Adenoviral Gene Therapy Vectors in Clinical Use—Basic Aspects with a Special Reference to Replication-Competent Adenovirus Formation and Its Impact on Clinical Safety
Abstract
:1. Introduction
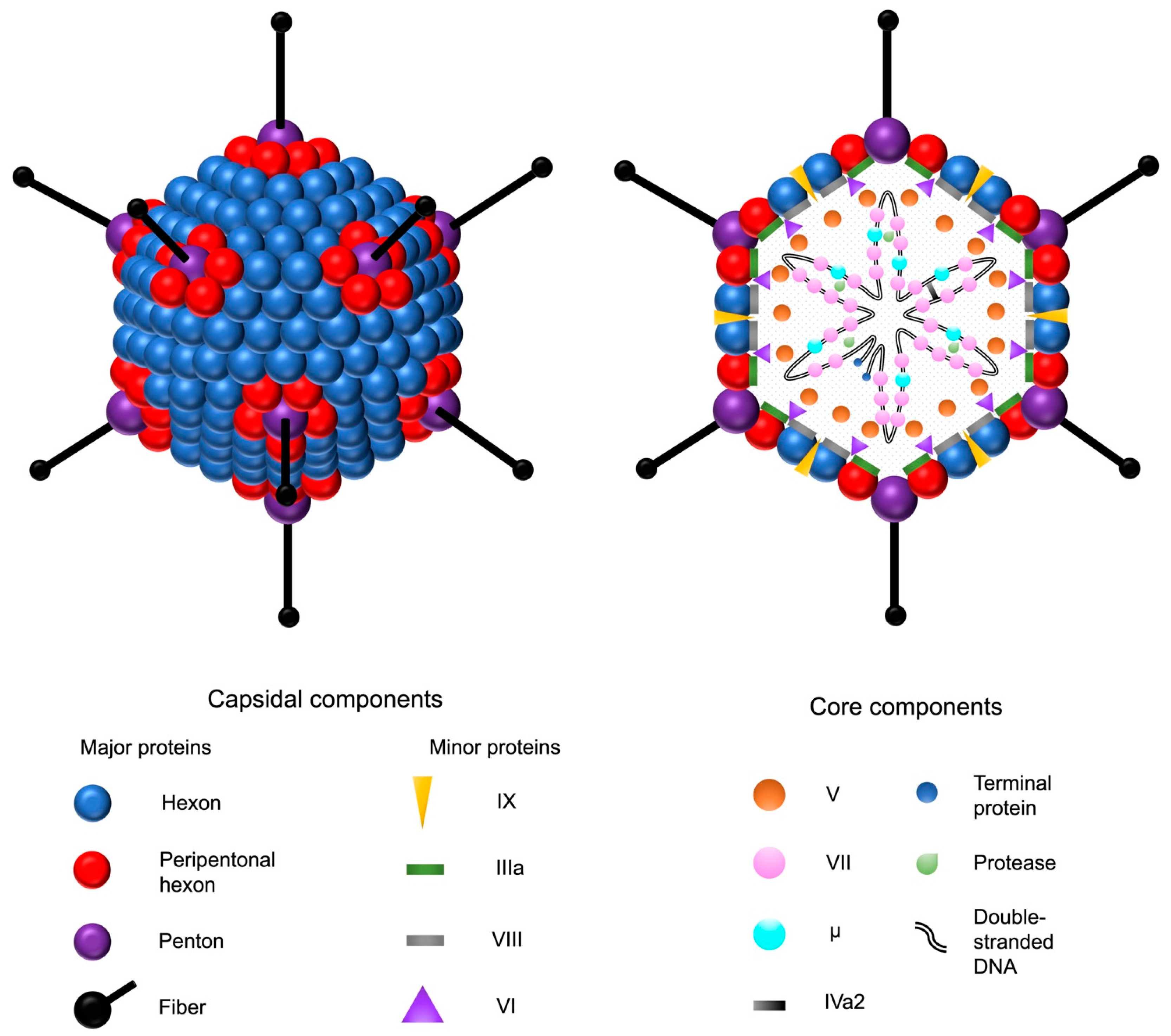
2. Adenoviruses
2.1. General Properties
2.2. Adenovirus Infection
2.3. Environmental Aspects
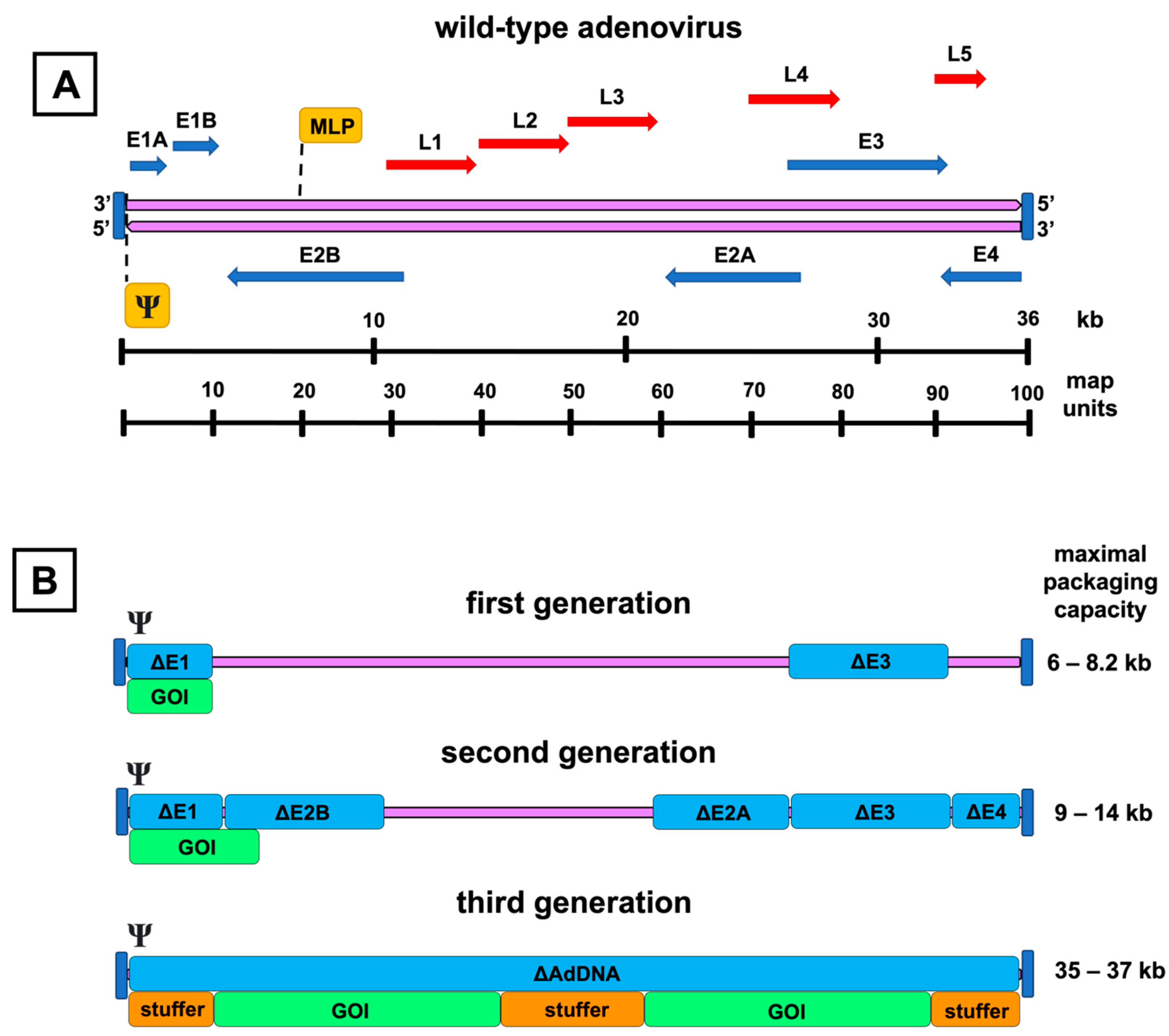
3. Adenovirus Vectors
3.1. First-Generation Adenovirus Vectors
3.2. Second-Generation Adenovirus Vectors
3.3. Third-Generation Adenovirus Vectors
4. Vector Manufacturing
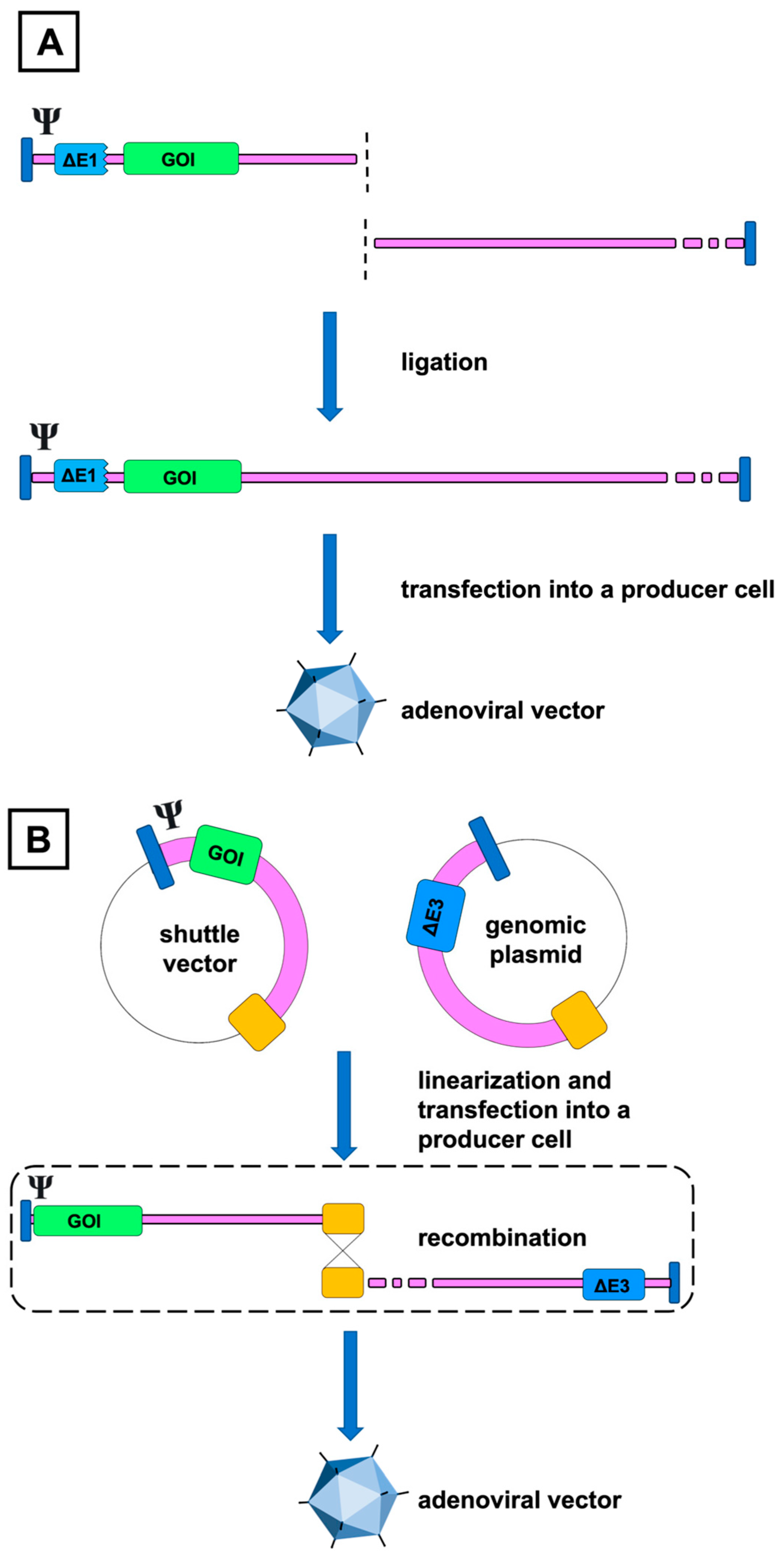
4.1. Construction Methods
4.1.1. In Vitro Ligation
4.1.2. Homologous Recombination
4.2. Vector Propagation
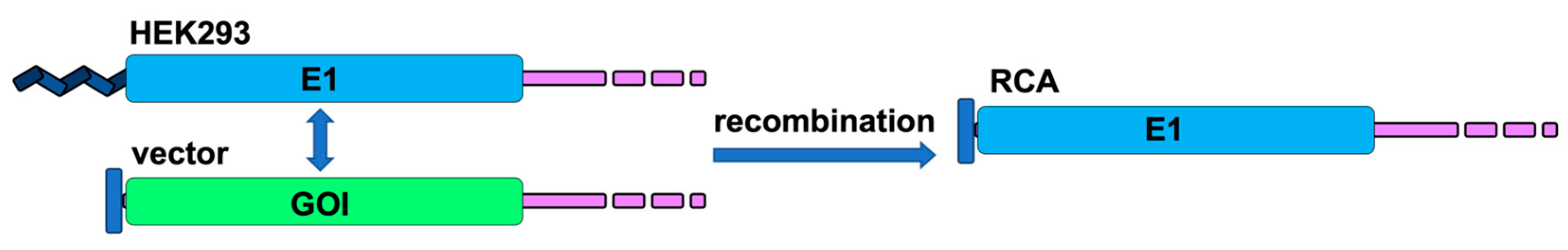
4.3. Formation of RCA
4.4. RCA Prevention
5. RCA and the Safety of Gene Therapy
5.1. Preclinical Data
5.2. RCA in Clinical Trials
5.3. Observations from Cardiovascular Studies
5.4. Safety Testing
5.5. Oncolytic Adenoviruses
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- High, K.A.; Roncarolo, M.G. Gene Therapy. N. Engl. J. Med. 2019, 381, 455–464. [Google Scholar] [CrossRef]
- Approved Cellular and Gene Therapy Products. Available online: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/approved-cellular-and-gene-therapy-products (accessed on 22 September 2022).
- Korpela, H.; Järveläinen, N.; Siimes, S.; Lampela, J.; Airaksinen, J.; Valli, K.; Turunen, M.; Pajula, J.; Nurro, J.; Ylä-Herttuala, S. Gene therapy for ischaemic heart disease and heart failure. J. Intern. Med. 2021, 290, 567–582. [Google Scholar] [CrossRef]
- Naldini, L. Ex vivo gene transfer and correction for cell based therapies. Nat. Rev. Genet. 2011, 12, 301–315. [Google Scholar] [CrossRef]
- Mendell, J.R.; Al-Zaidy, S.A.; Rodino-Klapac, L.R.; Goodspeed, K.; Gray, S.J.; Kay, C.N.; Boye, S.L.; Boye, S.E.; George, L.A.; Salabarria, S.; et al. Current Clinical Applications of In Vivo Gene Therapy with AAVs. Mol. Ther. 2021, 29, 464–488. [Google Scholar] [CrossRef]
- Gene Therapy Clinical Trials Worldwide—Provided by the Journal of Gene Medicine. Available online: https://a873679.fmphost.com/fmi/webd/GTCT (accessed on 22 September 2022).
- Aldhamen, Y.A.; Amalfitano, A. Methods to Mitigate Immune Responses to Adenoviral Vectors. In Adenoviral Vectors for Gene Therapy, 2nd ed.; Curiel, D.T., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 391–422. [Google Scholar]
- Mendonça, S.A.; Lorincz, R.; Boucher, P.; Curiel, D.T. Adenoviral vector vaccine platforms in the SARS-CoV-2 pandemic. NPJ Vaccines 2021, 6, 97. [Google Scholar] [CrossRef]
- Wold, W.S.; Toth, K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr. Gene Ther. 2013, 13, 421–433. [Google Scholar] [CrossRef]
- Raper, S.E.; Chirmule, N.; Lee, F.S.; Wivel, N.A.; Bagg, A.; Gao, G.P.; Wilson, J.M.; Batshaw, M.L. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 2003, 80, 148–158. [Google Scholar] [CrossRef]
- Rissanen, T.T.; Ylä-Herttuala, S. Current status of cardiovascular gene therapy. Mol. Ther. 2007, 15, 1233–1247. [Google Scholar] [CrossRef]
- Fallaux, F.J.; van der Eb, A.J.; Hoeben, R.C. Who’s afraid of replication-competent adenoviruses? Gene Ther. 1999, 6, 709–712. [Google Scholar] [CrossRef]
- Lochmüller, H.; Jani, A.; Huard, J.; Prescott, S.; Simoneau, M.; Massie, B.; Karpati, G.; Acsadi, G. Emergence of early region 1-containing replication-competent adenovirus in stocks of replication-defective adenovirus recombinants (ΔE1 + ΔE3) during multiple passages in 293 cells. Hum. Gene Ther. 1994, 5, 1485–1491. [Google Scholar] [CrossRef]
- Hermens, W.T.; Verhaagen, J. Adenoviral vector-mediated gene expression in the nervous system of immunocompetent Wistar and T cell-deficient nude rats: Preferential survival of transduced astroglial cells in nude rats. Hum. Gene Ther. 1997, 8, 1049–1063. [Google Scholar] [CrossRef]
- Imler, J.L.; Bout, A.; Dreyer, D.; Dieterlé, A.; Schultz, H.; Valerio, D.; Mehtali, M.; Pavirani, A. Trans-complementation of E1-deleted adenovirus: A new vector to reduce the possibility of codissemination of wild-type and recombinant adenoviruses. Hum. Gene Ther. 1995, 6, 711–721. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services Food and Drug Administration Center for Biologics Evaluation and Research. Guidance for Industry: Guidance for Human Somatic Cell Therapy and Gene Therapy. Available online: https://www.fda.gov/media/72402/download (accessed on 24 October 2022).
- U.S. Department of Health and Human Services Food and Drug Administration Center for Biologics Evaluation and Research. Guidance for Industry: Chemistry, Manufacturing, and Control (CMC) Information for Human Gene Therapy Investigational New Drug Applications (INDs). Available online: https://www.fda.gov/media/113760/download (accessed on 24 October 2022).
- Peltonen, H.; Mykkänen, M.; Hassinen, M. Gene Therapy Vector Contamination Assay. U.S. Patent 2019/0292609 A1, 26 September 2019. [Google Scholar]
- Berk, A.J. Adenoviridae: The viruses and their replication. In Fields Virology, 5th ed.; Fields, B.N., Knipe, D.M., Griffin, D.E., Eds.; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 2355–2394. [Google Scholar]
- Rhee, G.E.; Barouch, D.H. Adenoviruses. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 8th ed.; Bennett, J.E., Dolin, R., Blaser, J., Eds.; Elsevier Saunders: Philadelphia, PA, USA, 2015; pp. 1787–1793. [Google Scholar]
- Fox, J.P.; Brandt, C.D.; Wassermann, F.E.; Hall, C.E.; Spigland, I.; Kogon, A.; Elveback, L.R. The virus watch program: A continuing surveillance of viral infections in metropolitan New York families: Observations of adenovirus infections: Virus excretion patterns, antibody response, efficiency of surveillance, patterns of infection, and relation to illness. Am. J. Epidemiol. 1969, 89, 25–50. [Google Scholar] [CrossRef]
- Lessler, J.; Reich, N.G.; Brookmeyer, R.; Perl, T.M.; Nelson, K.E.; Cummings, D.A. Incubation periods of acute respiratory viral infections: A systematic review. Lancet Infect. Dis. 2009, 9, 291–300. [Google Scholar] [CrossRef]
- Mast, T.C.; Kierstead, L.; Gupta, S.B.; Nikas, A.A.; Kallas, E.G.; Novitsky, V.; Mbewe, B.; Pitisuttithum, P.; Schechter, M.; Vardas, E.; et al. International epidemiology of human pre-existing adenovirus (Ad) type-5, type-6, type-26 and type-36 neutralizing antibodies: Correlates of high Ad5 titers and implications for potential HIV vaccine trials. Vaccine 2010, 28, 950–957. [Google Scholar] [CrossRef]
- Lenaerts, L.; De Clercq, E.; Naesens, L. Clinical features and treatment of adenovirus infections. Rev. Med. Virol. 2008, 18, 357–374. [Google Scholar] [CrossRef]
- Subramanian, T.; Vijayalingam, S.; Chinnadurai, G. Genetic identification of adenovirus type 5 genes that influence viral spread. J. Virol. 2006, 80, 2000–2012. [Google Scholar] [CrossRef]
- Jogler, C.; Hoffmann, D.; Theegarten, D.; Grunwald, T.; Uberla, K.; Wildner, O. Replication properties of human adenovirus in vivo and in cultures of primary cells from different animal species. J. Virol. 2006, 80, 3549–3558. [Google Scholar] [CrossRef]
- Lichtenstein, D.L.; Wold, W.S. Experimental infections of humans with wild-type adenoviruses and with replication-competent adenovirus vectors: Replication, safety, and transmission. Cancer Gene Ther. 2004, 11, 819–829. [Google Scholar] [CrossRef]
- Harui, A.; Suzuki, S.; Kochanek, S.; Mitani, K. Frequency and stability of chromosomal integration of adenovirus vectors. J. Virol. 1999, 73, 6141–6146. [Google Scholar] [CrossRef]
- McAllister, R.M.; Nicolson, M.O.; Reed, G.; Kern, J.; Gilden, R.V.; Huebner, R.J. Transformation of rodent cells by adenovirus 19 and other group D adenoviruses. J. Natl. Cancer Inst. 1969, 43, 917–923. [Google Scholar] [PubMed]
- de Jong, P.J.; Valderrama, G.; Spigland, I.; Horwitz, M.S. Adenovirus isolates from urine of patients with acquired immunodeficiency syndrome. Lancet 1983, 1, 1293–1296. [Google Scholar] [CrossRef] [PubMed]
- Vora, G.J.; Lin, B.; Gratwick, K.; Meador, C.; Hansen, C.; Tibbetts, C.; Stenger, D.A.; Irvine, M.; Seto, D.; Purkayastha, A.; et al. Co-infections of adenovirus species in previously vaccinated patients. Emerg. Infect. Dis. 2006, 12, 921–930. [Google Scholar] [CrossRef]
- Gordon, Y.J.; Gordon, R.Y.; Romanowski, E.; Araullo-Cruz, T.P. Prolonged recovery of desiccated adenoviral serotypes 5, 8, and 19 from plastic and metal surfaces in vitro. Ophthalmology 1993, 100, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Rutala, W.A.; Peacock, J.E.; Gergen, M.F.; Sobsey, M.D.; Weber, D.J. Efficacy of hospital germicides against adenovirus 8, a common cause of epidemic keratoconjunctivitis in health care facilities. Antimicrob. Agents Chemother. 2006, 50, 1419–1424. [Google Scholar] [CrossRef]
- Gaydos, C.A.; Gaydos, J.C. Adenovirus vaccines in the U.S. military. Mil. Med. 1995, 160, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, M.J.; De Berry, P.; Sullivan, E.J.; Edwards, E.A.; Pierce, W.E.; Muldoon, R.L.; Jackson, G.G.; Peckinpaugh, R.O. Characteristics of vaccine-induced and natural infection with adenovirus type 4 in naval recruits. Am. J. Epidemiol. 1968, 88, 45–54. [Google Scholar] [CrossRef]
- Chinnadurai, G. Control of apoptosis by human adenovirus genes. Semin. Virol. 1998, 5, 399–408. [Google Scholar] [CrossRef]
- Steegenga, W.T.; Riteco, N.; Jochemsen, A.G.; Fallaux, F.J.; Bos, J.L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 1998, 16, 349–357. [Google Scholar] [CrossRef]
- Lichtenstein, D.L.; Toth, K.; Doronin, K.; Tollefson, A.E.; Wold, W.S.M. Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 2004, 23, 75–111. [Google Scholar] [CrossRef]
- Tollefson, A.E.; Scaria, A.; Hermiston, T.W.; Ryerse, J.S.; Wold, L.J.; Wold, W.S. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 1996, 70, 2296–2306. [Google Scholar] [CrossRef] [PubMed]
- Burgert, H.G.; Maryanski, J.L.; Kvist, S. “E3/19K” protein of adenovirus type 2 inhibits lysis of cytolytic T lymphocytes by blocking cell-surface expression of histocompatibility class I antigens. Proc. Natl. Acad. Sci. USA 1987, 84, 1356–1360. [Google Scholar] [CrossRef] [PubMed]
- Gooding, L.R.; Ranheim, T.S.; Tollefson, A.E.; Aquino, L.; Duerksen-Hughes, P.; Horton, T.M.; Wold, W.S. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus function together to protect many but not all mouse cell lines against lysis by tumor necrosis factor. J. Virol. 1991, 65, 4114–4123. [Google Scholar] [CrossRef] [PubMed]
- Benedict, C.A.; Norris, P.S.; Prigozy, T.I.; Bodmer, J.L.; Mahr, J.A.; Garnett, C.T.; Martinon, F.; Tschopp, J.; Gooding, L.R.; Ware, C.F. Three adenovirus E3 proteins cooperate to evade apoptosis by tumor necrosis factor-related apoptosis-inducing ligand receptor-1 and -2. J. Biol. Chem. 2001, 276, 3270–3278. [Google Scholar] [CrossRef]
- Chen, P.; Tian, J.; Kovesdi, I.; Bruder, J.T. Interaction of the adenovirus 14.7kDa protein with FLICE inhibits Fas ligand-induced apoptosis. J. Biol. Chem. 1998, 273, 5815–5820. [Google Scholar] [CrossRef]
- Krajcsi, P.; Dimitrov, T.; Hermiston, T.W.; Tollefson, A.E.; Ranheim, T.S.; Vande Pol, S.B.; Stephenson, A.H.; Wold, W.S. The adenovirus E3-14.7K protein and the E3-10.4K/14.5K complex of proteins, which independently inhibit tumor necrosis factor (TNF)-induced apoptosis, also independently inhibit TNF-induced release of arachidonic acid. J. Virol. 1996, 70, 4904–4913. [Google Scholar] [CrossRef]
- Swaminathan, S.; Thimmapaya, B. Regulation of adenovirus E2 transcription unit. Curr. Top. Microbiol. Immunol. 1995, 199, 177–194. [Google Scholar] [CrossRef]
- Huang, M.M.; Hearing, P. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 1989, 63, 2605. [Google Scholar] [CrossRef]
- Bridge, E.; Ketner, G. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 1989, 63, 631–638. [Google Scholar] [CrossRef]
- Lusky, M.; Christ, M.; Rittner, K.; Dieterle, A.; Dreyer, D.; Mourot, B.; Schultz, H.; Stoeckel, F.; Pavirani, A.; Mehtali, M. In vitro and in vivo biology of recombinant adenovirus vectors with E1, E1/E2A, or E1/E4 deleted. J. Virol. 1998, 72, 2022–2032. [Google Scholar] [CrossRef]
- O’Neal, W.K.; Zhou, H.; Morral, N.; Aguilar-Cordova, E.; Pestaner, J.; Langston, C.; Mull, B.; Wang, Y.; Beaudet, A.L.; Lee, B. Toxicological comparison of E2a-deleted and first-generation adenoviral vectors expressing alpha1-antitrypsin after systemic delivery. Hum. Gene. Ther. 1998, 9, 1587–1598. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Schneider, D.B.; Driscoll, R.M.; Vassalli, G.; Sassani, A.B.; Dichek, D.A. Second-generation adenoviral vectors do not prevent rapid loss of transgene expression and vector DNA from the arterial wall. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- Jornot, L.; Petersen, H.; Lusky, M.; Pavirani, A.; Moix, I.; Morris, M.A.; Rochat, T. Effects of first generation E1E3-deleted and second generation E1E3E4-deleted/modified adenovirus vectors on human endothelial cell death. Endothelium 2001, 8, 167–179. [Google Scholar] [CrossRef]
- Kovesdi, I.; Hedley, S.J. Adenoviral producer cells. Viruses 2010, 2, 1681–1703. [Google Scholar] [CrossRef]
- Guimet, A.; Hearin, P. Adenovirus Replication. In Adenoviral Vectors for Gene Therapy, 2nd ed.; Curiel, D.T., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 59–84. [Google Scholar]
- Segura, M.M.; Alba, R.; Bosch, A.; Chillón, M. Advances in helper-dependent adenoviral vector research. Curr. Gene Ther. 2008, 8, 222–235. [Google Scholar] [CrossRef]
- Fleury, S.; Driscoll, R.; Simeoni, E.; Dudler, J.; von Segesser, L.K.; Kappenberger, L.; Vassalli, G. Helper-dependent adenovirus vectors devoid of all viral genes cause less myocardial inflammation compared with first-generation adenovirus vectors. Basic Res. Cardiol. 2004, 99, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Brunetti-Pierri, N.; Ng, T.; Iannitti, D.; Cioffi, W.; Stapleton, G.; Law, M.; Breinholt, J.; Palmer, D.; Grove, N.; Rice, K.; et al. Transgene expression up to 7 years in nonhuman primates following hepatic transduction with helper-dependent adenoviral vectors. Hum. Gene. Ther. 2013, 24, 761–765. [Google Scholar] [CrossRef]
- Rastall, D.P.; Seregin, S.S.; Aldhamen, Y.A.; Kaiser, L.M.; Mullins, C.; Liou, A.; Ing, F.; Pereria-Hicks, C.; Godbehere-Roosa, S.; Palmer, D.; et al. Long-term, high-level hepatic secretion of acid α-glucosidase for Pompe disease achieved in non-human primates using helper-dependent adenovirus. Gene Ther. 2016, 23, 743–752. [Google Scholar] [CrossRef]
- Stow, N.D. Cloning of a DNA fragment from the left-hand terminus of the adenovirus type 2 genome and its use in site-directed mutagenesis. J. Virol. 1981, 37, 171–180. [Google Scholar] [CrossRef]
- Mizuguchi, H.; Kay, M.A.; Hayakawa, T. Approaches for generating recombinant adenovirus vectors. Adv. Drug Deliv. Rev. 2001, 52, 165–176. [Google Scholar] [CrossRef]
- Shyambabu, C.; Hitt, M.M. Adenoviral Vector Construction I: Mammalian Systems. In Adenoviral Vectors for Gene Therapy, 2nd ed.; Curiel, D.T., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 85–112. [Google Scholar]
- Mizuguchi, H.; Kay, M.A. Efficient construction of a recombinant adenovirus vector by an improved in vitro ligation method. Hum. Gene Ther. 1998, 9, 2577–2583. [Google Scholar] [CrossRef] [PubMed]
- Souza, D.W.; Armentano, D. Novel cloning method for recombinant adenovirus construction in Escherichia coli. Biotechniques 1999, 26, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Danthinne, X.; Werth, E. New tools for the generation of E1- and/or E3-substituted adenoviral vectors. Gene Ther. 2000, 7, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Haviv, Y.S. A simplified in vitro ligation approach to clone an E1B55k-deleted double-targeted conditionally-replicative adenovirus. Virol. J. 2009, 6, 18. [Google Scholar] [CrossRef]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977, 36, 59–74. [Google Scholar] [CrossRef]
- Bett, A.J.; Haddara, W.; Prevec, L.; Graham, F.L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc. Natl. Acad. Sci. USA 1994, 91, 8802–8806. [Google Scholar] [CrossRef]
- McGrory, W.J.; Bautista, D.S.; Graham, F.L. A simple technique for the rescue of early region I mutations into infectious human adenovirus type 5. Virology 1988, 163, 614–617. [Google Scholar] [CrossRef] [PubMed]
- Chartier, C.; Degryse, E.; Gantzer, M.; Dieterle, A.; Pavirani, A.; Mehtali, M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 1996, 70, 4805–4810. [Google Scholar] [CrossRef]
- Luo, J.; Deng, Z.L.; Luo, X.; Tang, N.; Song, W.X.; Chen, J.; Sharff, K.A.; Luu, H.H.; Haydon, R.C.; Kinzler, K.W.; et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2007, 2, 1236–1247. [Google Scholar] [CrossRef]
- Ketner, G.; Spencer, F.; Tugendreich, S.; Connelly, C.; Hieter, P. Efficient manipulation of the human adenovirus genome as an infectious yeast artificial chromosome clone. Proc. Natl. Acad. Sci. USA 1994, 91, 6186–6190. [Google Scholar] [CrossRef]
- Hokanson, C.A.; Dora, E.; Donahue, B.A.; Rivkin, M.; Finer, M.; Mendez, M.J. Hybrid yeast-bacteria cloning system used to capture and modify adenoviral and nonviral genomes. Hum. Gene Ther. 2003, 14, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.; Parks, R.J.; Cummings, D.T.; Evelegh, C.M.; Sankar, U.; Graham, F.L. A high-efficiency Cre/loxP-based system for construction of adenoviral vectors. Hum. Gene Ther. 1999, 10, 2667–2672. [Google Scholar] [CrossRef] [PubMed]
- Parks, R.J.; Chen, L.; Anton, M.; Sankar, U.; Rudnicki, M.A.; Graham, F.L. A helper-dependent adenovirus vector system: Removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. USA 1996, 93, 13565–13570. [Google Scholar] [CrossRef] [PubMed]
- Alba, R.; Bosch, A.; Chillon, M. Gutless adenovirus: Last-generation adenovirus for gene therapy. Gene Ther. 2005, 12 (Suppl. 1), S18–S27. [Google Scholar] [CrossRef]
- Tollefson, A.E.; Kuppuswamy, M.; Shashkova, E.V.; Doronin, K.; Wold, W.S. Preparation and titration of CsCl-banded adenovirus stocks. Methods Mol. Med. 2007, 130, 223–235. [Google Scholar] [CrossRef]
- Vellinga, J.; Smith, J.P.; Lipiec, A.; Majhen, D.; Lemckert, A.; van Ooij, M.; Ives, P.; Yallop, C.; Custers, J.; Havenga, M. Challenges in manufacturing adenoviral vectors for global vaccine product deployment. Hum. Gene Ther. 2014, 25, 318–327. [Google Scholar] [CrossRef]
- Lesch, H.P.; Heikkilä, K.M.; Lipponen, E.M.; Valonen, P.; Müller, A.; Räsänen, E.; Tuunanen, T.; Hassinen, M.M.; Parker, N.; Karhinen, M.; et al. Process Development of Adenoviral Vector Production in Fixed Bed Bioreactor: From Bench to Commercial Scale. Hum. Gene Ther. 2015, 26, 560–571. [Google Scholar] [CrossRef]
- Leinonen, H.M.; Lepola, S.; Lipponen, E.M.; Heikura, T.; Koponen, T.; Parker, N.; Ylä-Herttuala, S.; Lesch, H.P. Benchmarking of Scale-X Bioreactor System in Lentiviral and Adenoviral Vector Production. Hum. Gene Ther. 2020, 31, 376–384. [Google Scholar] [CrossRef]
- Hehir, K.M.; Armentano, D.; Cardoza, L.M.; Choquette, T.L.; Berthelette, P.B.; White, G.A.; Couture, L.A.; Everton, M.B.; Keegan, J.; Martin, J.M.; et al. Molecular characterization of replication-competent variants of adenovirus vectors and genome modifications to prevent their occurrence. J. Virol. 1996, 70, 8459–8467. [Google Scholar] [CrossRef]
- Fallaux, F.J.; Bout, A.; van der Velde, I.; van den Wollenberg, D.J.; Hehir, K.M.; Keegan, J.; Auger, C.; Cramer, S.J.; van Ormondt, H.; van der Eb, A.J.; et al. New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther. 1998, 9, 1909–1917. [Google Scholar] [CrossRef]
- Zhu, J.; Grace, M.; Casale, J.; Chang, A.T.; Musco, M.L.; Bordens, R.; Greenberg, R.; Schaefer, E.; Indelicato, S.R. Characterization of replication-competent adenovirus isolates from large-scale production of a recombinant adenoviral vector. Hum. Gene Ther. 1999, 10, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, K.T.; Curiel, D.T.; Engler, J.A.; Garver, R.I., Jr. Trans complementation of an E1A-deleted adenovirus with codelivered E1A sequences to make recombinant adenoviral producer cells. Hum. Gene Ther. 1994, 5, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Roitsch, C.; Achstetter, T.; Benchaibi, M.; Bonfils, E.; Cauet, G.; Gloeckler, R.; L’hôte, H.; Keppi, E.; Nguyen, M.; Spehner, D.; et al. Characterization and quality control of recombinant adenovirus vectors for gene therapy. J. Chromatogr. B. Biomed. Sci. Appl. 2001, 752, 263–280. [Google Scholar] [CrossRef]
- Schalk, J.A.; de Vries, C.G.; Orzechowski, T.J.; Rots, M.G. A rapid and sensitive assay for detection of replication-competent adenoviruses by a combination of microcarrier cell culture and quantitative PCR. J. Virol. Methods 2007, 145, 89–95. [Google Scholar] [CrossRef]
- Ishii-Watabe, A.; Uchida, E.; Iwata, A.; Nagata, R.; Satoh, K.; Fan, K.; Murata, M.; Mizuguchi, H.; Kawasaki, N.; Kawanishi, T.; et al. Detection of replication-competent adenoviruses spiked into recombinant adenovirus vector products by infectivity PCR. Mol. Ther. 2003, 8, 1009–1016. [Google Scholar] [CrossRef]
- Gao, M.; Yngve, E.; Yu, D.; Jin, C. A qPCR-Based Method for Quantification of RCA Contaminants in Oncolytic Adenovirus Products. Front. Mol. Biosci. 2022, 9, 883249. [Google Scholar] [CrossRef] [PubMed]
- Murakami, P.; Pungor, E.; Files, J.; Do, L.; van Rijnsoever, R.; Vogels, R.; Bout, A.; McCaman, M. A single short stretch of homology between adenoviral vector and packaging cell line can give rise to cytopathic effect-inducing, helper-dependent E1-positive particles. Hum. Gene Ther. 2002, 13, 909–920. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, S.H.; Cho, Y.S.; Park, K.; Kim, Y.H.; Lee, J.H. Development of a packaging cell line for propagation of replication-deficient adenovirus vector. Exp. Mol. Med. 2001, 33, 145–149. [Google Scholar] [CrossRef]
- Schiedner, G.; Hertel, S.; Kochanek, S. Efficient transformation of primary human amniocytes by E1 functions of Ad5: Generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 2000, 11, 2105–2116. [Google Scholar] [CrossRef]
- Xu, Q.; Arevalo, M.T.; Pichichero, M.E.; Zeng, M. A new complementing cell line for replication-incompetent E1-deleted adenovirus propagation. Cytotechnology 2006, 51, 133–140. [Google Scholar] [CrossRef]
- Sakhuja, K.; Reddy, P.S.; Ganesh, S.; Cantaniag, F.; Pattison, S.; Limbach, P.; Kayda, D.B.; Kadan, M.J.; Kaleko, M.; Connelly, S. Optimization of the generation and propagation of gutless adenoviral vectors. Hum. Gene Ther. 2003, 14, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Vellinga, J.; Uil, T.G.; de Vrij, J.; Rabelink, M.J.; Lindholm, L.; Hoeben, R.C. A system for efficient generation of adenovirus protein IX-producing helper cell lines. J. Gene Med. 2006, 8, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Parks, R.J. Adenovirus protein IX: A new look at an old protein. Mol. Ther. 2005, 11, 19–25. [Google Scholar] [CrossRef]
- Krougliak, V.; Graham, F.L. Development of cell lines capable of complementing E1, E4, and protein IX defective adenovirus type 5 mutants. Hum. Gene Ther. 1995, 6, 1575–1586. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Finer, M.H. Second-generation adenovirus vectors. Nat. Med. 1996, 2, 714–716. [Google Scholar] [CrossRef]
- Lee, D.; Liu, J.; Junn, H.J.; Lee, E.J.; Jeong, K.S.; Seol, D.W. No more helper adenovirus: Production of gutless adenovirus (GLAd) free of adenovirus and replication-competent adenovirus (RCA) contaminants. Exp. Mol. Med. 2019, 51, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Fechner, H.; Wang, X.; Wang, H.; Jansen, A.; Pauschinger, M.; Scherübl, H.; Bergelson, J.M.; Schultheiss, H.P.; Poller, W. Trans-complementation of vector replication versus Coxsackie-adenovirus-receptor overexpression to improve transgene expression in poorly permissive cancer cells. Gene Ther. 2000, 7, 1954–1968. [Google Scholar] [CrossRef]
- Speiseder, T.; Hofmann-Sieber, H.; Rodríguez, E.; Schellenberg, A.; Akyüz, N.; Dierlamm, J.; Spruss, T.; Lange, C.; Dobner, T. Efficient Transformation of Primary Human Mesenchymal Stromal Cells by Adenovirus Early Region 1 Oncogenes. J. Virol. 2016, 91, e01782-16. [Google Scholar] [CrossRef]
- Ginsberg, H.S.; Lundholm-Beauchamp, U.; Horswood, R.L.; Pernis, B.; Wold, W.S.; Chanock, R.M.; Prince, G.A. Role of early region 3 (E3) in pathogenesis of adenovirus disease. Proc. Natl. Acad. Sci. USA 1989, 86, 3823–3827. [Google Scholar] [CrossRef]
- Hartikainen, J.; Hassinen, I.; Hedman, A.; Kivelä, A.; Saraste, A.; Knuuti, J.; Husso, M.; Mussalo, H.; Hedman, M.; Rissanen, T.T.; et al. Adenoviral intramyocardial VEGF-DΔNΔC gene transfer increases myocardial perfusion reserve in refractory angina patients: A phase I/IIa study with 1-year follow-up. Eur. Heart J. 2017, 38, 2547–2555, Erratum in: Eur. Heart J. 2018, 39, 1652. [Google Scholar] [CrossRef]
- Leikas, A.J.; Laham-Karam, N.; Agtereek, E.; Peltonen, H.M.; Selander, T.; Korpisalo, P.; Holappa, L.; Hartikainen, J.E.K.; Heikura, T.; Ylä-Herttuala, S. Efficacy and Safety of Clinical-Grade Human Vascular Endothelial Growth Factor-DΔNΔC Gene Therapy Containing Residual Replication-Competent Adenoviruses. Hum. Gene Ther. 2021, 32, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Stacker, S.A.; Stenvers, K.; Caesar, C.; Vitali, A.; Domagala, T.; Nice, E.; Roufail, S.; Simpson, R.J.; Moritz, R.; Karpanen, T.; et al. Biosynthesis of vascular endothelial growth factor-D involves proteolytic processing which generates non-covalent homodimers. J. Biol. Chem. 1999, 274, 32127–32136. [Google Scholar] [CrossRef] [PubMed]
- Ylä-Herttuala, S.; Rissanen, T.T.; Vajanto, I.; Hartikainen, J. Vascular endothelial growth factors: Biology and current status of clinical applications in cardiovascular medicine. J. Am. Coll. Cardiol. 2007, 49, 1015–1026. [Google Scholar] [CrossRef]
- Rutanen, J.; Rissanen, T.T.; Markkanen, J.E.; Gruchala, M.; Silvennoinen, P.; Kivelä, A.; Hedman, A.; Hedman, M.; Heikura, T.; Ordén, M.R.; et al. Adenoviral catheter-mediated intramyocardial gene transfer using the mature form of vascular endothelial growth factor-D induces transmural angiogenesis in porcine heart. Circulation 2004, 109, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Nurro, J.; Halonen, P.J.; Kuivanen, A.; Tarkia, M.; Saraste, A.; Honkonen, K.; Lähteenvuo, J.; Rissanen, T.T.; Knuuti, J.; Ylä-Herttuala, S. AdVEGF-B186 and AdVEGF-DΔNΔC induce angiogenesis and increase perfusion in porcine myocardium. Heart 2016, 102, 1716–1720. [Google Scholar] [CrossRef] [PubMed]
- Pajula, J.; Lähteenvuo, J.; Lähteenvuo, M.; Honkonen, K.; Halonen, P.; Hätinen, O.P.; Kuivanen, A.; Heikkilä, M.; Nurro, J.; Hartikainen, J.; et al. Adenoviral VEGF-DΔN ΔC gene therapy for myocardial ischemia. Front. Bioeng. Biotechnol. 2022, 10, 999226. [Google Scholar] [CrossRef]
- Rissanen, T.T.; Markkanen, J.E.; Gruchala, M.; Heikura, T.; Puranen, A.; Kettunen, M.I.; Kholová, I.; Kauppinen, R.A.; Achen, M.G.; Stacker, S.A.; et al. VEGF-D is the strongest angiogenic and lymphangiogenic effector among VEGFs delivered into skeletal muscle via adenoviruses. Circ. Res. 2003, 92, 1098–1106. [Google Scholar] [CrossRef]
- Leikas, A.J.; Hassinen, I.; Hedman, A.; Kivelä, A.; Ylä-Herttuala, S.; Hartikainen, J.E.K. Long-term safety and efficacy of intramyocardial adenovirus-mediated VEGF-DΔNΔC gene therapy eight-year follow-up of phase I KAT301 study. Gene Ther. 2022, 29, 289–293. [Google Scholar] [CrossRef]
- Leikas, A.J.; Hassinen, I.; Kivelä, A.; Hedman, A.; Mussalo, H.; Ylä-Herttuala, S.; Hartikainen, J.E.K. Intramyocardial adenoviral vascular endothelial growth factor-D∆N∆C gene therapy does not induce ventricular arrhythmias. J. Gene Med. 2022, 24, e3437. [Google Scholar] [CrossRef]
- Mäkinen, K.; Manninen, H.; Hedman, M.; Matsi, P.; Mussalo, H.; Alhava, E.; Ylä-Herttuala, S. Increased vascularity detected by digital subtraction angiography after VEGF gene transfer to human lower limb artery: A randomized, placebo-controlled, double-blinded phase II study. Mol. Ther. 2002, 6, 127–133. [Google Scholar] [CrossRef]
- Hedman, M.; Hartikainen, J.; Syvänne, M.; Stjernvall, J.; Hedman, A.; Kivelä, A.; Vanninen, E.; Mussalo, H.; Kauppila, E.; Simula, S.; et al. Safety and feasibility of catheter-based local intracoronary vascular endothelial growth factor gene transfer in the prevention of postangioplasty and in-stent restenosis and in the treatment of chronic myocardial ischemia: Phase II results of the Kuopio Angiogenesis Trial (KAT). Circulation 2003, 107, 2677–2683. [Google Scholar] [CrossRef] [PubMed]
- Hedman, M.; Muona, K.; Hedman, A.; Kivelä, A.; Syvänne, M.; Eränen, J.; Rantala, A.; Stjernvall, J.; Nieminen, M.S.; Hartikainen, J.; et al. Eight-year safety follow-up of coronary artery disease patients after local intracoronary VEGF gene transfer. Gene Ther. 2009, 16, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Wirth, T.; Hedman, M.; Mäkinen, K.; Manninen, H.; Immonen, A.; Vapalahti, M.; Ylä-Herttuala, S. Safety profile of plasmid/liposomes and virus vectors in clinical gene therapy. Curr. Drug. Saf. 2006, 1, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Westphal, M.; Ylä-Herttuala, S.; Martin, J.; Warnke, P.; Menei, P.; Eckland, D.; Kinley, J.; Kay, R.; Ram, Z.; ASPECT Study Group. Adenovirus-mediated gene therapy with sitimagene ceradenovec followed by intravenous ganciclovir for patients with operable high-grade glioma (ASPECT): A randomised, open-label, phase 3 trial. Lancet Oncol. 2013, 14, 823–833. [Google Scholar] [CrossRef]
- Heise, C.; Sampson-Johannes, A.; Williams, A.; McCormick, F.; Von Hoff, D.D.; Kirn, D.H. ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat. Med. 1997, 3, 639–645. [Google Scholar] [CrossRef]
- Edwards, S.J.; Dix, B.R.; Myers, C.J.; Dobson-Le, D.; Huschtscha, L.; Hibma, M.; Royds, J.; Braithwaite, A.W. Evidence that replication of the antitumor adenovirus ONYX-015 is not controlled by the p53 and p14(ARF) tumor suppressor genes. J. Virol. 2002, 76, 12483–12490. [Google Scholar] [CrossRef]
- Yamamoto, M.; Curiel, D.T. Current issues and future directions of oncolytic adenoviruses. Mol. Ther. 2010, 18, 243–250. [Google Scholar] [CrossRef]
- Schenk-Braat, E.A.; van Mierlo, M.M.; Wagemaker, G.; Bangma, C.H.; Kaptein, L.C. An inventory of shedding data from clinical gene therapy trials. J. Gene Med. 2007, 9, 910–921. [Google Scholar] [CrossRef]
- Huebner, R.J.; Rowe, W.P.; Schatten, W.E.; Smith, R.R.; Thomas, L.B. Studies on the use of viruses in the treatment of carcinoma of the cervix. Cancer 1956, 9, 1211–1218. [Google Scholar] [CrossRef]

| Animal Model | Female NZW Rabbit; a Dose of 1 × 1011 vp in Ten 100 μL Injections in the Non-Ischemic Hindlimb | ||
|---|---|---|---|
| Group and administered preparation | AdVEGF-D low RCA | AdVEGF-D moderate RCA | AdVEGF-D high RCA |
| Sample size (n) | 5 | 5 | 6 |
| RCA level (per 3 × 1010 tested vp) | <10 | 10–100 | 100–200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leikas, A.J.; Ylä-Herttuala, S.; Hartikainen, J.E.K. Adenoviral Gene Therapy Vectors in Clinical Use—Basic Aspects with a Special Reference to Replication-Competent Adenovirus Formation and Its Impact on Clinical Safety. Int. J. Mol. Sci. 2023, 24, 16519. https://doi.org/10.3390/ijms242216519
Leikas AJ, Ylä-Herttuala S, Hartikainen JEK. Adenoviral Gene Therapy Vectors in Clinical Use—Basic Aspects with a Special Reference to Replication-Competent Adenovirus Formation and Its Impact on Clinical Safety. International Journal of Molecular Sciences. 2023; 24(22):16519. https://doi.org/10.3390/ijms242216519
Chicago/Turabian StyleLeikas, Aleksi J., Seppo Ylä-Herttuala, and Juha E. K. Hartikainen. 2023. "Adenoviral Gene Therapy Vectors in Clinical Use—Basic Aspects with a Special Reference to Replication-Competent Adenovirus Formation and Its Impact on Clinical Safety" International Journal of Molecular Sciences 24, no. 22: 16519. https://doi.org/10.3390/ijms242216519
APA StyleLeikas, A. J., Ylä-Herttuala, S., & Hartikainen, J. E. K. (2023). Adenoviral Gene Therapy Vectors in Clinical Use—Basic Aspects with a Special Reference to Replication-Competent Adenovirus Formation and Its Impact on Clinical Safety. International Journal of Molecular Sciences, 24(22), 16519. https://doi.org/10.3390/ijms242216519







