Nervous Necrosis Virus Modulation of European Sea Bass (Dicentrarchus labrax, L.) Immune Genes and Transcriptome towards Establishment of Virus Carrier State
Abstract
:1. Introduction
2. Results
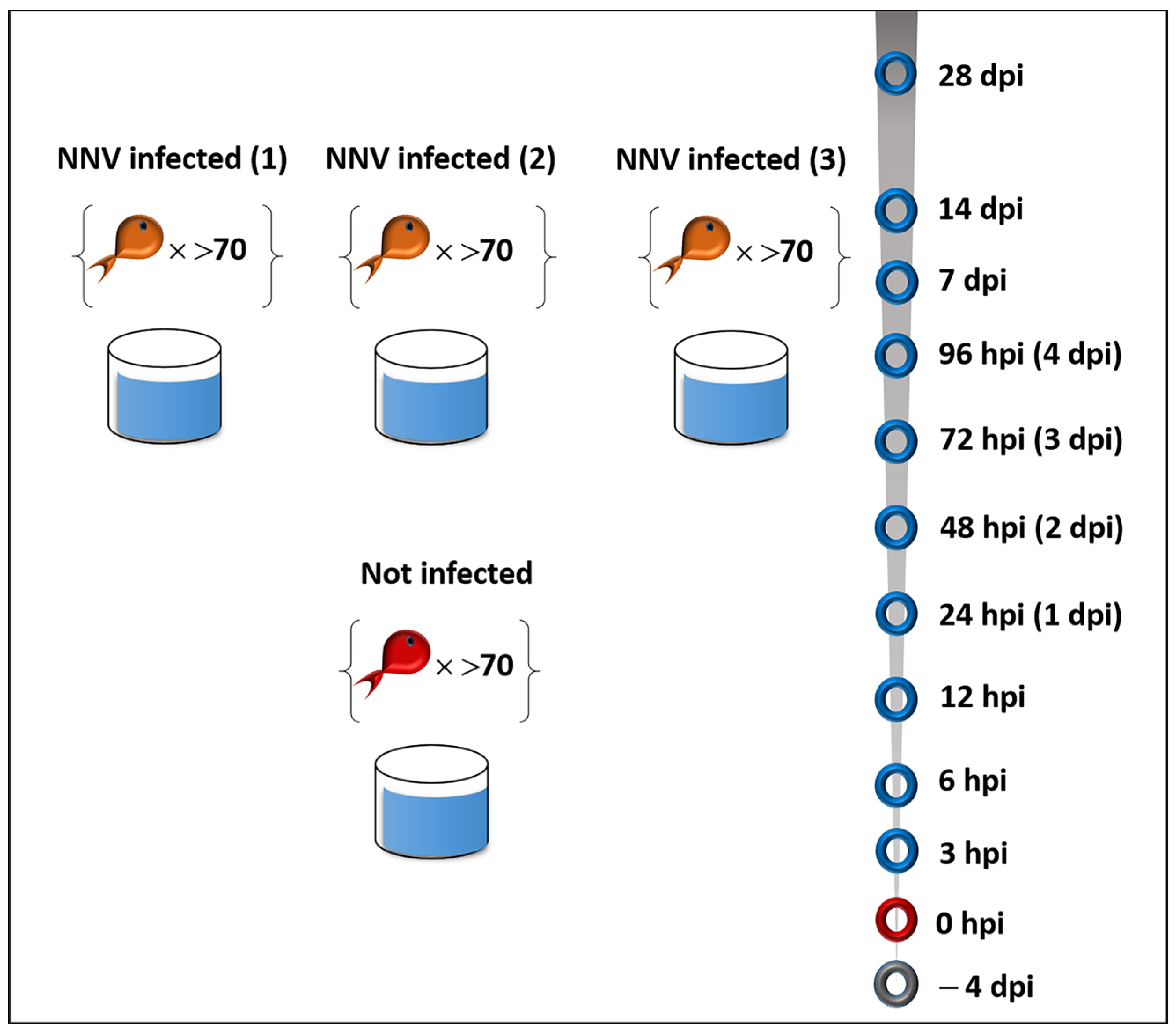
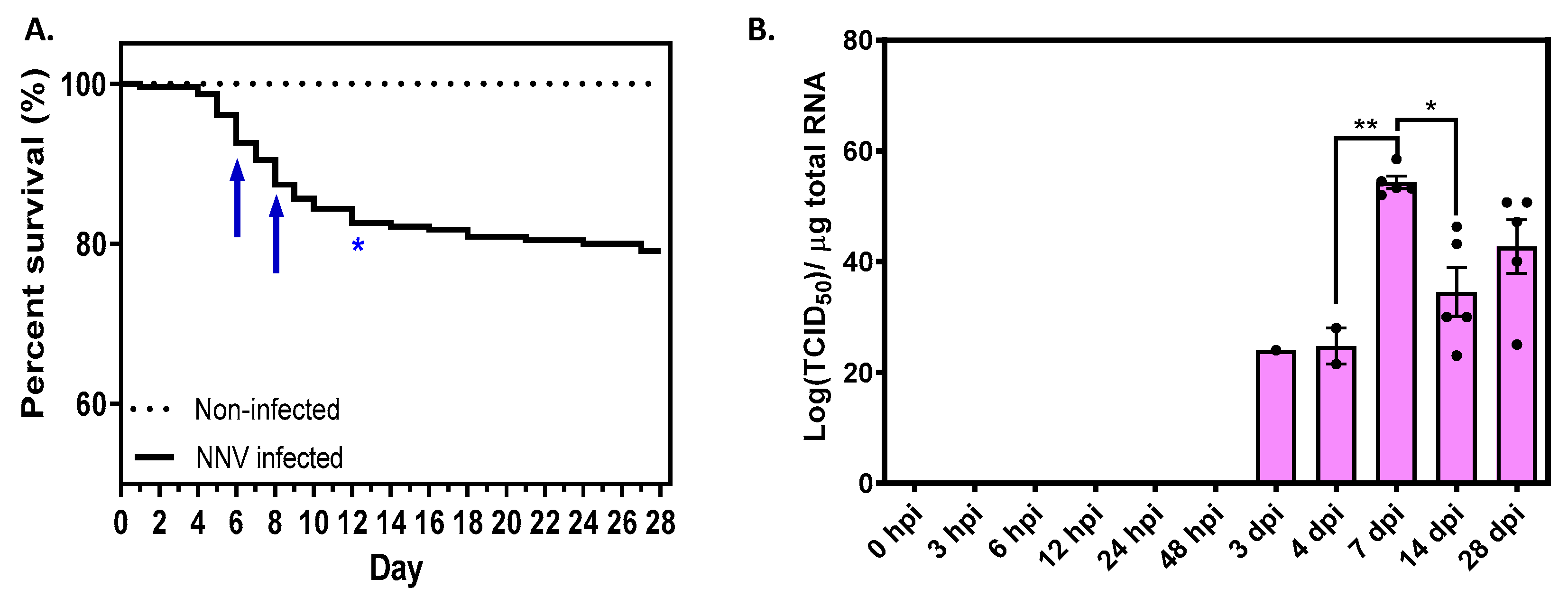
2.1. Cumulative Survival of D. labrax Challenged with NNV Reached 79% with Mortality Peak on Days 5–6
2.2. NNV Viral Load Remained High until the End of Experimentation
2.3. Biometrical Data Were Not Significantly Affected by NNV Infection
2.4. Haematological Parameters Were Significantly Affected by NNV Infection
2.5. Gene Expression of D. labrax Challenged with NNV
2.5.1. Interferon Pathway Related Genes Were Differentiated at Early Time Points and 14 dpi
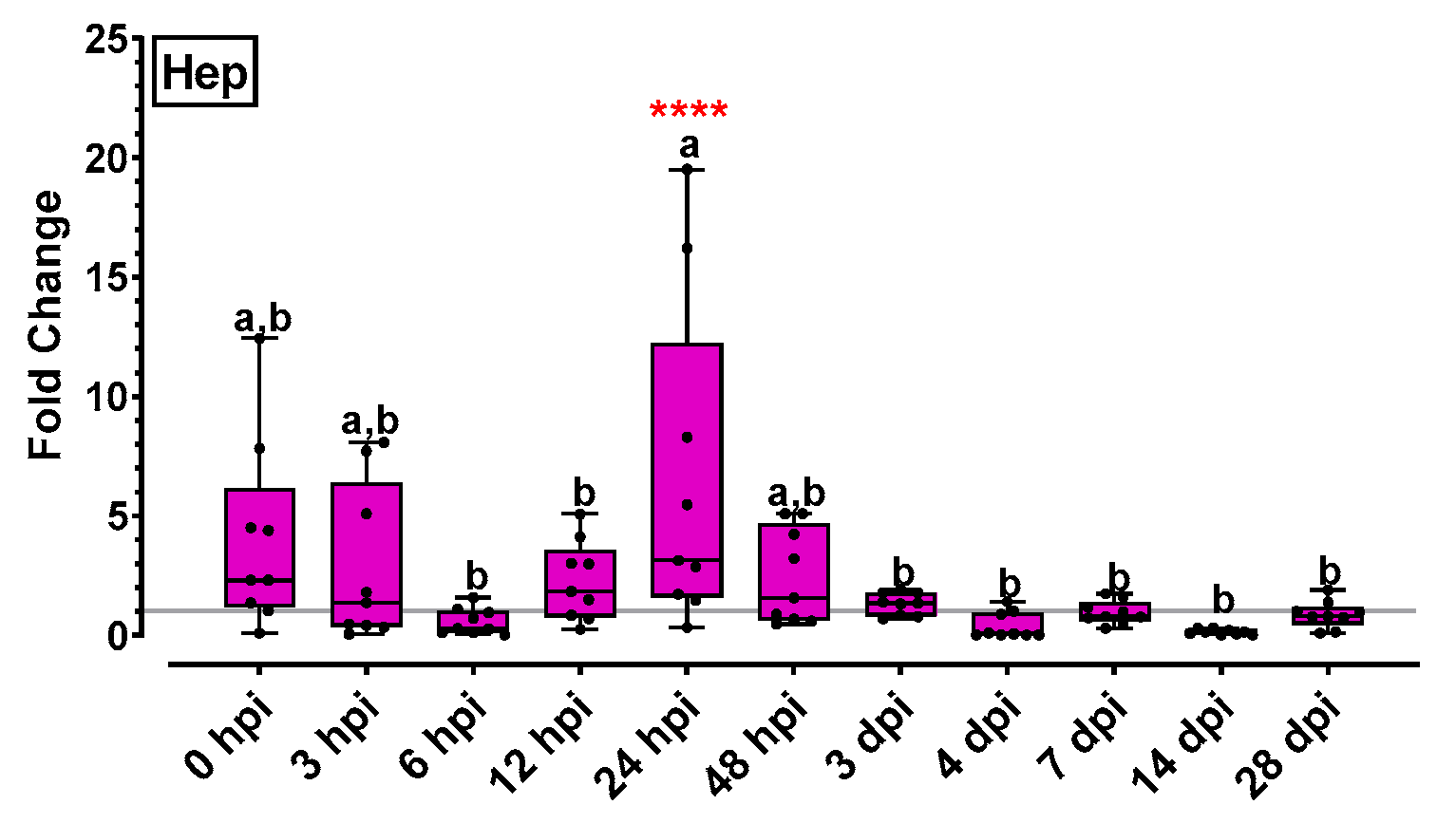
2.5.2. Cytokine Gene Expression Was Affected by NNV Infection Mostly in Late Time Points
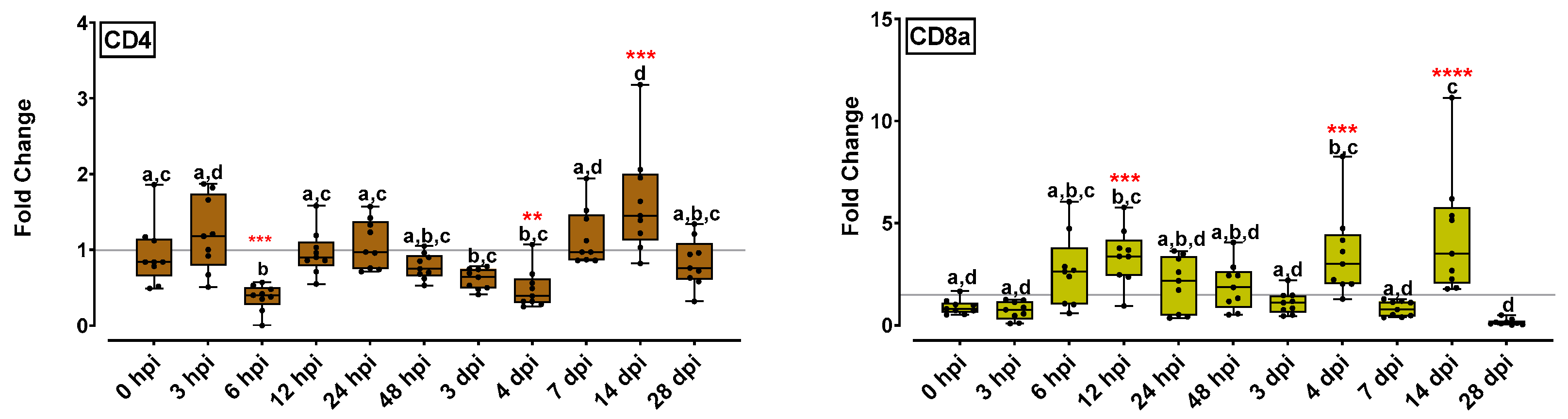
2.5.3. T-cell Marker Gene Expression Levels Were Fluctuated at Early Time Points but Were Up-Regulated at 14 dpi
2.5.4. Antimicrobial Peptide Gene Expression Was Not Significantly Affected by NNV Infection
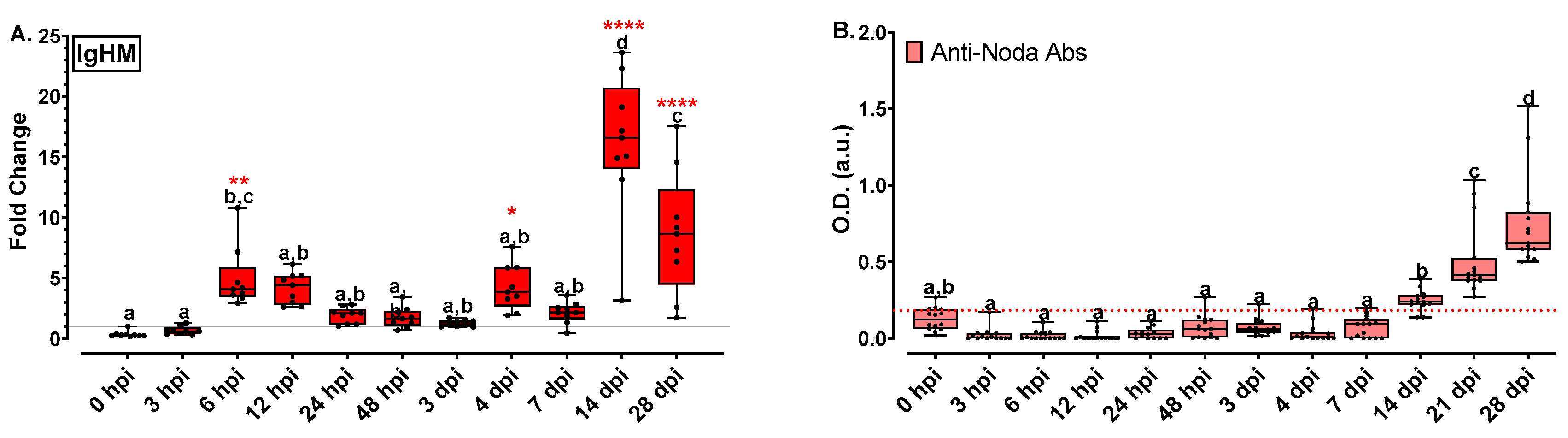
2.5.5. Immunoglobulin Analysis Revealed an Increasing Trend to Late Time Points
2.6. Transcriptome Analysis of NNV-Challenged D. labrax Head Kidney 14 dpi
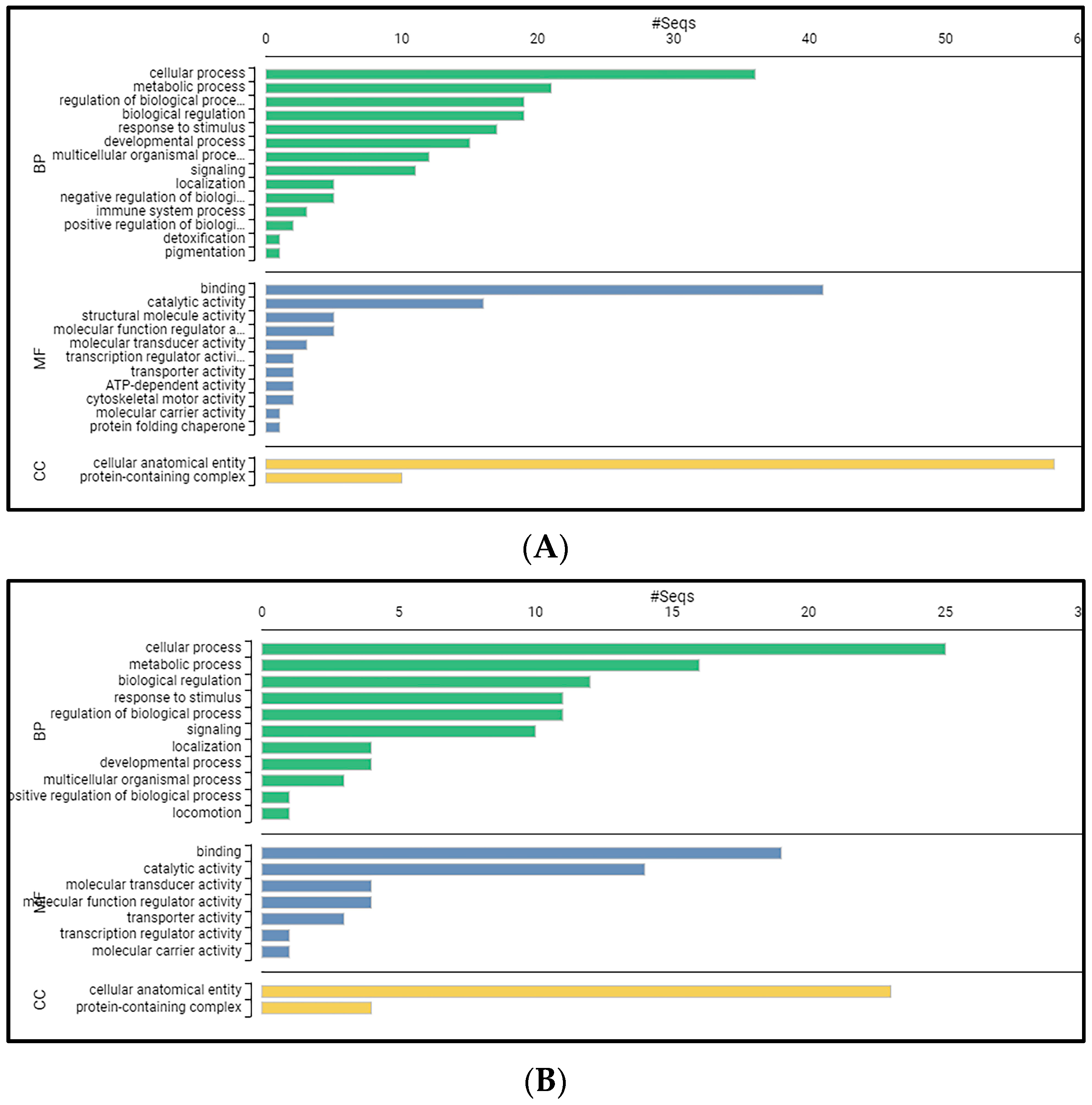
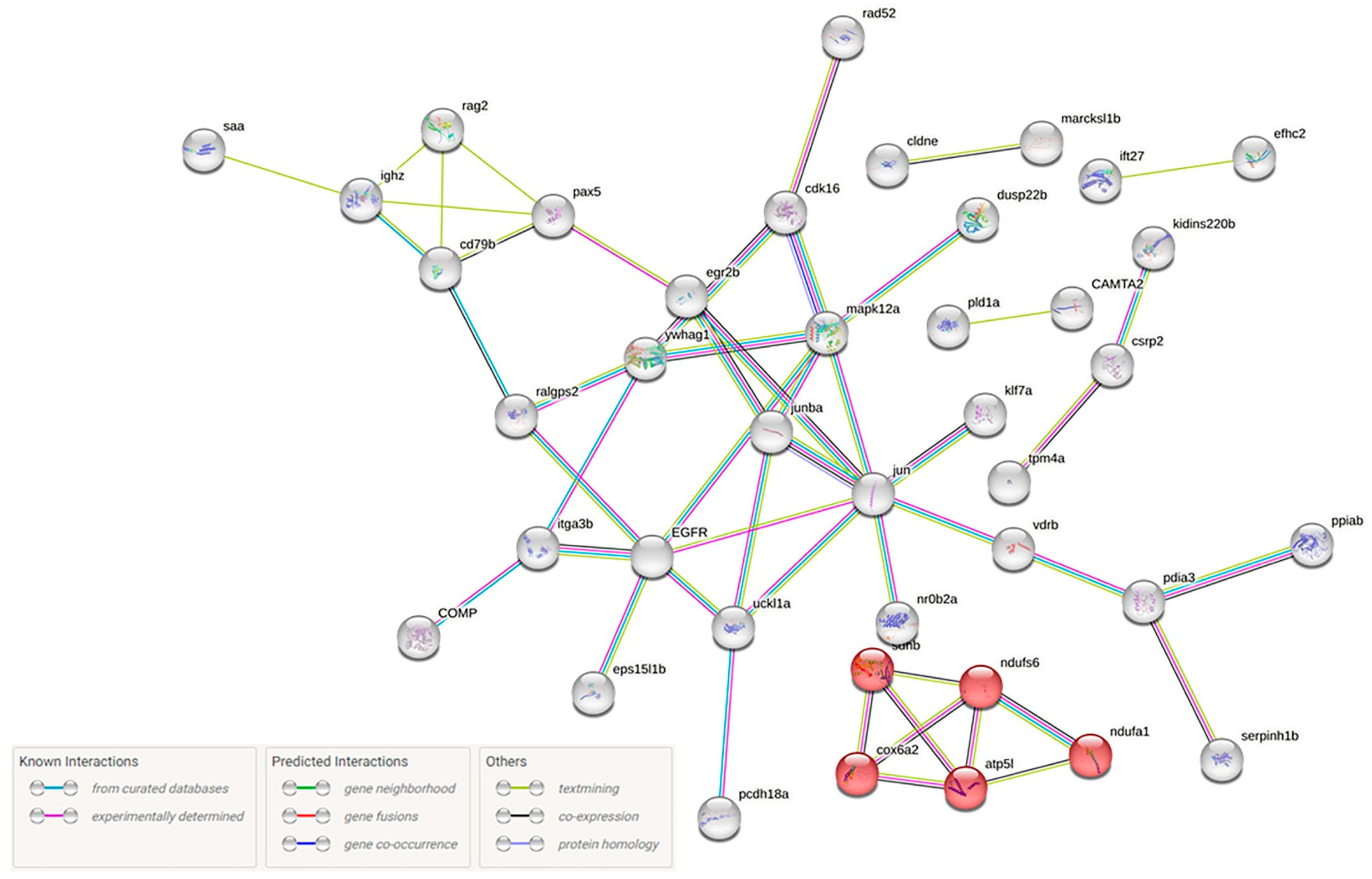
Differential Expression Analysis and GO Classification Revealed 133 Up- and 68 Down-Regulated Genes with Various Biological Functions
3. Discussion
4. Materials and Methods
4.1. Experimental Fish and NNV Challenge
4.2. Ethics Statement
4.3. Biometrical Data
4.4. Haematological Parameters
4.5. Measurement of Specific Anti-NNV and Total Sea Bass Serum Antibodies
4.5.1. Rabbit Anti-NNV Polyclonal Antibodies and Hyperimmune Sea Bass Anti-NNV Serum Preparation
4.5.2. Determination of Specific Anti-NNV Antibody Amount
4.6. RNA Isolation and Complementary DNA (cDNA) Synthesis
4.7. Gene Expression Analysis
4.8. NGS Sample Preparation
4.9. cDNA Library Construction, Sequencing and Transcriptome Mapping
4.10. Annotation of Differentially Expressed Genes, Functional Analysis and Construction of a Protein–Protein Interaction Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Toubanaki, D.K.; Efstathiou, A.; Karagouni, E. Transcriptomic Analysis of Fish Hosts Responses to Nervous Necrosis Virus. Pathogens 2022, 11, 201. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Thompson, S. The blue dimensions of aquaculture: A global synthesis. Sci. Total Environ. 2019, 652, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Moreno, P.; Gemez-Mata, J.; Garcia-Rosado, E.; Bejar, J.; Labella, A.M.; Souto, S.; Alonso, M.C. Differential immunogene expression profile of European sea bass (Dicentrarchus labrax, L.) in response to highly and low virulent NNV. Fish Shellfish Immunol. 2020, 106, 56–70. [Google Scholar] [CrossRef]
- Munday, B.L.; Kwang, J.; Moody, N. Betanodavirus infections of teleost fish: A review. J. Fish Dis. 2002, 25, 127–142. [Google Scholar] [CrossRef]
- Doan, Q.K.; Vandeputte, M.; Chatain, B.; Morin, T.; Allal, F. Viral encephalopathy and retinopathy in aquaculture: A review. J. Fish Dis. 2017, 40, 717–742. [Google Scholar] [CrossRef]
- Bandín, I.; Souto, S. Betanodavirus and VER Disease: A 30-year Research Review. Pathogens 2020, 9, 106. [Google Scholar] [CrossRef]
- Nishizawa, T.; Furuhashi, M.; Nagai, T.; Nakai, T.; Muroga, K. Genomic classification of fish nodaviruses by molecular phylogenetic analysis of the coat protein gene. Appl. Environ. Microbiol. 1997, 63, 1633–1636. [Google Scholar] [CrossRef]
- Costa, J.Z.; Thompson, K.D. Understanding the interaction between Betanodavirus and its host for the development of prophylactic measures for viral encephalopathy and retinopathy. Fish Shellfish Immunol. 2016, 53, 35–49. [Google Scholar] [CrossRef]
- Gonzalez-Silvera, D.; Guardiola, F.A.; Espinosa, C.; Chaves-Pozo, E.; Esteban, M.Á.; Cuesta, A. Recombinant nodavirus vaccine produced in bacteria and administered without purification elicits humoral immunity and protects European sea bass against infection. Fish Shellfish Immunol. 2019, 88, 458–463. [Google Scholar] [CrossRef]
- Tine, M.; Kuhl, H.; Gagnaire, P.A.; Louro, B.; Desmarais, E.; Martins, R.S.; Hecht, J.; Knaust, F.; Belkhir, K.; Klages, S.; et al. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat. Commun. 2014, 5, 5770. [Google Scholar] [CrossRef]
- Poisa-Beiro, L.; Dios, S.; Montes, A.; Aranguren, R.; Figueras, A.; Novoa, B. Nodavirus increases the expression of Mx and inflammatory cytokines in fish brain. Mol. Immunol. 2008, 45, 218–225. [Google Scholar] [CrossRef]
- Scapigliati, G.; Buonocore, F.; Randelli, E.; Casani, D.; Meloni, S.; Zarletti, G.; Tiberi, M.; Pietretti, D.; Boschi, I.; Manchado, M.; et al. Cellular and molecular immune responses of the sea bass (Dicentrarchus labrax) experimentally infected with betanodavirus. Fish Shellfish Immunol. 2010, 28, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Pozo, E.; Guardiola, F.A.; Meseguer, J.; Esteban, M.A.; Cuesta, A. Nodavirus infection induces a great innate cell-mediated cytotoxic activity in resistant, gilthead seabream, and susceptible, European sea bass, teleost fish. Fish Shellfish Immunol. 2012, 33, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Carballo, C.; Garcia-Rosado, E.; Borrego, J.J.; Alonso, M.C. SJNNV down-regulates RGNNV replication in European sea bass by the induction of the type I interferon system. Vet. Res. 2016, 47, 6. [Google Scholar] [CrossRef]
- Moreno, P.; Lopez-Jimena, B.; Randelli, E.; Scapigliati, G.; Buonocore, F.; Garcia-Rosado, E.; Borrego, J.J.; Alonso, M.C. Immuno-related gene transcription and antibody response in nodavirus (RGNNV and SJNNV)-infected European sea bass (Dicentrarchus labrax L.). Fish Shellfish Immunol. 2018, 78, 270–278. [Google Scholar] [CrossRef]
- Chaves-Pozo, E.; Bandín, I.; Olveira, J.G.; Esteve-Codina, A.; Gómez-Garrido, J.; Dabad, M.; Alioto, T.; Ángeles Esteban, M.; Cuesta, A. European sea bass brain DLB-1 cell line is susceptible to nodavirus: A transcriptomic study. Fish Shellfish Immunol. 2019, 86, 14–24. [Google Scholar] [CrossRef]
- Moreno, P.; Souto, S.; Leiva-Rebollo, R.; Borrego, J.J.; Bandín, I.; Alonso, M.C. Capsid amino acids at positions 247 and 270 are involved in the virulence of betanodaviruses to European sea bass. Sci. Rep. 2019, 9, 14068. [Google Scholar] [CrossRef]
- Vaz, M.; Pires, D.; Pires, P.; Simões, M.; Pombo, A.; Santos, P.; do Carmo, B.; Passos, R.; Costa, J.Z.; Thompson, K.D.; et al. Early Immune Modulation in European Seabass (Dicentrarchus labrax) Juveniles in Response to Betanodavirus Infection. Fishes 2022, 7, 63. [Google Scholar] [CrossRef]
- Tso, C.H.; Hung, Y.F.; Tan, S.P.; Lu, M.W. Identification of the STAT1 gene and the characterisation of its immune response to immunostimulants, including nervous necrosis virus (NNV) infection, in Malabar grouper (Epinephelus malabaricus). Fish Shellfish Immunol. 2013, 35, 1339–1348. [Google Scholar] [CrossRef]
- Lu, M.W.; Tso, C.H.; Periyasamy, T. Studies on Nervous Necrosis Virus Pathogenesis by Transcriptomic Approaches. In Marine OMICS: Principles and Applications; Kim, S.K., Ed.; CRC Press: Boca Raton, FL, USA, 2016; pp. 297–306. [Google Scholar]
- Lu, M.W.; Chao, Y.M.; Guo, T.C.; Santi, N.; Evensen, O.; Kasani, S.K.; Hong, J.R.; Wu, J.L. The interferon response is involved in nervous necrosis virus acute and persistent infection in zebrafish infection model. Mol. Immunol. 2008, 45, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Chi, S.C. Persistence of betanodavirus in Barramundi brain (BB) cell line involves the induction of Interferon response. Fish Shellfish Immunol. 2006, 21, 540–547. [Google Scholar] [CrossRef]
- Santi, N.; Song, H.; Vakharia, V.N.; Evensen, Ø. Infectious pancreatic necrosis virus VP5 is dispensable for virulence and persistence. J. Virol. 2005, 79, 9206–9216. [Google Scholar] [CrossRef]
- Gjessing, M.C.; Kvellestad, A.; Ottesen, K.; Falk, K. Nodavirus provokes subclinical encephalitis and retinochoroiditis in adult farmed Atlantic cod, Gadus morhua L. J. Fish Dis. 2009, 32, 421–431. [Google Scholar] [CrossRef]
- St-Hilaire, S.; Beevers, N.; Way, K.; Le Deuff, R.M.; Martin, P.; Joiner, C. Reactivation of koi herpesvirus infections in common carp Cyprinus carpio. Dis. Aquat. Organ. 2005, 67, 15–23. [Google Scholar] [CrossRef]
- Hershberger, P.K.; Gregg, J.L.; Grady, C.A.; Taylor, L.; Winton, J.R. Chronic and persistent viral hemorrhagic septicemia virus infections in Pacific herring. Dis. Aquat. Organ. 2010, 93, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Sutherland, B.J.; Koop, B.F.; Johnson, S.C.; Garver, K.A. Infectious hematopoietic necrosis virus (IHNV) persistence in Sockeye Salmon: Influence on brain transcriptome and subsequent response to the viral mimic poly(I:C). BMC Genom. 2015, 16, 634. [Google Scholar] [CrossRef]
- Tso, C.H.; Lu, M.W. Transcriptome profiling analysis of grouper during nervous necrosis virus persistent infection. Fish Shellfish Immunol. 2018, 76, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Rise, M.L.; Hall, J.R.; Rise, M.; Hori, T.S.; Browne, M.J.; Gamperl, A.K.; Hubert, S.; Kimball, J.; Bowman, S.; Johnson, S.C. Impact of asymptomatic nodavirus carrier state and intraperitoneal viral mimic injection on brain transcript expression in Atlantic cod (Gadus morhua). Physiol. Genom. 2010, 42, 266–280. [Google Scholar] [CrossRef]
- Baud, M.; Cabon, J.; Salomoni, A.; Toffan, A.; Panzarin, V.; Bigarré, L. First generic one step real-time Taqman RT-PCR targeting the RNA1 of betanodaviruses. J. Virol. Methods. 2015, 211, 1–7. [Google Scholar] [CrossRef]
- Fazio, F. Fish hematology analysis as an important tool of aquaculture: A review. Aquaculture 2019, 500, 237–242. [Google Scholar] [CrossRef]
- Ray, S.D.; Roy, D.; Pal, S.; Homechaudhuri, S. Effects of Beta Glucan as Immunostimulant on Labeo rohita Challenged with a Bacterial Pathogen Aeromonas Hydrophila. Int. J. Innov. Stud. Aquat. Biol. Fish. 2016, 2, 10–19. [Google Scholar] [CrossRef]
- Wu, M.S.; Chen, C.W.; Lin, C.H.; Tzeng, C.S.; Chang, C.Y. Differential expression profiling of orange-spotted grouper larvae, Epinephelus coioides (Hamilton), that survived a betanodavirus outbreak. J. Fish Dis. 2012, 35, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yi, L.; Feng, S.; Liu, X.; Asim, M.; Zhou, Y.; Lan, J.; Jiang, S.; Tu, J.; Lin, L. Transcriptomic profiles of striped snakehead fish cells (SSN-1) infected with red-spotted grouper nervous necrosis virus (RGNNV) with an emphasis on apoptosis pathway. Fish Shellfish Immunol. 2017, 60, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Munang’andu, H.M.; Evensen, Ø. A Review of Intra- and Extracellular Antigen Delivery Systems for Virus Vaccines of Finfish. J. Immunol. Res. 2015, 2015, 960859. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Zhu, Z.; Ge, H.; Zheng, L.; Huang, Z.; Wu, S. Immunity to nervous necrosis virus infections of orange-spotted grouper (Epinephelus coioides) by vaccination with virus-like particles. Fish Shellfish Immunol. 2016, 56, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Sarropoulou, E.; Sepulcre, P.; Poisa-Beiro, L.; Mulero, V.; Meseguer, J.; Figueras, A.; Novoa, B.; Terzoglou, V.; Reinhardt, R.; Magoulas, A.; et al. Profiling of infection specific mRNA transcripts of the European seabass Dicentrarchus labrax. BMC Genom. 2009, 10, 157. [Google Scholar] [CrossRef]
- Labella, A.M.; Garcia-Rosado, E.; Bandín, I.; Dopazo, C.P.; Castro, D.; Alonso, M.C.; Borrego, J.J. Transcriptomic Profiles of Senegalese Sole Infected With Nervous Necrosis Virus Reassortants Presenting Different Degree of Virulence. Front. Immunol. 2018, 9, 1626. [Google Scholar] [CrossRef]
- Valero, Y.; Boughlala, B.; Arizcun, M.; Patel, S.; Fiksdal, I.U.; Esteban, M.Á.; De Juan, J.; Meseguer, J.; Chaves-Pozo, E.; Cuesta, A. Genes related to cell-mediated cytotoxicity and interferon response are induced in the retina of European sea bass upon intravitreal infection with nodavirus. Fish Shellfish Immunol. 2018, 74, 627–636. [Google Scholar] [CrossRef]
- Gan, Z.; Chen, S.N.; Huang, B.; Zou, J.; Nie, P. Fish type I and type II interferons: Composition, receptor usage, production and function. Rev. Aquac. 2019, 12, 773–804. [Google Scholar] [CrossRef]
- Valero, Y.; Morcillo, P.; Meseguer, J.; Buonocore, F.; Esteban, M.A.; Chaves-Pozo, E.; Cuesta, A. Characterization of the IFN pathway in the teleost fish gonad against vertically transmitted viral nervous necrosis virus. J. Gen. Virol. 2015, 96, 2176–2187. [Google Scholar] [CrossRef]
- Novel, P.; Fernández-Trujillo, M.A.; Gallardo-Gálvez, J.B.; Cano, I.; Manchado, M.; Buonocore, F.; Randelli, E.; Scapigliati, G.; Alvarez, M.C.; Béjar, J. Two Mx genes identified in European sea bass (Dicentrarchus labrax) respond differently to VNNV infection. Vet. Immunol. Immunopathol. 2013, 153, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, R.; Kurcheti, P.P.; Mushtaq, Z.; Jeena, K.; Naik, T.V. Interferon-regulatory factors, IRF3 and IRF7 in Asian seabass, Lates calcarifer: Characterization, ontogeny and transcriptional modulation upon challenge with nervous necrosis virus. Fish Shellfish Immunol. 2019, 89, 468–476. [Google Scholar] [CrossRef]
- Zou, J.; Secombes, C.J. The Function of Fish Cytokines. Biology 2016, 5, 23. [Google Scholar] [CrossRef]
- Teles, M.; Reyes-López, F.E.; Fierro-Castro, C.; Tort, L.; Soares, A.M.V.M.; Oliveira, M. Modulation of immune genes mRNA levels in mucosal tissues and DNA damage in red blood cells of Sparus aurata by gold nanoparticles. Mar. Pollut. Bull. 2018, 133, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Reyes-López, F.E.; Aerts, J.; Vallejos-Vidal, E.; Ampe, B.; Dierckens, K.; Tort, L.; Bossier, P. Modulation of Innate Immune-Related Genes and Glucocorticoid Synthesis in Gnotobiotic Full-Sibling European Sea Bass (Dicentrarchus labrax) Larvae Challenged With Vibrio anguillarum. Front. Immunol. 2018, 9, 914. [Google Scholar] [CrossRef] [PubMed]
- De Domenico, I.; Zhang, T.Y.; Koening, C.L.; Branch, R.W.; London, N.; Lo, E.; Daynes, R.A.; Kushner, J.P.; Li, D.; Ward, D.M.; et al. Hepcidin mediates transcriptional changes that modulate acute cytokine-induced inflammatory responses in mice. J. Clin. Investig. 2010, 120, 2395–2405. [Google Scholar] [CrossRef]
- Snell, L.M.; Osokine, I.; Yamada, D.H.; De la Fuente, J.R.; Elsaesser, H.J.; Brooks, D.G. Overcoming CD4 Th1 Cell Fate Restrictions to Sustain Antiviral CD8 T Cells and Control Persistent Virus Infection. Cell Rep. 2016, 16, 3286–3296. [Google Scholar] [CrossRef]
- Rauta, P.R.; Nayak, B.; Das, S. Immune system and immune responses in fish and their role in comparative immunity study: A model for higher organisms. Immunol. Lett. 2012, 148, 23–33. [Google Scholar] [CrossRef]
- Langevin, C.; Levraud, J.P.; Boudinot, P. Fish antiviral tripartite motif (TRIM) proteins. Fish Shellfish Immunol. 2019, 86, 724–733. [Google Scholar] [CrossRef]
- Krasnov, A.; Kileng, Ø.; Skugor, S.; Jørgensen, S.M.; Afanasyev, S.; Timmerhaus, G.; Sommer, A.I.; Jensen, I. Genomic analysis of the host response to nervous necrosis virus in Atlantic cod (Gadus morhua) brain. Mol. Immunol. 2013, 54, 443–452. [Google Scholar] [CrossRef]
- Yu, Y.; Huang, X.; Zhang, J.; Liu, J.; Hu, Y.; Yang, Y.; Cai, J.; Huang, Y.; Qin, Q. Fish TRIM16L exerts negative regulation on antiviral immune response against grouper iridoviruses. Fish Shellfish Immunol. 2016, 59, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yao, L.; Chen, X.; Li, M.; Yi, M.; Jia, K. Functional characterization of myosin III light chain b of sea perch (Lateolabrax japonicus) in response to fish virus infection. Aquaculture 2022, 550, 737840. [Google Scholar] [CrossRef]
- Lehmann, M.J.; Sherer, N.M.; Marks, C.B.; Pypaert, M.; Mothes, W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J. Cell Biol. 2005, 170, 317–325. [Google Scholar] [CrossRef]
- Kumar, P.S.; Radhakrishnan, A.; Mukherjee, T.; Khamaru, S.; Chattopadhyay, S.; Chattopadhyay, S. Understanding the role of Ca2+ via transient receptor potential (TRP) channel in viral infection: Implications in developing future antiviral strategies. Virus Res. 2022, 323, 198992. [Google Scholar] [CrossRef]
- Zhao, J.Z.; Xu, L.M.; Ren, G.M.; Shao, Y.Z.; Liu, Q.; Teng, C.B.; Lu, T.Y. Comparative transcriptome analysis of rainbow trout gonadal cells (RTG-2) infected with U and J genogroup infectious hematopoietic necrosis virus. Front. Microbiol. 2023, 13, 1109606. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.A.; Gopikrishna, G.; Baranski, M.; Katneni, V.K.; Shekhar, M.S.; Shanmugakarthik, J.; Jothivel, S.; Gopal, C.; Ravichandran, P.; Gitterle, T.; et al. QTL for white spot syndrome virus resistance and the sex-determining locus in the Indian black tiger shrimp (Penaeus monodon). BMC Genom. 2014, 15, 731. [Google Scholar] [CrossRef]
- Shi, H.; Yan, X.; Xu, X.; Ruan, L. Molecular cloning and characterization of a cDNA encoding extracellular signal-regulated kinase from Litopenaeus vannamei. Fish Shellfish Immunol. 2012, 33, 813–820. [Google Scholar] [CrossRef]
- Toubanaki, D.K.; Margaroni, M.; Karagouni, E. Development of a Novel Allele-Specific PCR Method for Rapid Assessment of Nervous Necrosis Virus Genotypes. Curr. Microbiol. 2015, 71, 529–539. [Google Scholar] [CrossRef]
- Toubanaki, D.K.; Margaroni, M.; Prapas, A.; Karagouni, E. Development of a Nanoparticle-based Lateral Flow Strip Biosensor for Visual Detection of Whole Nervous Necrosis Virus Particles. Sci. Rep. 2020, 10, 6529. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A Simple Method of Estimating Fifty Per Cent Endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- García Beltrán, J.M.; Silvera, D.G.; Ruiz, C.E.; Campo, V.; Chupani, L.; Faggio, C.; Esteban, M.Á. Effects of dietary Origanum vulgare on gilthead seabream (Sparus aurata L.) immune and antioxidant status. Fish Shellfish Immunol. 2020, 99, 452–461. [Google Scholar] [CrossRef]
- Pérez-Sánchez, J.; Benedito-Palos, L.; Estensoro, I.; Petropoulos, Y.; Calduch-Giner, J.A.; Browdy, C.L.; Sitjà-Bobadilla, A. Effects of dietary NEXT ENHANCE®150 on growth performance and expression of immune and intestinal integrity related genes in gilthead sea bream (Sparus aurata L.). Fish Shellfish Immunol. 2015, 44, 117–128. [Google Scholar] [CrossRef]
- Rigos, G.; Karagouni, E.; Kyriazis, I.; Athanasiou, E.; Grigorakis, K.; Kotou, E.; Katharios, P. In vitro and in vivo evaluation of quinine in gilthead sea bream, Sparus aurata naturally infected with the ciliate Cryptocaryon irritans. Aquaculture 2013, 416–417, 185–191. [Google Scholar] [CrossRef]
- Bakopoulos, V.; Volpatti, D.; Adams, A.; Galeotti, M.; Richards, R. Qualitative differences in the immune response of rabbit, mouse and sea bass, Dicentrarchus labrax, L. to Photobacterium damsela subsp. piscicida, the causative agent of fish Pasteurellosis. Fish Shellfish Immunol. 1997, 7, 161–174. [Google Scholar] [CrossRef]
- Bakopoulos, V.; Volpatti, D.; Papapanagiotou, E.; Richards, R.; Galleotti, M.; Adams, A. Development of an ELISA to detect Pasteurella piscicida in culture and in ‘spiked’ fish tissue. Aquaculture 1997, 153, 359–366. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Moulos, P.; Hatzis, P. Systematic integration of RNA-Seq statistical algorithms for accurate detection of differential gene expression patterns. Nucleic Acids Res. 2015, 43, e25. [Google Scholar] [CrossRef]
- Fanidis, D.; Moulos, P. Integrative, normalization-insusceptible statistical analysis of RNA-Seq data, with improved differential expression and unbiased downstream functional analysis. Brief. Bioinform. 2021, 22, bbaa156. [Google Scholar] [CrossRef]
- von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef]
- Kane, M.; Golovkina, T. Common threads in persistent viral infections. J. Virol. 2010, 84, 4116–4123. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.N.; Kaur, V.I.; Dhawan, A.; Jassal, G. Estimation of length-weight relationship and condition factor of spotted snakehead Channa punctata (Bloch) under different feeding regimes. Springerplus 2013, 2, 436. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Gupta, D.K.; Sharma, P.K. Condition factor and organosomatic indices of parasitized Rattus rattus as indicators of host health. J. Parasit. Dis. 2017, 41, 21–28. [Google Scholar] [CrossRef] [PubMed]



| Gene ID | Description | Gene Symbol | Log2FC | p-Value |
|---|---|---|---|---|
| DLAgn_00266700 | tripartite motif-containing protein 16-like | - | 4.7558 | 0.0013 |
| DLAgn_00246320 | myosin heavy chain | MYHZ1.1 | 3.8984 | 1.08 × 108 |
| DLAgn_00231310 | uncharacterized protein loc101466409 isoform x1 | - | 3.4739 | 0.0009 |
| DLAgn_00244650 | myosin heavy chain | MMYHL1 | 3.3833 | 0.0012 |
| DLAgn_00216380 | myosin heavy chain | MYHZ1.1 | 2.9484 | 0.0048 |
| DLAgn_00096300 | myosin-binding protein h-like | MYBPHA | 2.5482 | 0.0069 |
| DLAgn_00252300 | ubiquitin carboxyl-terminal hydrolase 35 | - | 2.5146 | 0.0209 |
| DLAgn_00118970 | kallikrein-8 | KLK8 | 2.4557 | 0.0139 |
| DLAgn_00266850 | proteinase-activated receptor 4 | F2RL3 | 2.4476 | 0.0256 |
| DLAgn_00232470 | uncharacterized protein iii | - | 2.4018 | 0.0231 |
| DLAgn_00159040 | serum amyloid a | SAA | 2.3315 | 0.0479 |
| DLAgn_00234950 | myosin heavy chain | MMYHL1 | 2.3037 | 0.0134 |
| DLAgn_00206120 | cysteine and glycine-rich protein 2 | CSRP2 | 2.3026 | 0.0231 |
| DLAgn_00080910 | peptidyl-prolyl cis-trans isomerase-like | PPIAB | 2.2970 | 0.0046 |
| DLAgn_00045390 | solute carrier family 22 member 5-like | SLC22A5 | 2.2270 | 0.0062 |
| DLAgn_00251790 | myosin heavy chain | MYHZ1.1 | 2.1442 | 0.0026 |
| DLAgn_00247100 | endonuclease domain-containing 1 | - | 1.9130 | 0.0097 |
| DLAgn_00062850 | tetranectin-like | TETN | 1.9005 | 0.0451 |
| DLAgn_00167280 | leptin | LEP | 1.9005 | 0.0480 |
| DLAgn_00266930 | histone-lysine n-methyltransferase mll3 | MLL3A | 1.8973 | 0.0008 |
| Gene ID | Description | Gene Symbol | Log2FC | p-Value |
| DLAgn_00049910 | transient receptor potential cation channel subfamily m member 2-like | TRPM2 | −2.3655 | 0.0033 |
| DLAgn_00227410 | mitogen-activated protein kinase-binding protein 1-like | MAPKBP1 | −2.1043 | 0.0159 |
| DLAgn_00218080 | kinase d-interacting substrate of 220 kda-like | KIDINS220B | −2.0489 | 0.0033 |
| DLAgn_00251680 | butyrophilin subfamily 1 member a1-like | MOG | −1.9534 | 0.0018 |
| DLAgn_00221860 | cytochrome p450 2j2-like | CYP2AD2 | −1.8864 | 0.0052 |
| DLAgn_00018460 | calmin delta | CLMN | −1.8694 | 0.0159 |
| DLAgn_00249930 | zinc finger protein 850-like | - | −1.6659 | 0.0016 |
| DLAgn_00231790 | hippocampus abundant transcript 1 | HIAT1 | −1.6471 | 0.0191 |
| DLAgn_00099340 | hemoglobin beta chain | - | −1.6142 | 0.0469 |
| DLAgn_00063590 | cub and sushi domain-containing protein 3-like | CSMD3 | −1.6049 | 0.0113 |
| DLAgn_00130980 | solute carrier family 26 member 10 | SLC26A10 | −1.4112 | 0.0057 |
| DLAgn_00142520 | sodium calcium exchanger 1-like | SLC8A4A | −1.3812 | 0.0221 |
| DLAgn_00241570 | sterile alpha motif domain-containing protein 9-like | SAMD9L | −1.3797 | 0.0175 |
| DLAgn_00218000 | tgf-beta receptor type-1-like | TGFBR1 | −1.3389 | 0.0068 |
| DLAgn_00151800 | ral gef with ph domain and sh3 binding motif 2 | RALGPS2 | −1.3248 | 0.0051 |
| DLAgn_00017450 | piezo-type mechanosensitive ion channel component 1 | PIEZO1 | −1.2886 | 0.0361 |
| DLAgn_00203250 | sperm acrosome membrane-associated protein 4 | SPACA4 | −1.2872 | 0.0402 |
| DLAgn_00252660 | general transcription factor ii-i repeat domain-containing protein 2-like | - | −1.2695 | 0.0088 |
| DLAgn_00134630 | plexin-partial | PLXNB1 | −1.2363 | 0.0020 |
| DLAgn_00227470 | v-set domain-containing t-cell activation inhibitor 1-like | - | −1.2264 | 0.0325 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toubanaki, D.K.; Efstathiou, A.; Tzortzatos, O.-P.; Valsamidis, M.-A.; Papaharisis, L.; Bakopoulos, V.; Karagouni, E. Nervous Necrosis Virus Modulation of European Sea Bass (Dicentrarchus labrax, L.) Immune Genes and Transcriptome towards Establishment of Virus Carrier State. Int. J. Mol. Sci. 2023, 24, 16613. https://doi.org/10.3390/ijms242316613
Toubanaki DK, Efstathiou A, Tzortzatos O-P, Valsamidis M-A, Papaharisis L, Bakopoulos V, Karagouni E. Nervous Necrosis Virus Modulation of European Sea Bass (Dicentrarchus labrax, L.) Immune Genes and Transcriptome towards Establishment of Virus Carrier State. International Journal of Molecular Sciences. 2023; 24(23):16613. https://doi.org/10.3390/ijms242316613
Chicago/Turabian StyleToubanaki, Dimitra K., Antonia Efstathiou, Odysseas-Panagiotis Tzortzatos, Michail-Aggelos Valsamidis, Leonidas Papaharisis, Vasileios Bakopoulos, and Evdokia Karagouni. 2023. "Nervous Necrosis Virus Modulation of European Sea Bass (Dicentrarchus labrax, L.) Immune Genes and Transcriptome towards Establishment of Virus Carrier State" International Journal of Molecular Sciences 24, no. 23: 16613. https://doi.org/10.3390/ijms242316613
APA StyleToubanaki, D. K., Efstathiou, A., Tzortzatos, O.-P., Valsamidis, M.-A., Papaharisis, L., Bakopoulos, V., & Karagouni, E. (2023). Nervous Necrosis Virus Modulation of European Sea Bass (Dicentrarchus labrax, L.) Immune Genes and Transcriptome towards Establishment of Virus Carrier State. International Journal of Molecular Sciences, 24(23), 16613. https://doi.org/10.3390/ijms242316613







