BAP31-Mediated miR-206/133b Cluster Promotes Transendothelial Migration and Metastasis of Colorectal Cancer
Abstract
:1. Introduction
2. Results
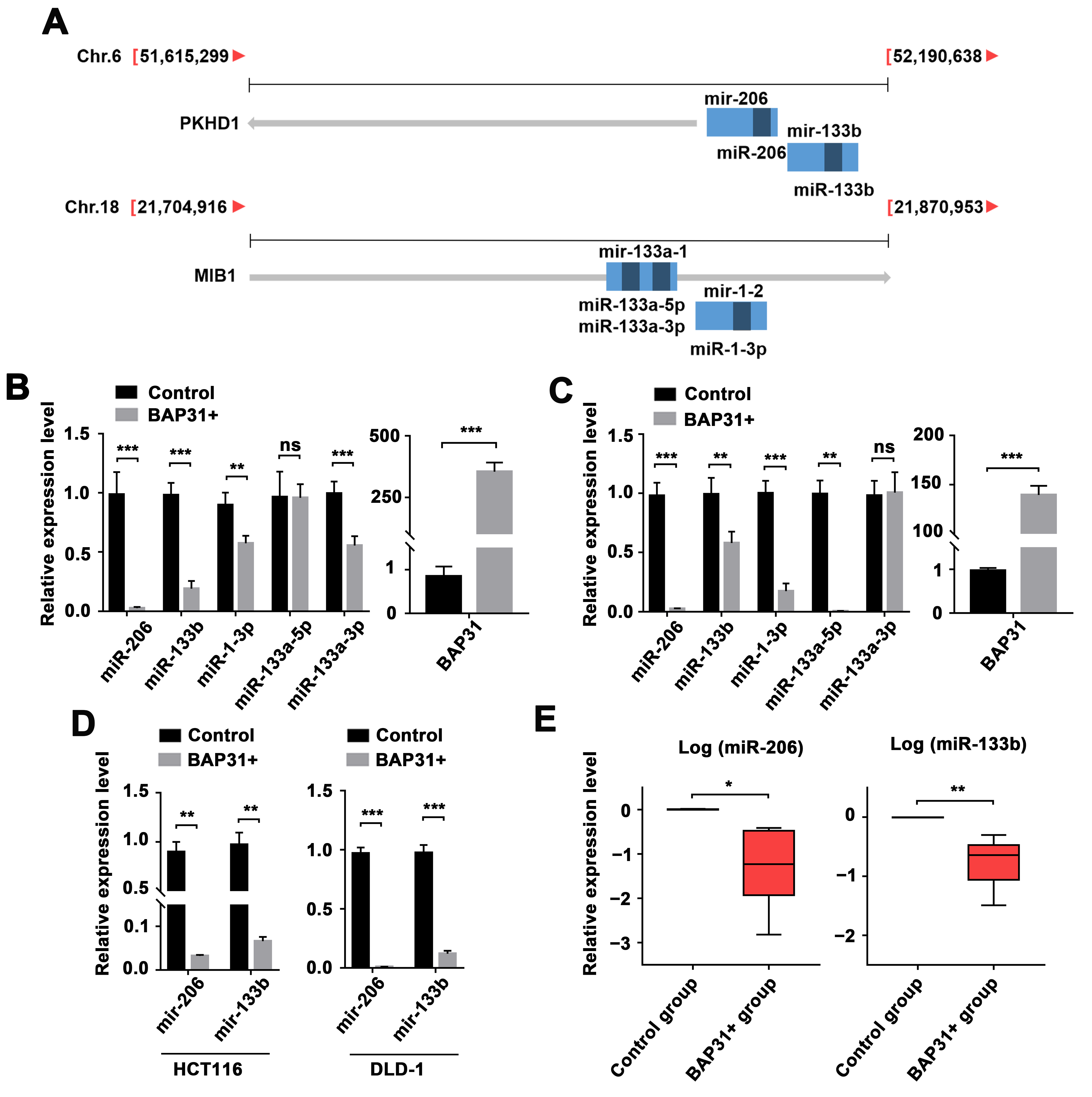
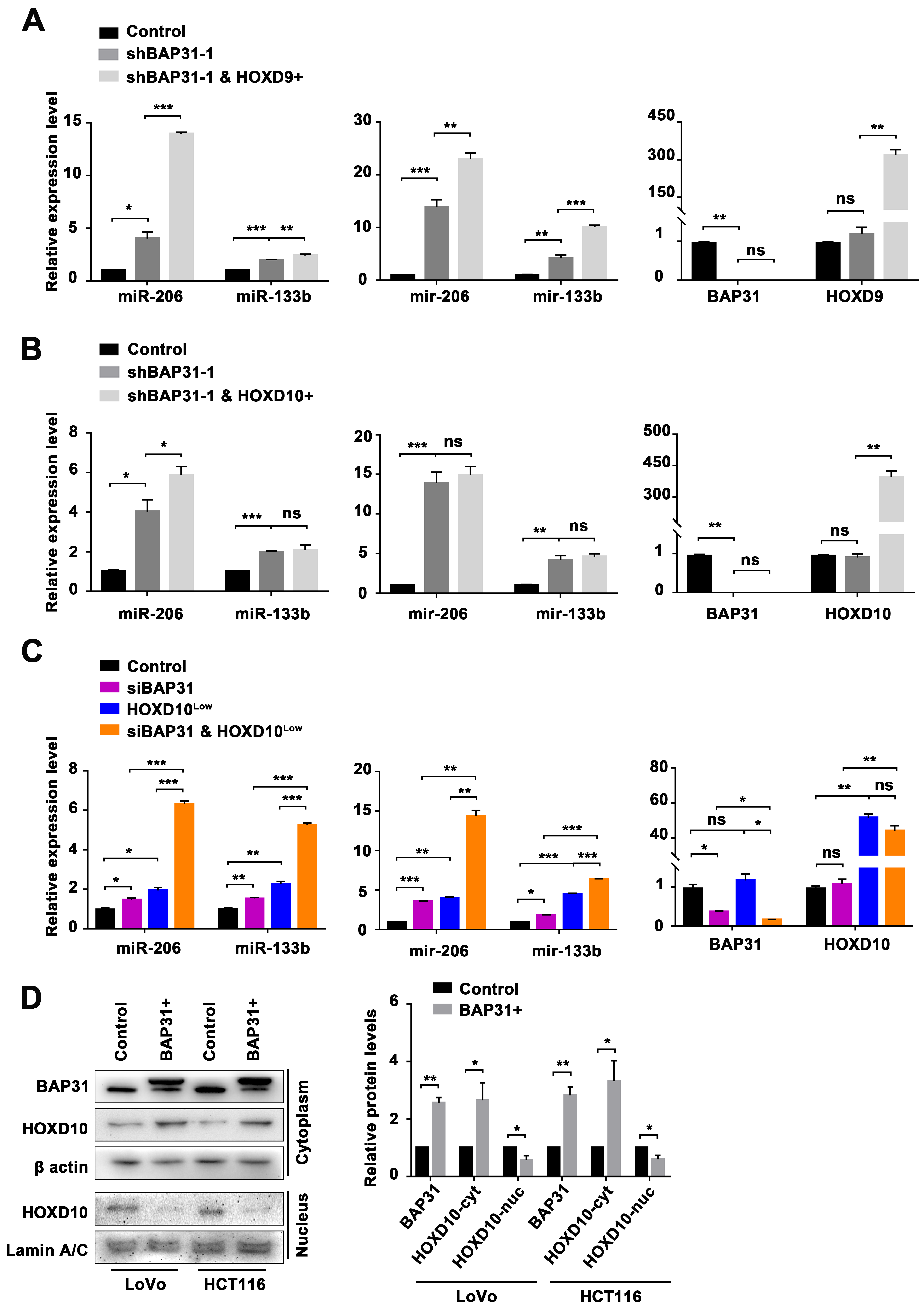
2.1. BAP31-Mediated Regulation of the miR-206/133b Cluster
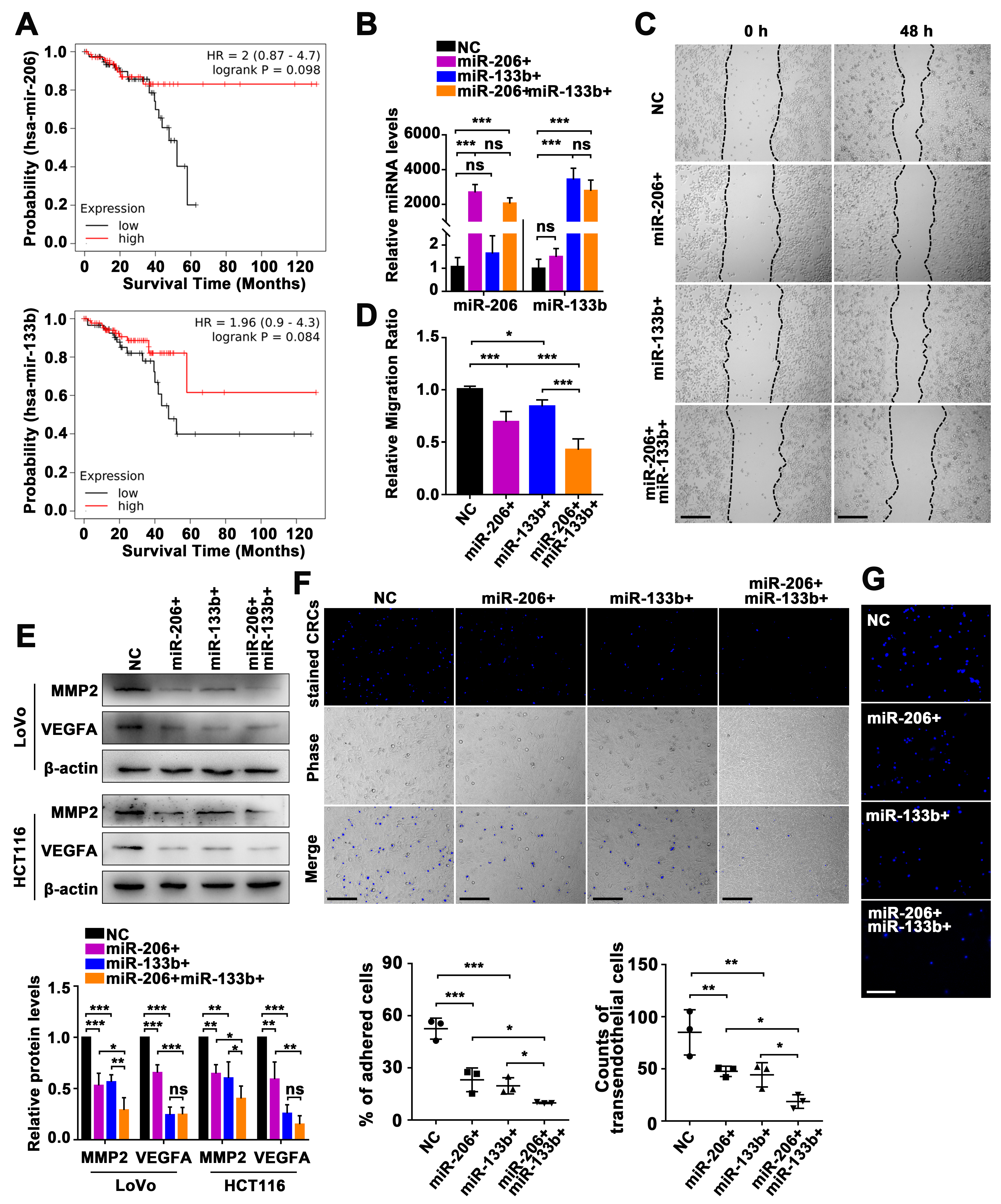
2.2. Manipulation of the miR-206/133b Cluster on CRC Cell Transendothelial Migration
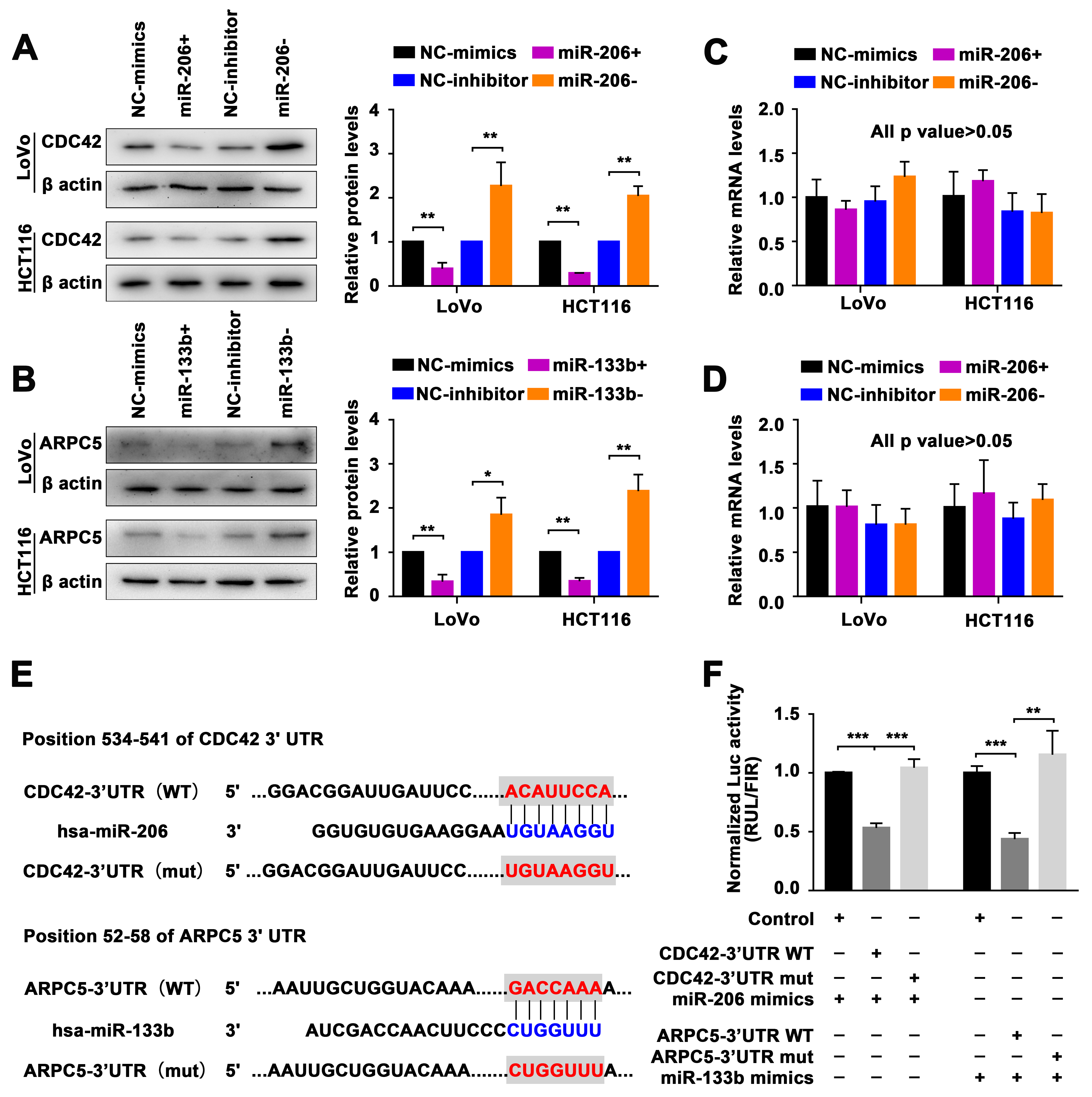
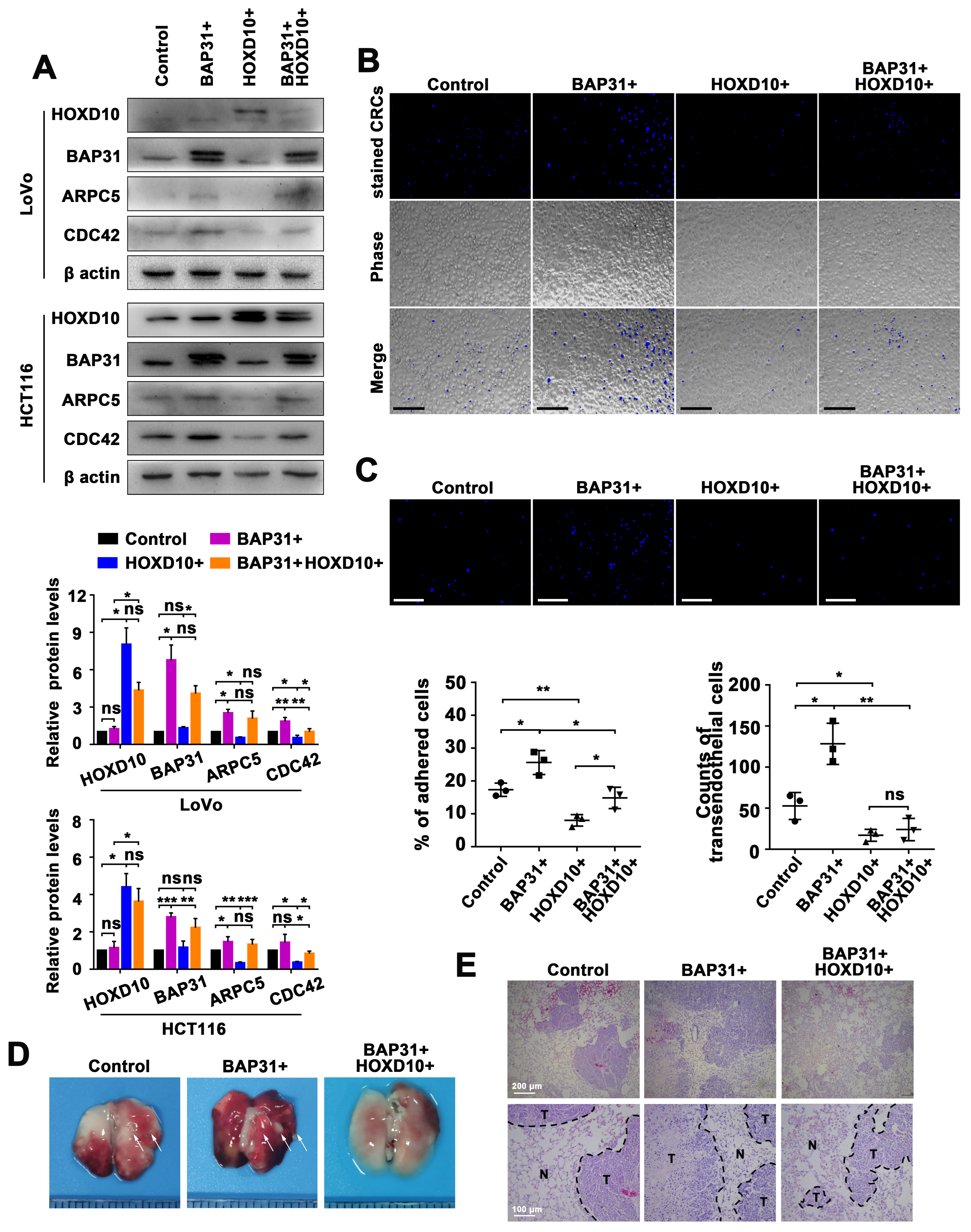
2.3. Identification of the miR-206/133b Cluster-Targeted Genes in CRC Cells
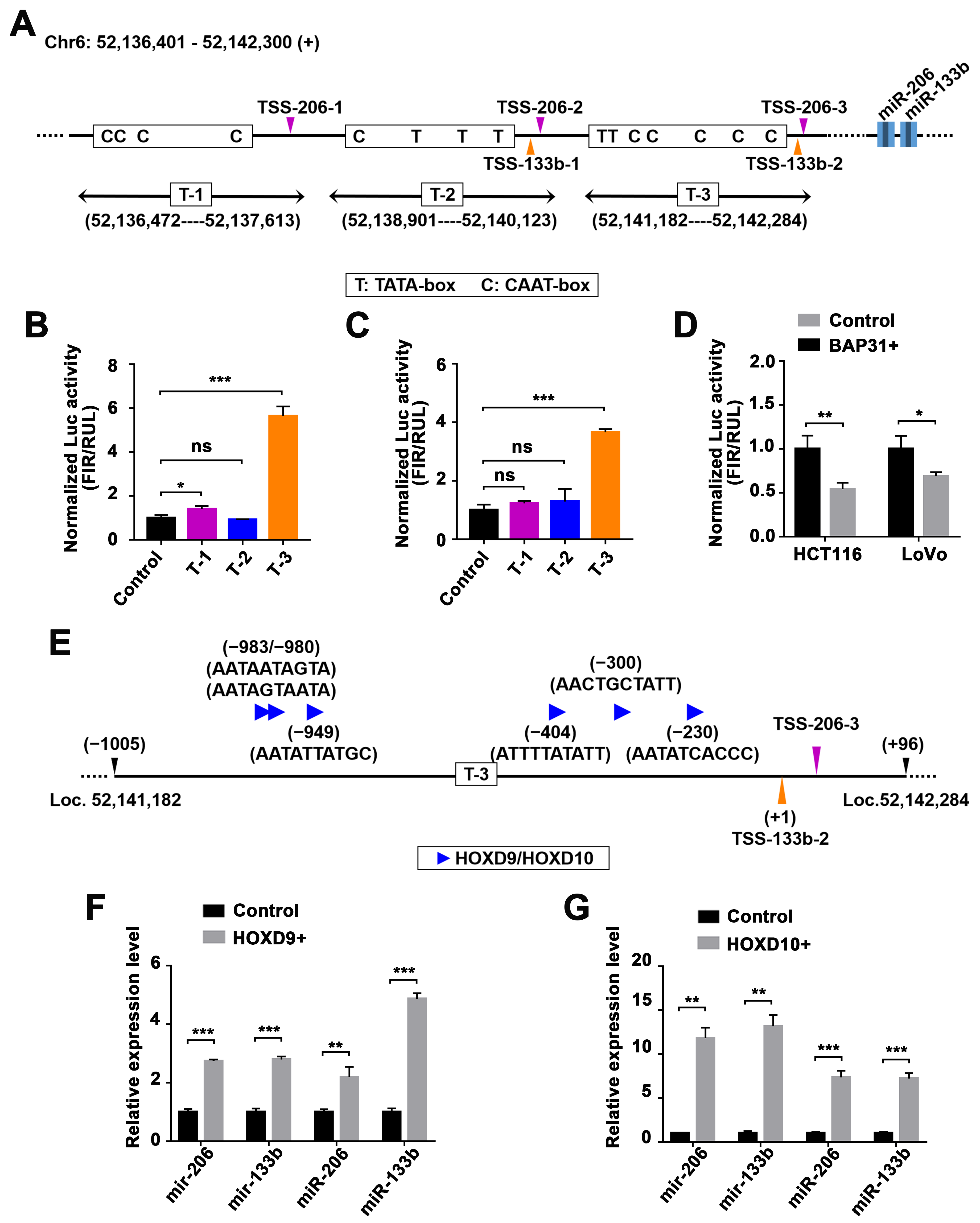
2.4. Identification of miR-206/133b Cluster Transcription Factors
2.5. BAP31-Mediated Transcriptional Activation of the miR-206/133b Cluster by HOXD10
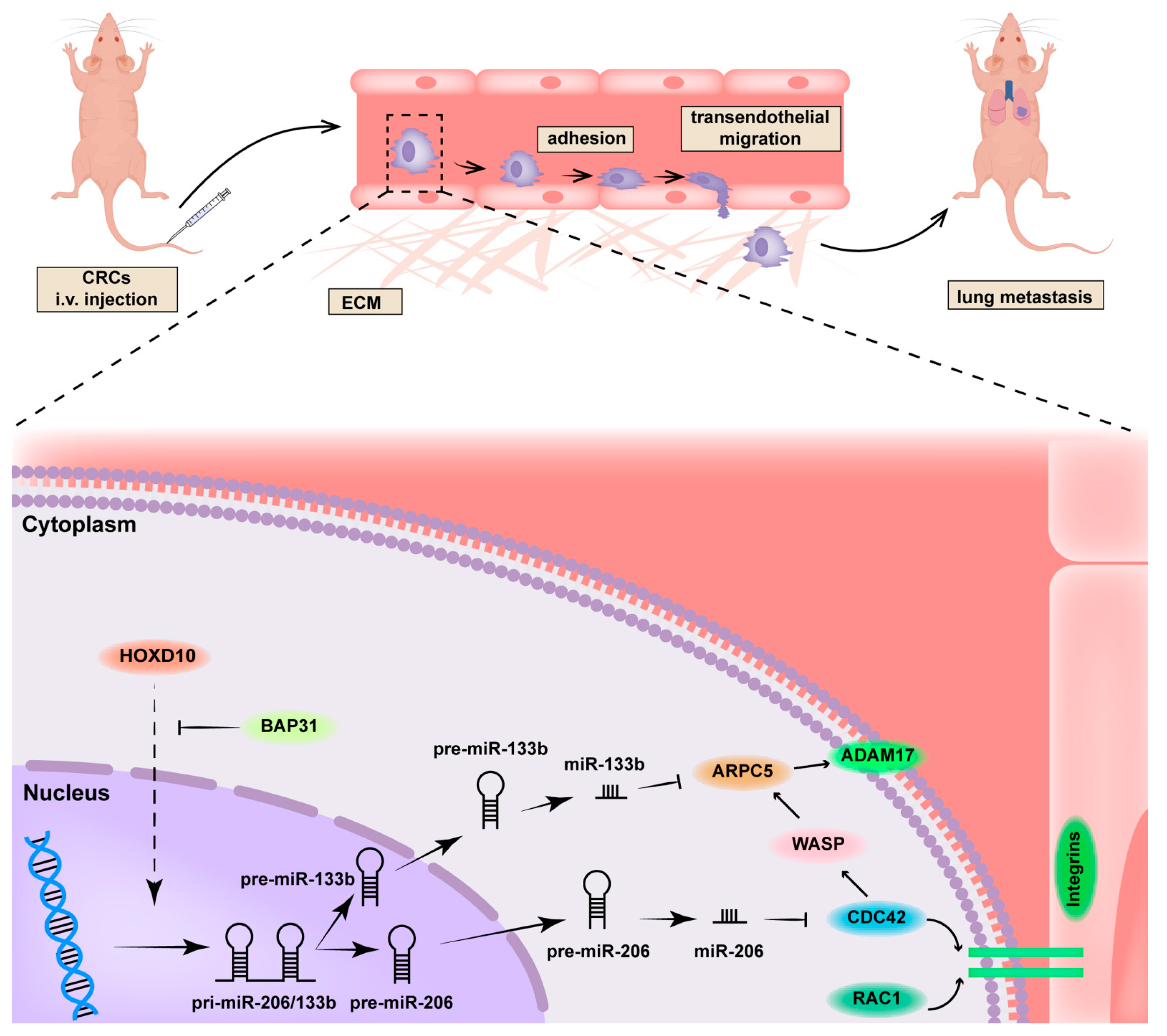
2.6. Manipulation of the BAP31/HOXD10 Axis in the Tumor Metastasis of CRC
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. RNA Analysis
4.3. Western Blot Analysis
4.4. Wound Healing Assay
4.5. In Vitro Adhesion and Transendothelial Migration Assay
4.6. Luciferase Reporter Assay
4.7. Animal Experiments
4.8. Hematoxylin–Eosin Staining
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, J.; Guo, H.; Jiang, H.; Namusamba, M.; Wang, C.; Lan, T.; Wang, T.; Wang, B. A BAP31 intrabody induces gastric cancer cell death by inhibiting p27(kip1) proteasome degradation. Int. J. Cancer 2019, 144, 2051–2062. [Google Scholar] [CrossRef] [PubMed]
- Dang, E.; Yang, S.; Song, C.; Jiang, D.; Li, Z.; Fan, W.; Sun, Y.; Tao, L.; Wang, J.; Liu, T.; et al. BAP31, a newly defined cancer/testis antigen, regulates proliferation, migration, and invasion to promote cervical cancer progression. Cell Death Dis. 2018, 9, 791. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Zhang, Y.; Cao, P. Inhibition of BAP31 expression inhibits cervical cancer progression by suppressing metastasis and inducing intrinsic and extrinsic apoptosis. Biochem. Biophys. Res. Commun. 2019, 508, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Karuniawati, H.; Jairoun, A.A.; Urbi, Z.; Ooi, J.; John, A.; Lim, Y.C.; Kibria, K.M.K.; Mohiuddin, A.K.M.; Ming, L.C.; et al. Colorectal Cancer: A Review of Carcinogenesis, Global Epidemiology, Current Challenges, Risk Factors, Preventive and Treatment Strategies. Cancers 2022, 14, 1732. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, F.; Fan, L.; Wei, Y.; Li, H.; Sun, Y.; Yang, A.; Jin, B.; Song, C.; Yang, K. BAP31, a promising target for the immunotherapy of malignant melanomas. J. Exp. Clin. Cancer Res. 2015, 34, 36. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Han, B.; Bai, Y.; Ma, X.Y.; Ji, Z.N.; Xiong, Y.; Miao, S.K.; Zhang, Y.Y.; Zhou, L.M. MiR-451a suppressing BAP31 can inhibit proliferation and increase apoptosis through inducing ER stress in colorectal cancer. Cell Death Dis. 2019, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Malvezzi, M.; Carioli, G.; Bertuccio, P.; Boffetta, P.; Levi, F.; La Vecchia, C.; Negri, E. European cancer mortality predictions for the year 2018 with focus on colorectal cancer. Ann. Oncol. 2018, 29, 1016–1022. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef]
- Hussen, B.M.; Hidayat, H.J.; Salihi, A.; Sabir, D.K.; Taheri, M.; Ghafouri-Fard, S. MicroRNA: A signature for cancer progression. Biomed. Pharmacother. 2021, 138, 111528. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xu, P.; Zou, Q.; Luo, H.; Yu, W. An epigenetic perspective on tumorigenesis: Loss of cell identity, enhancer switching, and NamiRNA network. Semin. Cancer Biol. 2022, 83, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Yin, G.; Zhang, Y.; Zhu, L.; Huang, H.; Lv, K. Recent advances in the functional explorations of nuclear microRNAs. Front. Immunol. 2023, 14, 1097491. [Google Scholar] [CrossRef]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.; Xi, Y.; Zhang, L.; Ding, C.; Luo, H.; Li, Y.; et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; Adiga, D.; Vethil Jishnu, P.; Chakrabarty, S.; Satyamoorthy, K. Cluster miRNAs and cancer: Diagnostic, prognostic and therapeutic opportunities. Wiley Interdiscip. Rev. RNA 2020, 11, e1563. [Google Scholar] [CrossRef]
- Mendes, N.D.; Freitas, A.T.; Sagot, M.F. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res. 2009, 37, 2419–2433. [Google Scholar] [CrossRef]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin structure analyses identify miRNA promoters. Genes. Dev. 2008, 22, 3172–3183. [Google Scholar] [CrossRef]
- Cui, W.; Meng, W.; Zhao, L.; Cao, H.; Chi, W.; Wang, B. TGF-beta-induced long non-coding RNA MIR155HG promotes the progression and EMT of laryngeal squamous cell carcinoma by regulating the miR-155-5p/SOX10 axis. Int. J. Oncol. 2019, 54, 2005–2018. [Google Scholar] [CrossRef] [PubMed]
- Saini, H.K.; Griffiths-Jones, S.; Enright, A.J. Genomic analysis of human microRNA transcripts. Proc. Natl. Acad. Sci. USA 2007, 104, 17719–17724. [Google Scholar] [CrossRef]
- Chien, C.H.; Sun, Y.M.; Chang, W.C.; Chiang-Hsieh, P.Y.; Lee, T.Y.; Tsai, W.C.; Horng, J.T.; Tsou, A.P.; Huang, H.D. Identifying transcriptional start sites of human microRNAs based on high-throughput sequencing data. Nucleic Acids Res. 2011, 39, 9345–9356. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Chen, L.; Wang, J.; Li, J.; Wingender, E. Identifying cell-specific microRNA transcriptional start sites. Bioinformatics 2016, 32, 2403–2410. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Tang, R.; Xu, X.; Wang, Z.; Xu, Q.; Chen, L.; Wingender, E.; Li, J.; Zhang, C.; Wang, J. mirTrans: A resource of transcriptional regulation on microRNAs for human cell lines. Nucleic Acids Res. 2018, 46, D168–D174. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhang, G.; Wang, Y.; Xu, K.; Chen, L.; Luo, G.; Xu, J.; Li, H.; Pei, D.; Zhao, X.; et al. CircGNB1 facilitates the malignant phenotype of GSCs by regulating miR-515-5p/miR-582-3p-XPR1 axis. Cancer Cell Int. 2023, 23, 132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, C.; Li, R.; Liu, J.; Wang, J.; Wang, T.; Wang, B. The BAP31/miR-181a-5p/RECK axis promotes angiogenesis in colorectal cancer via fibroblast activation. Front. Oncol. 2023, 13, 1056903. [Google Scholar] [CrossRef]
- Li, X.; Shahid, M.Q.; Wu, J.; Wang, L.; Liu, X.; Lu, Y. Comparative Small RNA Analysis of Pollen Development in Autotetraploid and Diploid Rice. Int. J. Mol. Sci. 2016, 17, 499. [Google Scholar] [CrossRef]
- Nagy, A.; Munkacsy, G.; Gyorffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Pidikova, P.; Reis, R.; Herichova, I. miRNA Clusters with Down-Regulated Expression in Human Colorectal Cancer and Their Regulation. Int. J. Mol. Sci. 2020, 21, 4633. [Google Scholar] [CrossRef]
- Ren, X.L.; He, G.Y.; Li, X.M.; Men, H.; Yi, L.Z.; Lu, G.F.; Xin, S.N.; Wu, P.X.; Li, Y.L.; Liao, W.T.; et al. MicroRNA-206 functions as a tumor suppressor in colorectal cancer by targeting FMNL2. J. Cancer Res. Clin. Oncol. 2016, 142, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.T.; Qian, F.; Fang, K.; Lin, K.Y.; Wang, W.T.; Chen, Y.Q. miR-133b, a muscle-specific microRNA, is a novel prognostic marker that participates in the progression of human colorectal cancer via regulation of CXCR4 expression. Mol. Cancer 2013, 12, 164. [Google Scholar] [CrossRef]
- Bandres, E.; Cubedo, E.; Agirre, X.; Malumbres, R.; Zarate, R.; Ramirez, N.; Abajo, A.; Navarro, A.; Moreno, I.; Monzo, M.; et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 2006, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Matouskova, P.; Hanouskova, B.; Skalova, L. MicroRNAs as Potential Regulators of Glutathione Peroxidases Expression and Their Role in Obesity and Related Pathologies. Int. J. Mol. Sci. 2018, 19, 1199. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Mitra, R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics 2009, 25, 2625–2631. [Google Scholar] [CrossRef]
- Qian, J.; Zhang, Z.; Liang, J.; Ge, Q.; Duan, X.; Ma, F.; Li, F. The full-length transcripts and promoter analysis of intergenic microRNAs in Drosophila melanogaster. Genomics 2011, 97, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Farre, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Rosello, L.; Alba, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Wu, X.; Li, R.; Song, Q.; Zhang, C.; Jia, R.; Han, Z.; Zhou, L.; Sui, H.; Liu, X.; Zhu, H.; et al. JMJD2C promotes colorectal cancer metastasis via regulating histone methylation of MALAT1 promoter and enhancing beta-catenin signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 435. [Google Scholar] [CrossRef]
- Ambros, V. microRNAs: Tiny regulators with great potential. Cell 2001, 107, 823–826. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Talukder, A.; Cha, M.; Li, X.; Hu, H. Computational annotation of miRNA transcription start sites. Brief. Bioinform. 2021, 22, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Sylvestre, Y.; De Guire, V.; Querido, E.; Mukhopadhyay, U.K.; Bourdeau, V.; Major, F.; Ferbeyre, G.; Chartrand, P. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2007, 282, 2135–2143. [Google Scholar] [CrossRef] [PubMed]
- Benhamou, D.; Labi, V.; Novak, R.; Dai, I.; Shafir-Alon, S.; Weiss, A.; Gaujoux, R.; Arnold, R.; Shen-Orr, S.S.; Rajewsky, K.; et al. A c-Myc/miR17-92/Pten Axis Controls PI3K-Mediated Positive and Negative Selection in B Cell Development and Reconstitutes CD19 Deficiency. Cell Rep. 2016, 16, 419–431. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Carlier, M.F.; Ducruix, A.; Pantaloni, D. Signalling to actin: The Cdc42-N-WASP-Arp2/3 connection. Chem. Biol. 1999, 6, R235–R240. [Google Scholar] [CrossRef]
- Qu, G.; Zhang, Y.; Duan, H.; Tang, C.; Yang, G.; Chen, D.; Xu, Y. ARPC5 is transcriptionally activated by KLF4, and promotes cell migration and invasion in prostate cancer via up-regulating ADAM17: ARPC5 serves as an oncogene in prostate cancer. Apoptosis 2023, 28, 783–795. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Wang, C.; Wu, Y.; Liu, J.; Wang, T.; Wang, B. BAP31-Mediated miR-206/133b Cluster Promotes Transendothelial Migration and Metastasis of Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 16740. https://doi.org/10.3390/ijms242316740
Zhang Q, Wang C, Wu Y, Liu J, Wang T, Wang B. BAP31-Mediated miR-206/133b Cluster Promotes Transendothelial Migration and Metastasis of Colorectal Cancer. International Journal of Molecular Sciences. 2023; 24(23):16740. https://doi.org/10.3390/ijms242316740
Chicago/Turabian StyleZhang, Qi, Changli Wang, Yufei Wu, Jingjing Liu, Tianyi Wang, and Bing Wang. 2023. "BAP31-Mediated miR-206/133b Cluster Promotes Transendothelial Migration and Metastasis of Colorectal Cancer" International Journal of Molecular Sciences 24, no. 23: 16740. https://doi.org/10.3390/ijms242316740




