, 61, 101–110. [Google Scholar] [CrossRef]
- Ponnusamy, S.; Zinjarde, S.S.; Bhargava, S.Y.; Kumar, A.R. Potent α-amylase inhibitory activity of Indian Ayurvedic medicinal plants. BMC Complement. Altern. Med. 2011, 11, 5. [Google Scholar] [CrossRef]
- Ponnusamy, S.; Ravindran, R.; Zinjarde, S.; Bhargava, S.; Ameeta, R.K. Evaluation of Traditional Indian Antidiabetic Medicinal Plants for Human Pancreatic Amylase Inhibitory Effect in Vitro. Evid.-Based Complement. Altern. Med. 2011, 2011, 515647. [Google Scholar] [CrossRef]
- Kasabri, V.; Afifi, F.U.; Hamdan, I. Evaluation of the acute antihyperglycemic effects of four selected indigenous plants from Jordan used in traditional medicine. Pharm. Biol. 2011, 49, 687–695. [Google Scholar] [CrossRef]
- Kasabri, V.; Afifi, F.U.; Hamdan, I. In vitro and in vivo acute antihyperglycemic effects of five selected indigenous plants from Jordan used in traditional medicine. J. Ethnopharmacol. 2011, 133, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Afifi, F.U.; Kasabri, V.; Litescu, S.C.; Abaza, I.M. In vitro and in vivo comparison of the biological activities of two traditionally and widely used Arum species from Jordan: Arum dioscoridis Sibth & Sm. and Arum palaestinum Boiss. Nat. Prod. Res. 2016, 30, 1777–1786. [Google Scholar] [CrossRef] [PubMed]
- Peres, M.A.; Macpherson, L.M.D.; Weyant, R.J.; Daly, B.; Venturelli, R.; Mathur, M.R.; Listl, S.; Celeste, R.K.; Guarnizo-Herreño, C.C.; Kearns, C.; et al. Oral diseases: A global public health challenge. Lancet 2019, 394, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Vos, T.; Abajobir, A.A.; Abbafati, C.; Abbas, K.M.; Abate, K.H.; Abd-Allah, F.; Abdulle, A.M.; Abebo, T.A.; Abera, S.F.; Aboyans, V.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1211–1259. [Google Scholar] [CrossRef]
- Scannapieco, F.A.; Solomon, L.; Wadenya, R.O. Emergence in Human Dental Plaque and Host Distribution of Amylase-binding Streptococci. J. Dent. Res. 1994, 73, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Gwynn, J.P.; Douglas, C.W.I. Comparison of amylase-binding proteins in oral streptococci. FEMS Microbiol. Lett. 1994, 124, 373–379. [Google Scholar] [CrossRef][Green Version]
- Arya, V.; Taneja, L. Inhibition of salivary amylase by black tea in high-caries and low-caries index children: A comparative in vivo study. AYU (An Int. Q. J. Res. Ayurveda) 2015, 36, 278. [Google Scholar] [CrossRef]
- Zhang, J.; Kashket, S. Inhibition of Salivary Amylase by Black and Green Teas and Their Effects on the Intraoral Hydrolysis of Starch. Caries Res. 1998, 32, 233–238. [Google Scholar] [CrossRef]
- Homoki, J.; Gyémánt, G.; Balogh, P.; Stündl, L.; Bíró-Molnár, P.; Paholcsek, M.; Váradi, J.; Ferenc, F.; Kelentey, B.; Nemes, J.; et al. Sour cherry extract inhibits human salivary α-amylase and growth of Streptococcus mutans (a pilot clinical study). Food Funct. 2018, 9, 4008–4016. [Google Scholar] [CrossRef]
- Forester, S.C.; Gu, Y.; Lambert, J.D. Inhibition of starch digestion by the green tea polyphenol, (−)-epigallocatechin-3-gallate. Mol. Nutr. Food Res. 2012, 56, 1647–1654. [Google Scholar] [CrossRef]
- Barroso, H.; Ramalhete, R.; Domingues, A.; Maci, S. Inhibitory activity of a green and black tea blend on Streptococcus mutans. J. Oral Microbiol. 2018, 10, 1481322. [Google Scholar] [CrossRef] [PubMed]
- Aschauer, H.; Vértesy, L.; Nesemann, G.; Braunitzer, G. The primary structure of the alpha-amylase inhibitor Hoe 467A from Streptomyces tendae 4158. A new class of inhibitors. Hoppe. Seylers. Z. Physiol. Chem. 1983, 364, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, O.; Vértesy, L.; Braunitzer, G. The Primary Structure of α-Amylase Inhibitor Z-2685 from Streptomyces parvullus FH-1641: Sequence Homology between Inhibitor and α-Amylase. Biol. Chem. Hoppe. Seyler. 1985, 366, 1161–1168. [Google Scholar] [CrossRef]
- Murai, H.; Hara, S.; Ikenaka, T.; Goto, A.; Arai, M.; Murao, S. Amino acid sequence of protein α-amylase inhibitor from Streptomyces griseosporeus YM-25. J. Biochem. 1985, 97, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Vértesy, L.; Tripier, D. Isolation and structure elucidation of an alpha-amylase inhibitor, AI-3688, from Streptomyces aureofaciens. FEBS Lett. 1985, 185, 187–190. [Google Scholar] [CrossRef]
- Hirayama, K.; Takahashi, R.; Akashi, S.; Fukuhara, K.I.; Oouchi, N.; Murai, A.; Arai, M.; Murao, S.; Tanaka, K.; Nojima, I. Primary Structure of Paim I, an α-Amylase Inhibitor from Streptomyces corchorushii, Determined by the Combination of Edman Degradation and Fast Atom Bombardment Mass Spectrometry. Biochemistry 1987, 26, 6483–6488. [Google Scholar] [CrossRef]
- Katsuyama, K.; Iwata, N.; Shimazu, A. Purification and primary structure of proteinous alpha-amylase inhibitor from Streptomyces chartreusis. Biosci. Biotechnol. Biochem. 1992, 56, 1949–1954. [Google Scholar] [CrossRef]
- Sumitani, J.i.; Kawaguchi, T.; Hattori, N.; Murao, S.; Arai, M. Molecular Cloning and Expression of Proteinaceous α-Amylase Inhibitor Gene from Streptomyces nitrosporeus in Escherichia coli. Biosci. Biotechnol. Biochem. 1993, 57, 1243–1248. [Google Scholar] [CrossRef]
- Pflugrath, J.W.; Wiegand, G.; Huber, R.; Vértesy, L. Crystal structure determination, refinement and the molecular model of the α-amylase inhibitor Hoe-467A. J. Mol. Biol. 1986, 189, 383–386. [Google Scholar] [CrossRef]
- Akashi, S.; Hirayama, K.; Seino, T.; Ozawa, S.i.; Fukuhara, K.-i.; Oouchi, N.; Murai, A.; Arai, M.; Murao, S.; Tanaka, K.; et al. A determination of the positions of disulphide bonds in Paim I, α-amylase inhibitor from Streptomyces corchorushii, using fast atom bombardment mass spectrometry. Biomed. Environ. Mass Spectrom. 1988, 15, 541–546. [Google Scholar] [CrossRef]
- Miyagawa, M.; Fukuhara, K.I.; Katayanagi, K.; Ishimaru, K.; Morikawa, K.; Matsuzaki, T.; Sato, Y.; Hara, S.; Ikenaka, T.; Arai, M.; et al. Crystallization and preliminary crystallographic data of the α-amylase inhibitors, haim I and paim I. J. Biochem. 1994, 115, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Yoshlda, M.; Nakai, T.; Fukuhara, K.; Saitoh, S.; Yoshikawa, W.; Kobayashi, Y.; Nakamura, H. Three-dimensional structure of an α-amylase inhibitor HAIM as determined by nuclear magnetic resonance methods. J. Biochem. 1990, 108, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Rehm, S.; Han, S.; Hassani, I.; Sokocevic, A.; Jonker, H.R.A.; Engels, J.W.; Schwalbe, H. The high resolution NMR structure of parvulustat (Z-2685) from Streptomyces parvulus FH-1641: Comparison with tendamistat from Streptomyces tendae 4158. ChemBioChem 2009, 10, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, G.; Epp, O.; Huber, R. The crystal structure of porcine pancreatic alpha-amylase in complex with the microbial inhibitor Tendamistat. J. Mol. Biol. 1995, 247, 99–110. [Google Scholar] [CrossRef]
- Vértesy, L.; Oeding, V.; Bender, R.; Zepf, K.; Nesemann, G. Tendamistat (HOE 467), a tight-binding α-amylase inhibitor from Streptomyces tendae 4158: Isolation, biochemical properties. Eur. J. Biochem. 1984, 141, 505–512. [Google Scholar] [CrossRef]
- Rupp, W.; Grigoleit, H.G.; Grötsch, H. Effect on salivary enzymes of alpha-amylase inhibitor HOE 467. Lancet 1983, 321, 1103–1104. [Google Scholar] [CrossRef]
- Meyer, B.H.; Müller, F.O.; Kruger, J.B.; Grigoleit, H.G. Inhibition of starch absorption by alpha-amylase inactivator given with food. Lancet 1983, 321, 934. [Google Scholar] [CrossRef]
- Gonçalves, O.; Dintinger, T.; Blanchard, D.; Tellier, C. Functional mimicry between anti-Tendamistat antibodies and α-amylase. J. Immunol. Methods 2002, 269, 29–37. [Google Scholar] [CrossRef]
- Long, C.M.; Virolle, M.J.; Chang, S.Y.; Chang, S.; Bibb, M.J. alpha-Amylase gene of Streptomyces limosus: Nucleotide sequence, expression motifs, and amino acid sequence homology to mammalian and invertebrate alpha-amylases. J. Bacteriol. 1987, 169, 5745–5754. [Google Scholar] [CrossRef] [PubMed]
- Franco, O.L.; Rigden, D.J.; Melo, F.R.; Grossi-de-Sá, M.F. Plant α-amylase inhibitors and their interaction with insect α-amylases: Structure, function and potential for crop protection. Eur. J. Biochem. 2002, 269, 397–412. [Google Scholar] [CrossRef]
- Rane, A.S.; Joshi, R.S.; Giri, A.P. Molecular determinant for specificity: Differential interaction of α-amylases with their proteinaceous inhibitors. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129703. [Google Scholar] [CrossRef] [PubMed]
- Svensson, B.; Fukuda, K.; Nielsen, P.K.; Bønsager, B.C. Proteinaceous α-amylase inhibitors. Biochim. Biophys. Acta Proteins Proteom. 2004, 1696, 145–156. [Google Scholar] [CrossRef]
- Lioi, L.; Galasso, I.; Lanave, C.; Daminati, M.G.; Bollini, R.; Sparvoli, F. Evolutionary analysis of the APA genes in the Phaseolus genus: Wild and cultivated bean species as sources of lectin-related resistance factors? Theor. Appl. Genet. 2007, 115, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.; Chrispeels, M.J. A lectin gene encodes the alpha-amylase inhibitor of the common bean. Proc. Natl. Acad. Sci. USA 1989, 86, 7885–7889. [Google Scholar] [CrossRef]
- Santimone, M.; Koukiekolo, R.; Moreau, Y.; Le Berre, V.; Rougé, P.; Marchis-Mouren, G.; Desseaux, V. Porcine pancreatic α-amylase inhibition by the kidney bean (Phaseolus vulgaris) inhibitor (α-AI1) and structural changes in the α-amylase inhibitor complex. Biochim. Biophys. Acta Proteins Proteom. 2004, 1696, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, E.R.; Whitaker, J.R. Some Aspects of the Mechanism of Complexation of Red Kidney Bean α-Amylase Inhibitor and α-Amylase. Biochemistry 1984, 23, 1783–1791. [Google Scholar] [CrossRef]
- Grossi de Sa, M.F.; Mirkov, T.E.; Ishimoto, M.; Colucci, G.; Bateman, K.S.; Chrispeels, M.J. Molecular characterization of a bean α-amylase inhibitor that inhibits the α-amylase of the Mexican bean weevil Zabrotes subfasciatus. Planta 1997, 203, 295–303. [Google Scholar] [CrossRef]
- Young, N.M.; Thibault, P.; Watson, D.C.; Chrispeels, M.J. Post-translational processing of two α-amylase inhibitors and an arcelin from the common bean, Phaseolus vulgaris. FEBS Lett. 1999, 446, 203–206. [Google Scholar] [CrossRef]
- Bompard-Gilles, C.; Rousseau, P.; Rouge, P.; Payan, F. Substrate mimicry in the active center of a mammalian α-amylase: Structural analysis of an enzyme-inhibitor complex. Structure 1996, 4, 1441–1452. [Google Scholar] [CrossRef]
- Nahoum, V.; Roux, G.; Anton, V.; Rougé, P.; Puigserver, A.; Bischoff, H.; Henrissat, B.; Payan, F. Crystal structures of human pancreatic α-amylase in complex with carbohydrate and proteinaceous inhibitors. Biochem. J. 2000, 346, 201–208. [Google Scholar] [CrossRef]
- Le Berre-Anton, V.; Nahoum, V.; Payan, F.; Rougé, P. Molecular basis for the specific binding of different α-amylase inhibitors from Phaseolus vulgaris seeds to the active site of α-amylase. Plant Physiol. Biochem. 2000, 38, 657–665. [Google Scholar] [CrossRef]
- Choudhury, A.; Maeda, K.; Murayama, R.; DiMagno, E.P. Character of a wheat amylase inhibitor preparation and effects on fasting human pancreaticobiliary secretions and hormones. Gastroenterology 1996, 111, 1313–1320. [Google Scholar] [CrossRef] [PubMed]
- Oneda, H.; Lee, S.; Inouye, K. Inhibitory Effect of 0.19 α-Amylase Inhibitor from Wheat Kernel on the Activity of Porcine Pancreas α-Amylase and Its Thermal Stability. J. Biochem. 2004, 135, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Goff, D.J.; Kull, F.J. The inhibition of human salivary α-amylase by type II α-amylase inhibitor from Triticum aestivum is competitive, slow and tight-binding. J. Enzym. Inhib. Med. Chem. 1995, 9, 163–170. [Google Scholar] [CrossRef]
- Sen, S.; Dutta, S.K. Cloning, expression and characterization of biotic stress inducible Ragi bifunctional inhibitor (RBI) gene from Eleusine coracana Gaertn. J. Plant Biochem. Biotechnol. 2012, 21, 66–76. [Google Scholar] [CrossRef]
- Oda, Y.; Matsunaga, T.; Fukuyama, K.; Miyazaki, T.; Morimoto, T. Tertiary and quaternary structures of 0.19 α-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 Å resolution. Biochemistry 1997, 36, 13503–13511. [Google Scholar] [CrossRef]
- Strobl, S.; Miihlhahn, P.; Bernstein, R.; Wiltscheck, R.; Maskos, K.; Wunderlich, M.; Huber, R.; Holak, T.A.; Glockshuber, R. Determination of the Three-Dimensional Structure of the Bifunctional α-Amylase/Trypsin Inhibitor from Ragi Seeds by NMR Spectroscopy. Biochemistry 1995, 34, 8281–8293. [Google Scholar] [CrossRef]
- Alam, N.; Gourinath, S.; Dey, S.; Srinivasan, A.; Singh, T.P. Substrate—Inhibitor interactions in the kinetics of α-amylase inhibition by ragi α-amylase/trypsin inhibitor (RATI) and its various N-terminal fragments. Biochemistry 2001, 40, 4229–4233. [Google Scholar] [CrossRef]
- Lankisch, M.; Layer, P.; Rizza, R.A.; DiMagno, E.P. Acute postprandial gastrointestinal and metabolic effects of wheat amylase inhibitor (WAI) in normal, obese, and diabetic humans. Pancreas 1998, 17, 176–181. [Google Scholar] [CrossRef]
- Kodama, T.; Miyazaki, T.; Kitamura, I.; Suzuki, Y.; Namba, Y.; Sakurai, J.; Torikai, Y.; Inoue, S. Effects of single and long-term administration of wheat albumin on blood glucose control: Randomized controlled clinical trials. Eur. J. Clin. Nutr. 2005, 59, 384–392. [Google Scholar] [CrossRef]
- Salcedo, G.; Snchez-Monge, R.; Garca-Casado, G.; Armentia, A.; Gomez, L.; Barber, D. The Cereal α-Amylase/Trypsin Inhibitor Family Associated with Bakers’ asthma and Food Allergy. In Plant Food Allergens; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2007; pp. 70–86. [Google Scholar]
- Junker, Y.; Zeissig, S.; Kim, S.J.; Barisani, D.; Wieser, H.; Leffler, D.A.; Zevallos, V.; Libermann, T.A.; Dillon, S.; Freitag, T.L.; et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J. Exp. Med. 2012, 209, 2395–2408. [Google Scholar] [CrossRef]
- Zevallos, V.F.; Raker, V.; Tenzer, S.; Jimenez-Calvente, C.; Ashfaq-Khan, M.; Rüssel, N.; Pickert, G.; Schild, H.; Steinbrink, K.; Schuppan, D. Nutritional Wheat Amylase-Trypsin Inhibitors Promote Intestinal Inflammation via Activation of Myeloid Cells. Gastroenterology 2017, 152, 1100–1113.e12. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Suarez, M.G.; Murray, J.A. Nonceliac Gluten and Wheat Sensitivity. Clin. Gastroenterol. Hepatol. 2020, 18, 1913–1922.e1. [Google Scholar] [CrossRef] [PubMed]
- Chokshi, D. Toxicity studies of Blockal, a dietary supplement containing Phase 2 Starch Neutralizer (Phase 2), a standardized extract of the common white kidney bean (Phaseolus vulgaris). Int. J. Toxicol. 2006, 25, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Obiro, W.C.; Zhang, T.; Jiang, B. The nutraceutical role of the Phaseolus vulgaris α-amylase inhibitor. Br. J. Nutr. 2008, 100, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.L.; Udani, J.K. A proprietary alpha-amylase inhibitor from white bean (Phaseolus vulgaris): A review of clinical studies on weight loss and glycemic control. Nutr. J. 2011, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Harikumar, K.B.; Jesil, A.M.; Sabu, M.C.; Kuttan, R. A preliminary assessment of the acute and subchronic toxicity profile of Phase2: An α-amylase inhibitor. Int. J. Toxicol. 2005, 24, 95–102. [Google Scholar] [CrossRef]
- Micheli, L.; Lucarini, E.; Trallori, E.; Avagliano, C.; de Caro, C.; Russo, R.; Calignano, A.; Ghelardini, C.; Pacini, A.; Di Cesare Mannelli1, L. Phaseolus vulgaris L. Extract: Alpha-amylase inhibition against metabolic syndrome in mice. Nutrients 2019, 11, 1778. [Google Scholar] [CrossRef]
- Tysoe, C.; Williams, L.K.; Keyzers, R.; Nguyen, N.T.; Tarling, C.; Wicki, J.; Goddard-Borger, E.D.; Aguda, A.H.; Perry, S.; Foster, L.J.; et al. Potent Human α-Amylase Inhibition by the β-Defensin-like Protein Helianthamide. ACS Cent. Sci. 2016, 2, 154–161. [Google Scholar] [CrossRef]
- Torres, A.M.; Kuchel, P.W. The β-defensin-fold family of polypeptides. Toxicon 2004, 44, 581–588. [Google Scholar] [CrossRef]
- Shafee, T.M.A.; Lay, F.T.; Phan, T.K.; Anderson, M.A.; Hulett, M.D. Convergent evolution of defensin sequence, structure and function. Cell. Mol. Life Sci. 2017, 74, 663–682. [Google Scholar] [CrossRef] [PubMed]
- Tysoe, C.; Withers, S.G. Structural Dissection of Helianthamide Reveals the Basis of Its Potent Inhibition of Human Pancreatic α-Amylase. Biochemistry 2018, 57, 5384–5387. [Google Scholar] [CrossRef]
- Sintsova, O.; Gladkikh, I.; Chausova, V.; Monastyrnaya, M.; Anastyuk, S.; Chernikov, O.; Yurchenko, E.; Aminin, D.; Isaeva, M.; Leychenko, E.; et al. Peptide fingerprinting of the sea anemone Heteractis magnifica mucus revealed neurotoxins, Kunitz-type proteinase inhibitors and a new β-defensin α-amylase inhibitor. J. Proteom. 2018, 173, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Sintsova, O.; Gladkikh, I.; Kalinovskii, A.; Zelepuga, E.; Monastyrnaya, M.; Kim, N.; Shevchenko, L.; Peigneur, S.; Tytgat, J.; Kozlovskaya, E.; et al. Magnificamide, a β-Defensin-like peptide from the mucus of the sea anemone heteractis magnifica, is a strong inhibitor of Mammalian α-Amylases. Mar. Drugs 2019, 17, 542. [Google Scholar] [CrossRef] [PubMed]
- Sintsova, O.; Popkova, D.; Kalinovskii, A.; Rasin, A.; Borozdina, N.; Shaykhutdinova, E.; Klimovich, A.; Menshov, A.; Kim, N.; Anastyuk, S.; et al. Control of postprandial hyperglycemia by oral administration of the sea anemone mucus-derived α-amylase inhibitor (magnificamide). Biomed. Pharmacother. 2023, 168, 115743. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Wang, L.; Zhai, J.; Han, N.; Liu, Z.; Li, S.; Yin, J. Characterization of antioxidant, α-glucosidase and tyrosinase inhibitors from the rhizomes of Potentilla anserina L. and their structure–activity relationship. Food Chem. 2021, 336, 127714. [Google Scholar] [CrossRef]
- Le, T.-K.-D.; Danova, A.; Aree, T.; Duong, T.-H.; Koketsu, M.; Ninomiya, M.; Sawada, Y.; Kamsri, P.; Pungpo, P.; Chavasiri, W. α-Glucosidase Inhibitors from the Stems of Knema globularia. J. Nat. Prod. 2022, 85, 776–786. [Google Scholar] [CrossRef]
- Sgariglia, M.A.; Garibotto, F.M.; Soberón, J.R.; Angelina, E.L.; Andujar, S.A.; Vattuone, M.A. Study of polyphenols from Caesalpinia paraguariensis as α-glucosidase inhibitors: Kinetics and structure-activity relationship. New J. Chem. 2022, 46, 11044–11055. [Google Scholar] [CrossRef]
- Sheikh, Y.; Chanu, M.B.; Mondal, G.; Manna, P.; Chattoraj, A.; Chandra Deka, D.; Chandra Talukdar, N.; Chandra Borah, J. Procyanidin A2, an anti-diabetic condensed tannin extracted from Wendlandia glabrata, reduces elevated G-6-Pase and mRNA levels in diabetic mice and increases glucose uptake in CC1 hepatocytes and C1C12 myoblast cells. RSC Adv. 2019, 9, 17211–17219. [Google Scholar] [CrossRef]
- Sokočević, A.; Han, S.; Engels, J.W. Biophysical characterization of α-amylase inhibitor Parvulustat (Z-2685) and comparison with Tendamistat (HOE-467). Biochim. Biophys. Acta Proteins Proteom. 2011, 1814, 1383–1393. [Google Scholar] [CrossRef]
- Popkova, D.; Otstavnykh, N.; Sintsova, O.; Baldaev, S.; Kalina, R.; Gladkikh, I.; Isaeva, M.; Leychenko, E. Bioprospecting of Sea Anemones (Cnidaria, Anthozoa, Actiniaria) for β-Defensin-like α-Amylase Inhibitors. Biomedicines 2023, 11, 2682. [Google Scholar] [CrossRef] [PubMed]
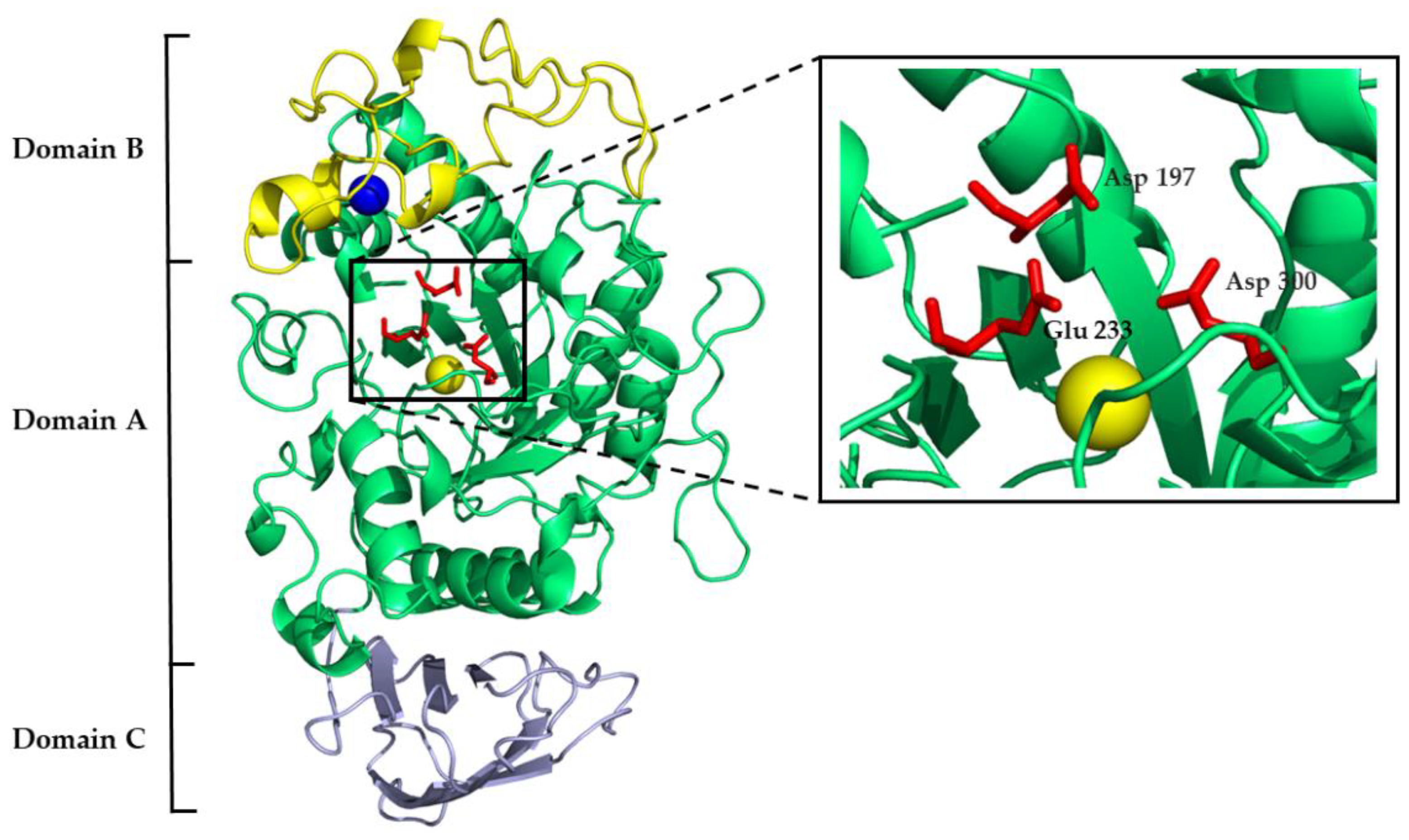
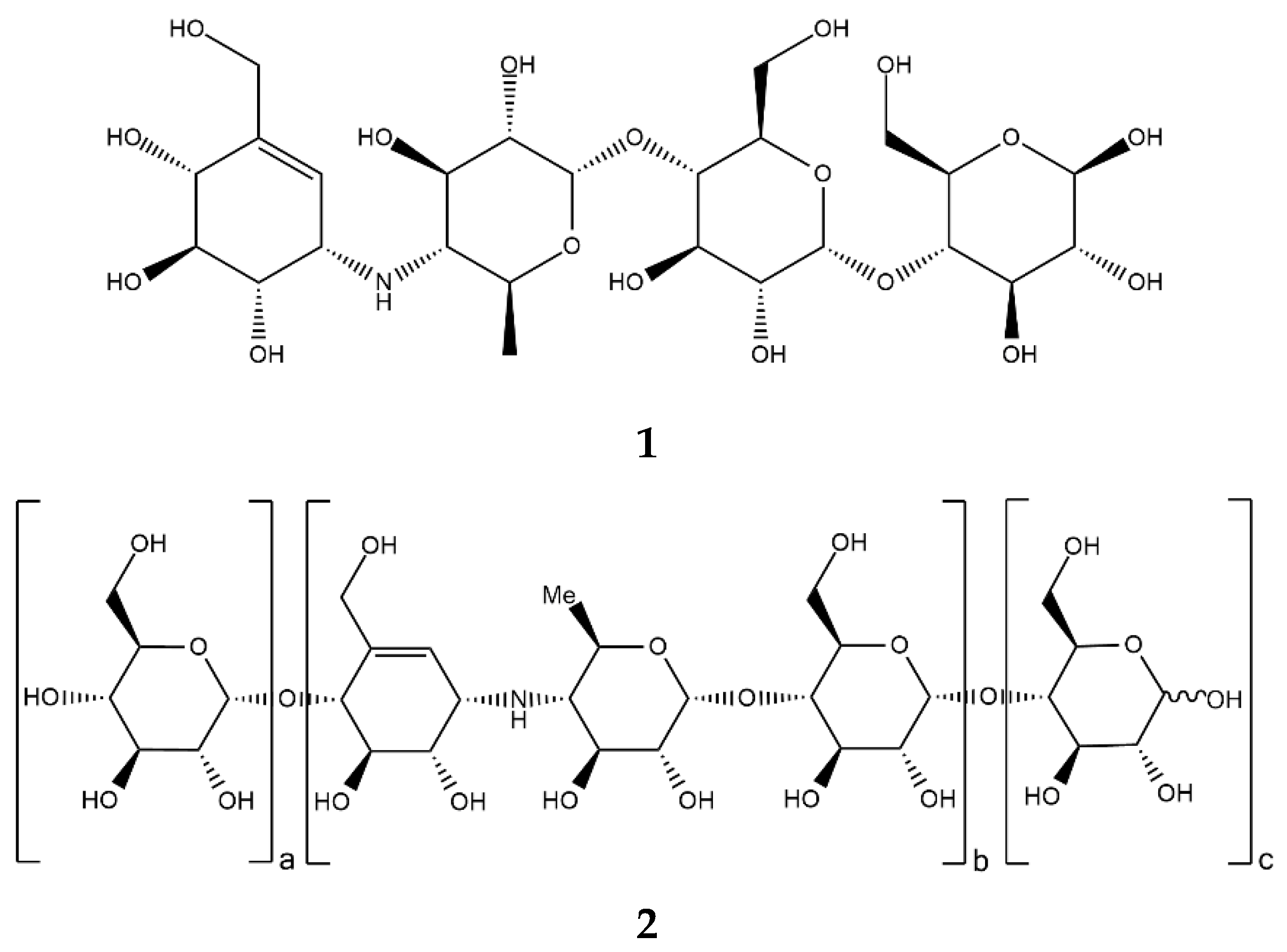


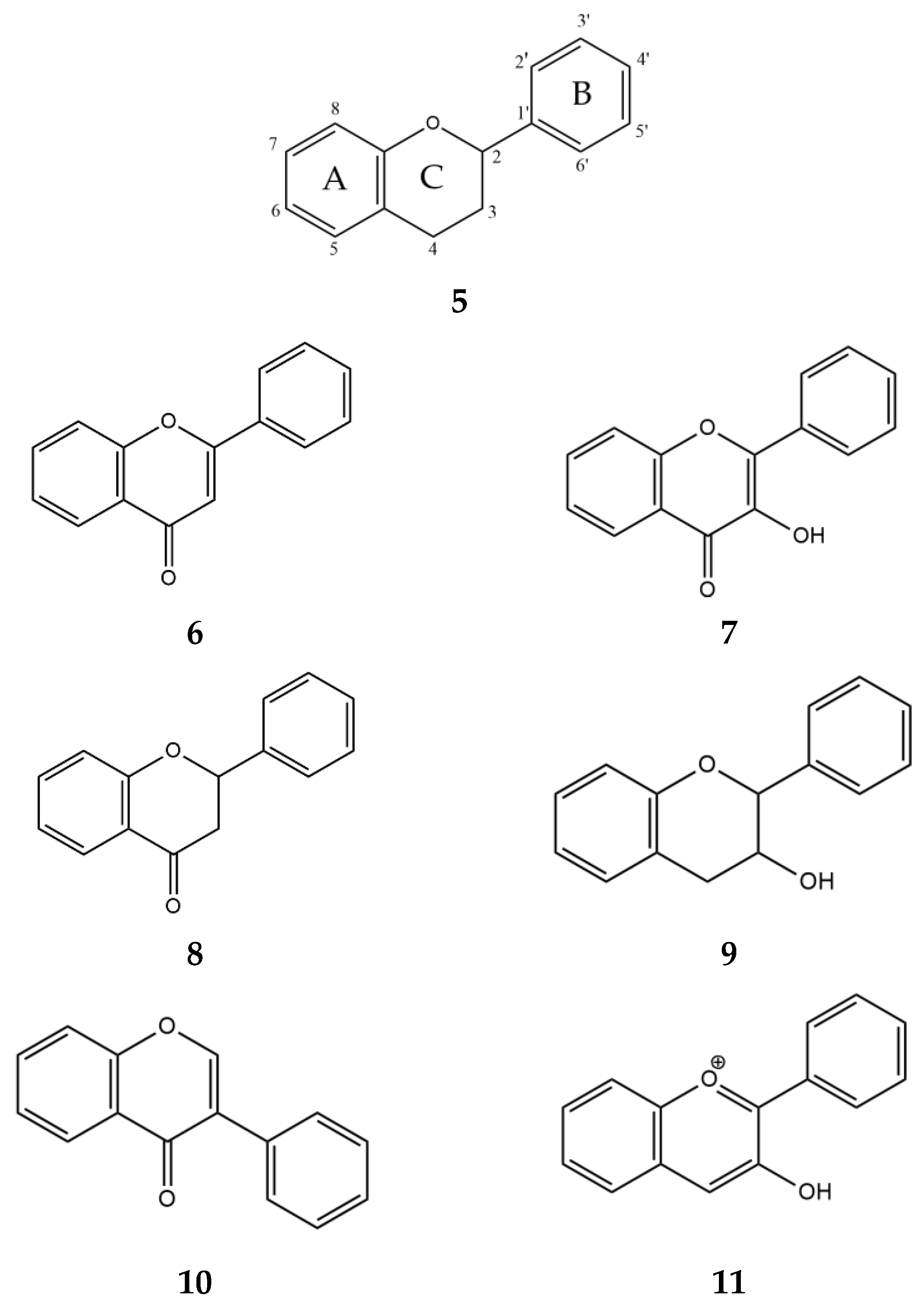

| Mammal | Name | Sequence Identity, % | a.a. | Molecular Weight, kDa |
|---|---|---|---|---|
| Human | 1A (salivary) | 100 | 496 | 55.9 |
| 2A (pancreatic) | 97 | 496 | 55.9 | |
| 2B (carcinoid) | 98 | 496 | 55.9 | |
| Sus scrofa | Pancreatic | 86 | 496 | 55.4 |
| Mus musculus | 1 (salivary and liver) | 85 | 496 | 55.9 |
| 2 (pancreatic) | 84 | 493 | 55.6 | |
| Rattus norvegicus | Pancreatic | 84 | 493 | 55.5 |
| Structural Class/Origin | Source | Name | Target | IC50/Ki, M | Ref. |
|---|---|---|---|---|---|
| Non-proteinaceous α-amylase inhibitors | |||||
| Actinomycete | Actinoplanes strain SE 50/110 | Acarbose | PPA | 0.8 × 10−6 | [31] |
| Flavonoids | Potentilla anserine (rhizome) | Quercetin-3-O-α-L-rhamnopyranoside-2″-gallate | α-glucosidase, α-amylase | 1.05 × 10−6 | [137] |
| Knema globularia (stem) | Calodenin A | 0.4 × 10−6 | [138] | ||
| Caesalpinia paraguariensis (bark) | (−) epigallocatechin-gallate | 5.20 × 10−6 | [139] | ||
| Tannins | Wendlandia glabrata | Procyanidin A2 | 0.47 × 10−6 | [140] | |
| Rubus chingii Hu | Chingiitannin A | 4.52 × 10−6 | |||
| Proteinaceous α-amylase inhibitors | |||||
| Microbial | Streptomyces tendae | Tendamistat | HPA | 9 × 10−12 – 2 × 10−10 | [95] |
| Streptomyces parvulus | Parvulustat | PPA | 2.8 × 10−11 | [141] | |
| Legume lectin-like | Phaseolus vulgaris | α-AI1 (heterodimer) | PPA | 3 × 10−11 | [106] |
| HPA | + | [110] | |||
| Cereal-type | Triticum aestivum | 0.19 (monomer) | PPA | 5.73 × 10−8 | [113] |
| HSA | 2.9 × 10−10 | [114] | |||
| Eleusine coracana | RBI | PPA | 1.8 × 10−10 | [115] | |
| β-defensin-like | Stichodactyla helianthus | Helianthamide | PPA | 1 × 10−10 | [130] |
| HSA | 1 × 10−11 | [130] | |||
| Heteractis magnifica | Magnificamide | PPA | 1.7 × 10−10 | [135] | |
| HSA | 7.7 × 10−9 | [135] | |||
| HPA | 3.1 × 10−9 | [136] | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalinovskii, A.P.; Sintsova, O.V.; Gladkikh, I.N.; Leychenko, E.V. Natural Inhibitors of Mammalian α-Amylases as Promising Drugs for the Treatment of Metabolic Diseases. Int. J. Mol. Sci. 2023, 24, 16514. https://doi.org/10.3390/ijms242216514
Kalinovskii AP, Sintsova OV, Gladkikh IN, Leychenko EV. Natural Inhibitors of Mammalian α-Amylases as Promising Drugs for the Treatment of Metabolic Diseases. International Journal of Molecular Sciences. 2023; 24(22):16514. https://doi.org/10.3390/ijms242216514
Chicago/Turabian StyleKalinovskii, Aleksandr P., Oksana V. Sintsova, Irina N. Gladkikh, and Elena V. Leychenko. 2023. "Natural Inhibitors of Mammalian α-Amylases as Promising Drugs for the Treatment of Metabolic Diseases" International Journal of Molecular Sciences 24, no. 22: 16514. https://doi.org/10.3390/ijms242216514
APA StyleKalinovskii, A. P., Sintsova, O. V., Gladkikh, I. N., & Leychenko, E. V. (2023). Natural Inhibitors of Mammalian α-Amylases as Promising Drugs for the Treatment of Metabolic Diseases. International Journal of Molecular Sciences, 24(22), 16514. https://doi.org/10.3390/ijms242216514







