Abstract
Propolis is a gelatinous substance processed by western worker bees from the resin of plant buds and mixed with the secretions of the maxillary glands and beeswax. Propolis has extensive biological activities and antitumor effects. There have been few reports about the antitumor effect of propolis against human cutaneous squamous cell carcinoma (CSCC) A431 cells and its potential mechanism. CCK-8 assays, label-free proteomics, RT–PCR, and a xenograft tumor model were employed to explore this possibility. The results showed that the inhibition rate of A431 cell proliferation by the ethanol extract of propolis (EEP) was dose-dependent, with an IC50 of 39.17 μg/mL. There were 193 differentially expressed proteins in the EEP group compared with the control group (p < 0.05), of which 103 proteins (53.37%) were upregulated, and 90 proteins (46.63%) were downregulated. The main three activated and suppressed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were extracellular matrix (ECM)-receptor interaction, amoebiasis, cell adhesion molecules (CAMs), nonalcoholic fatty liver disease (NAFLD), retrograde endocannabinoid signaling, and Alzheimer’s disease. The tumor volume of the 100 mg/kg EEP group was significantly different from that of the control group (p < 0.05). These results provide a theoretical basis for the potential treatment of human CSCC A431 cell tumors using propolis.
1. Introduction
Cutaneous squamous cell carcinoma (CSCC) is a malignant tumor originating from keratinocytes of the epidermis or appendages and the primary cause of death among nonmelanoma skin tumors [1]. In recent years, approximately 15–35 people in every 100,000 people have been diagnosed with CSCC, with an increase of 2–4% every year, which has become a worldwide threat to public health [2,3]. CSCC is mostly diagnosed in elderly patients, and the ratio of men to women is approximately 2~3:1. Most CSCC occurs in body parts, such as the head and face, exposed to ultraviolet light [4]. Currently, surgery, radiotherapy, and chemotherapy are the main treatment strategies. Surgery is an effective way to remove tumors, but it is not easy to detect cancer at an early stage. Chemotherapy and radiography treatments have side effects, such as damaging normal cells and drug resistance, which reduce the quality of life of patients. The search for alternative treatments is urgently needed.
Propolis (bee glue) is a kind of viscous substance processed by Western honeybee workers that collect mucilage, gums, resins, and lattice from plants, such as Pinus spp., Prunus spp., Acacia spp., Betula pendula, Aesculus hippocastanum, Salix alba, Baccharis dracunculifolia, and Dalbergia ecastaphyllum (L.) Taub., and the collections are mixed with the secretion of worker maxillary glands and beeswax [5,6]. The chemical composition of propolis, mainly including flavonoids, flavonols, flavanones, dihydroflavonoids, and phenylpropane derivatives, varies depending on the plant, geographical origin, and harvest season [7]. There are significant differences in color, active components, and harvesting tools of poplar propolis, red and green propolis [6]. Green Brazilian propolis is derived mainly from the leaf resin of Baccharis dracunculifolia, and red propolis is from the resin of Dalbergia ecastophyllum, while poplar propolis (brown color) is from the resin of Populus nigra L. [6]. The main active components of green propolis were derivatives of phenylpropanoids and diterpenes, chlorophyll, and small amounts of flavonoids, and those of red and poplar propolis were flavonoids [7]. Propolis has a wide range of biological activities, such as antibacterial, antifungal, antiviral, antiparasitic, antioxidant, antitumor, anti-inflammatory, anti-ulcer, and antidiabetic effects [5,8].
The antitumor activity of propolis has attracted much attention. In recent years, the antitumor effects of propolis on cancer and the relevant mechanisms have been reported in in vitro studies on colorectal, lung, breast, melanoma, gastric, lymphoma, tongue, and skin [7,8,9,10,11,12,13,14,15]. Many active ingredients in propolis, such as flavonoids and caffeic acid phenylethyl ester, inhibit tumor activity. Among them, caffeic acid phenylethyl ester has a specific and targeted killing effect on docetaxel-resistant prostate cancer cells [10]. Flavonoids block the cell cycle and then inhibit the proliferation of a human breast cancer cell line (MCF-7) [11]. There are selective antitumor effects of propolis on normal gingival fibroblasts and tongue cancer cells [12]. Red propolis has a cytotoxic effect on the breast cancer MDA MB-231 cell line, which is related to the PI3K/Akt and ERK1/2 pathways [13]. Propolis reduces mitochondrial membrane potential and changes the expressions of specific tumor suppressors (miR-34, miR-15a, and miR-16-5p) and carcinogenic (miR-21) miRNAs by increasing the levels of proapoptotic proteins (p21, Bax, p53, p53 Ser46, and p53 Ser15) in the MCF-7 cell line [14]. Brazilian propolis reduces the proliferation of a head and neck squamous cell carcinoma (HNSCC) cell line [15]. The antitumor effect of poplar propolis on CSCC A431 cells and its potential mechanism are unclear.
In this study, Cell Counting Kit-8 (CCK-8) assays, label-free proteomics, RT–PCR, and a xenograft tumor nude mouse model were employed to determine the effect of propolis on the proliferation, differentially expressed proteins, related pathways in A431 cells, and tumor volume in an animal xenograft tumor model.
2. Results
2.1. Components of Ethanol Extract of Propolis
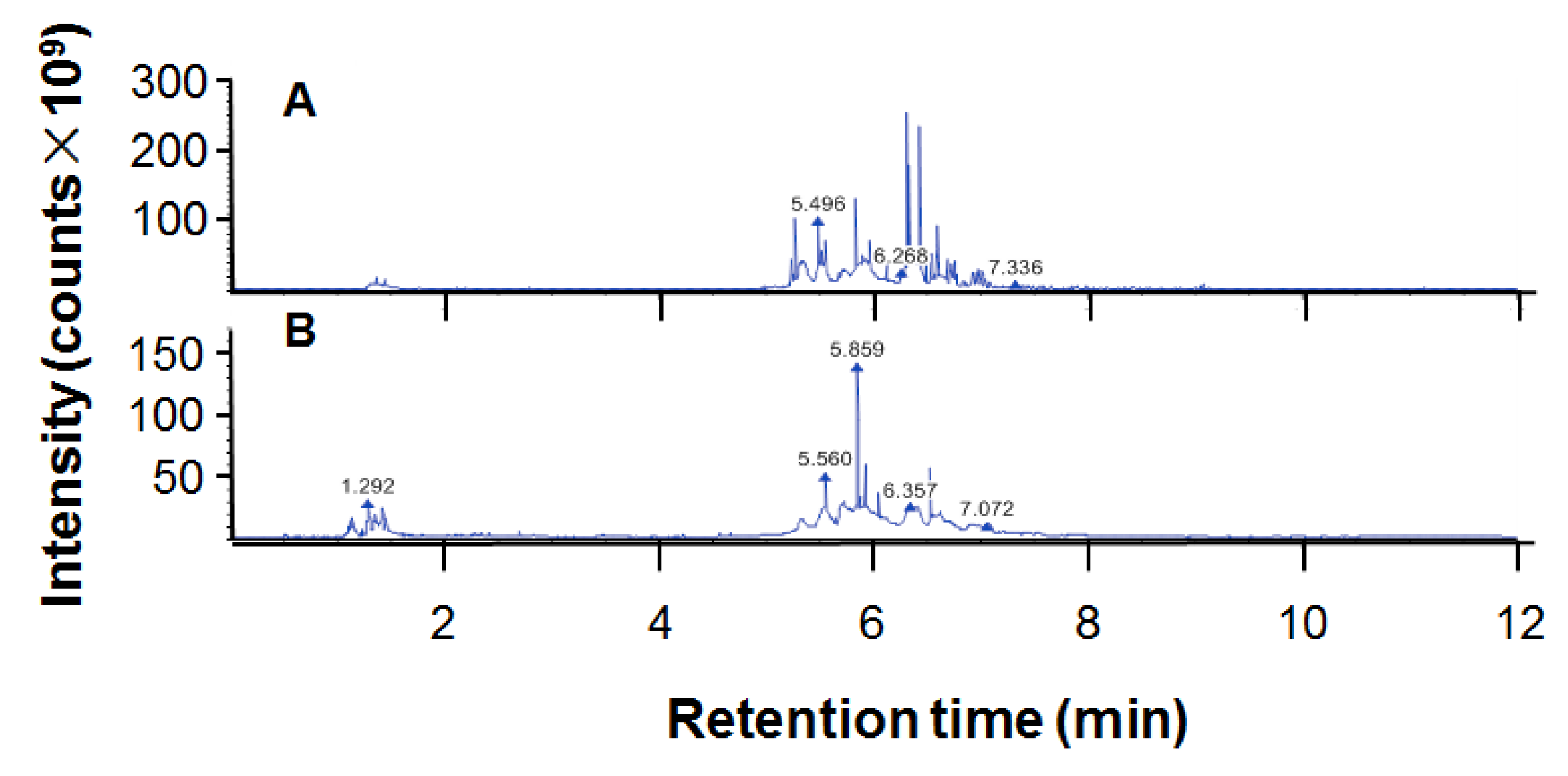
The total flavonoid content of ethanol extract of propolis (EEP) was 32.04 ± 0.89/100 g. The chromatogram for UPHLC-MS/MS of propolis is shown in Figure 1. The 214 chemical components of EEP dissolved in methanol are presented in Table 1.
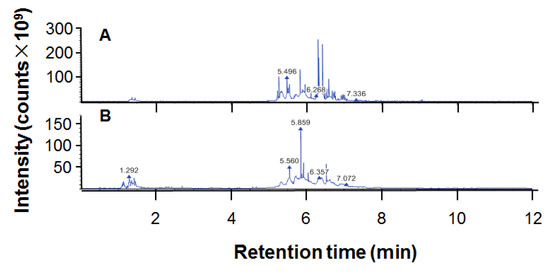
Figure 1.
The chromatogram of ethanol extract of propolis (EEP) for UPHLC-MS/MS: (A) negative polarity mode; (B) positive polarity mode.

Table 1.
Chemical components in ethanol extract of propolis (EEP) (full matched mzCloud).
2.2. The Antitumor Effect of Ethanol Extract of Propolis (EEP)
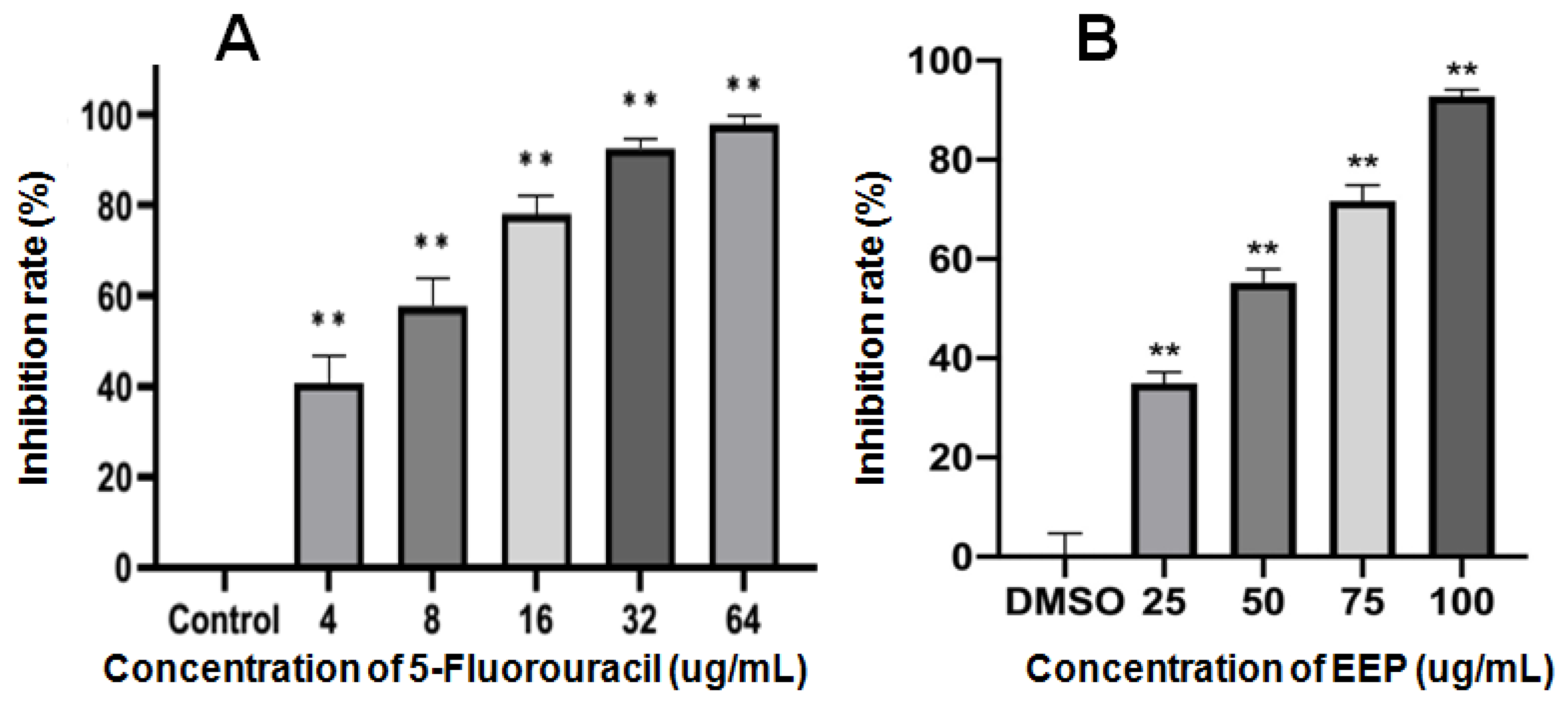
There were no significant differences between the viability of A431 cells in the solvent control group and the blank control group. EEP inhibited the proliferation of A431 cells. DMSO (0.05%) had no effect on A431 cells. The inhibition rates are shown in Figure 2. The IC50 of 5-FU and EEP against A431 cells for 48 h was 6.57 and 39.17 µg/mL, respectively.

Figure 2.
Inhibitory rates of 5-Fluorouracil (A) and ethanol extract of propolis (EEP) (B) against the proliferation of A431 cells for 48 h (** indicates significant differences compared with the solvent control group. p < 0.01).
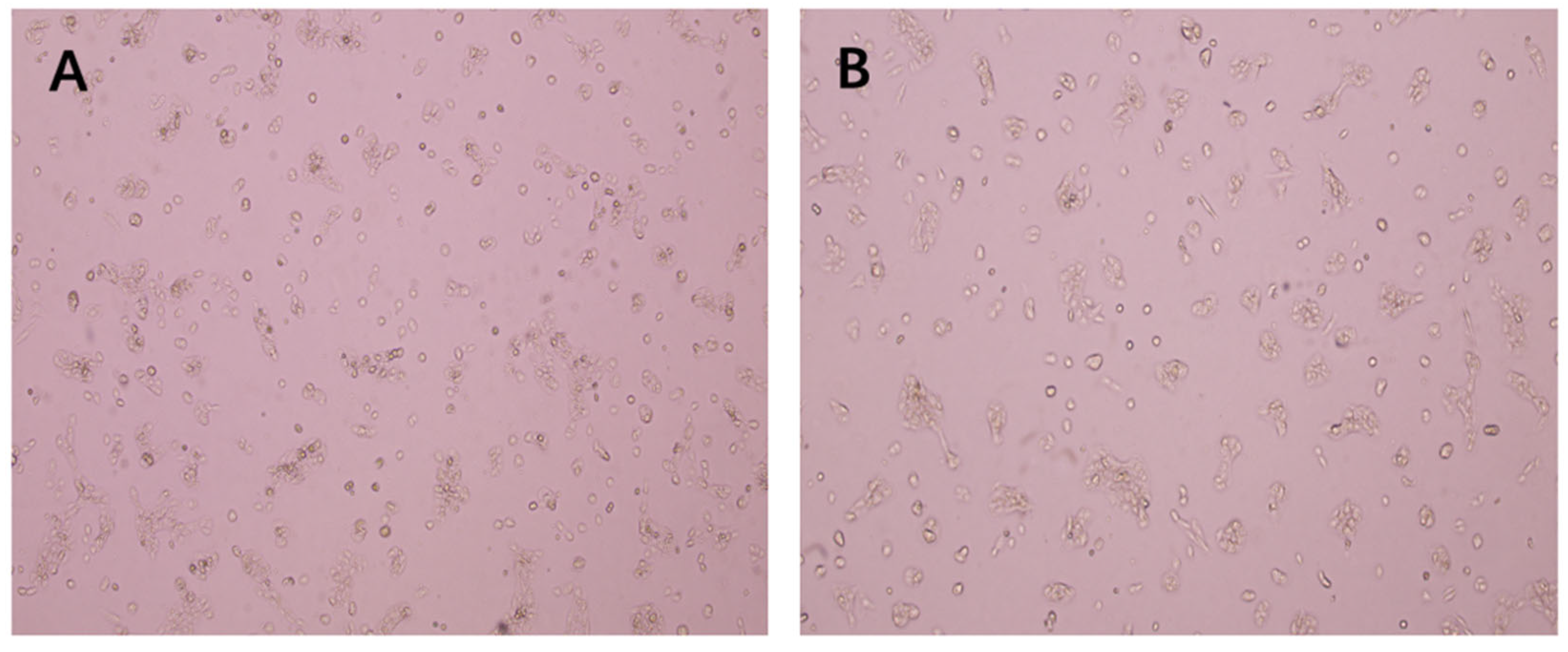
The morphology of A431 cells after EEP treatment for 48 h is shown in Figure 3. The growth of the cells in the control group was normal, and the treated cells were irregular in shape and distributed in sheets. The number of cells treated with EEP in the microscope field of view decreased, and the cell adhesion ability was weakened with some floating cells.

Figure 3.
Effect of ethanol extract of propolis (EEP) on the morphology of A431 cells (100×): (A): Control, (B) IC50 EEP group.
2.3. The Differentially Expressed Proteins
There were 103 upregulated and 90 downregulated differentially expressed proteins (DEPs) between the EEP group and the control group (screened by FC > 2.0 or FC < 0.5 and p < 0.05). Partial DEPs (p < 0.01) are shown in Table 2.

Table 2.
Partial significant (p < 0.01) differentially expressed proteins (DEPs) between the ethanol extract of propolis (EEP) group and the control group (screened by FC > 2.0 or FC < 0.5 and p < 0.05).
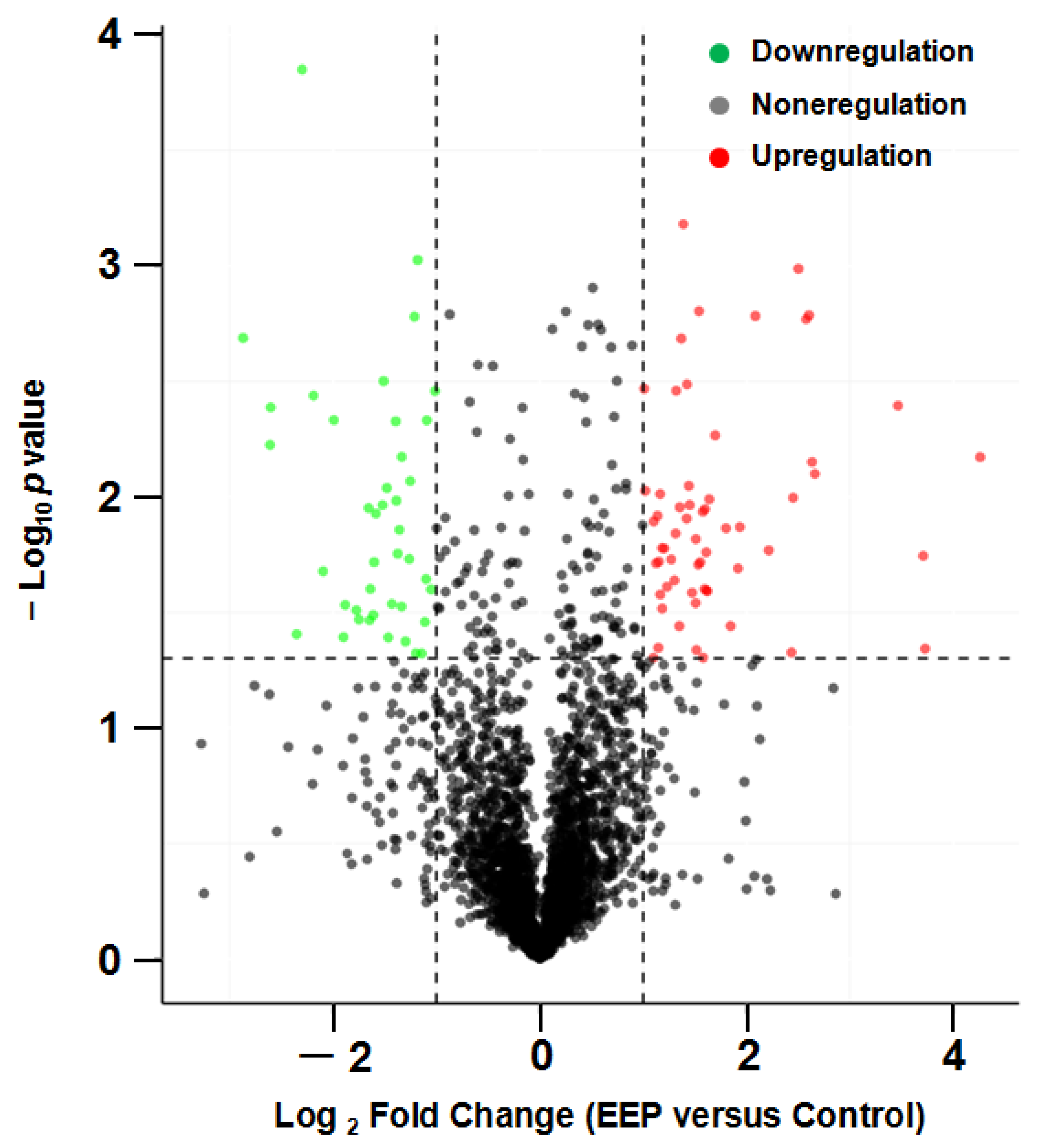
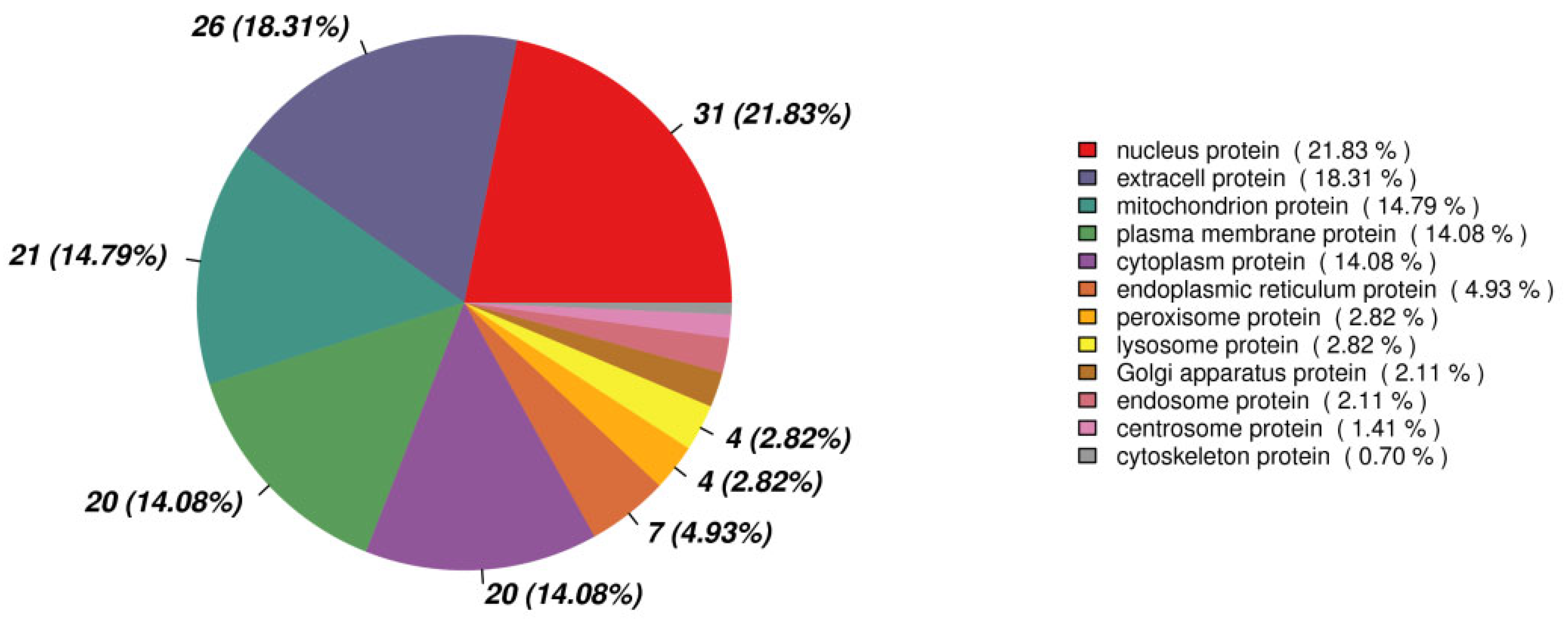
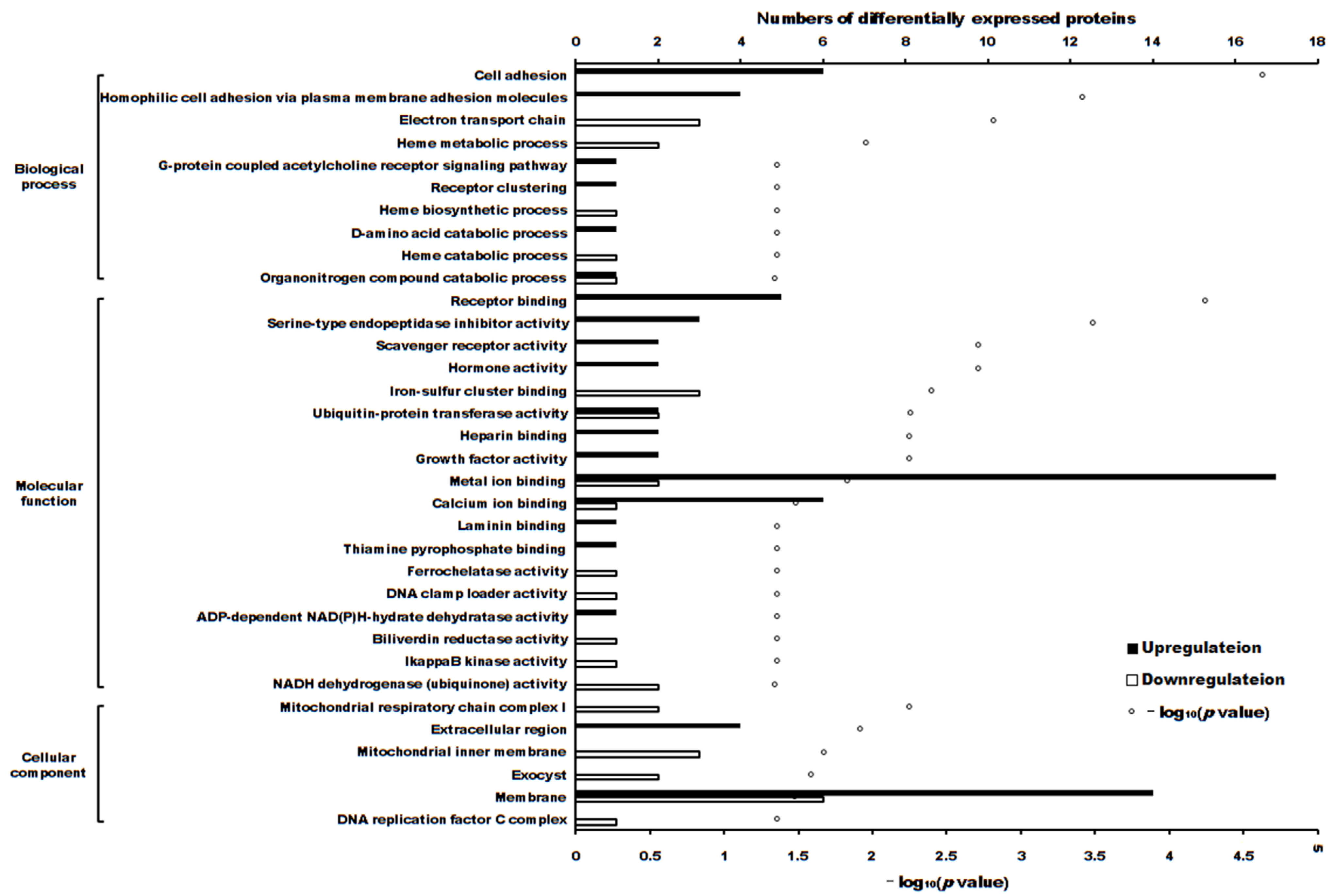
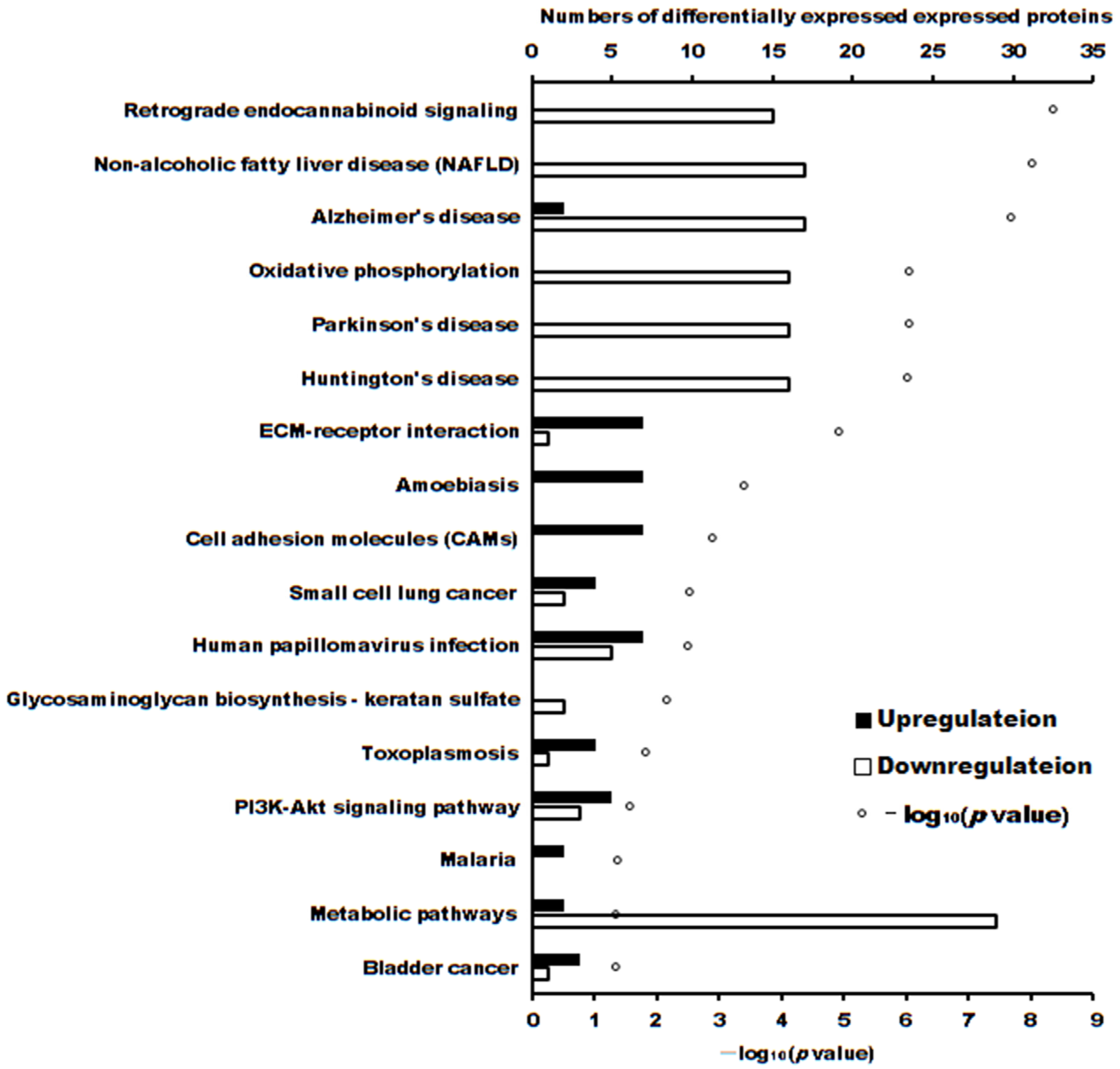
The volcano plot of proteins in the two groups is shown in Figure 4. The subcellular localization of the DEPs is shown in Figure 5. The DEPs subjected to Gene Ontology (p < 0.05) and Kyoto Encyclopedia of Genes and Genomes (p < 0.05) analyses are shown in Figure 6 and Figure 7, respectively.

Figure 4.
Volcano plot of proteins in A431 cells treated with ethanol extract of propolis (EEP) versus control cells.
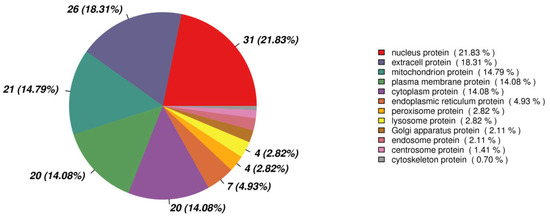
Figure 5.
The subcellular localization of the differentially expressed proteins.
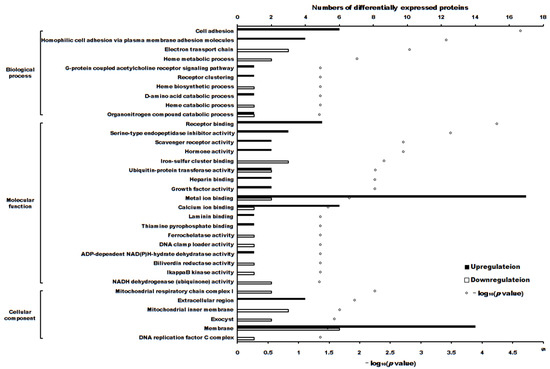
Figure 6.
The differentially expressed proteins enriched in Gene Ontology terms (p < 0.05).
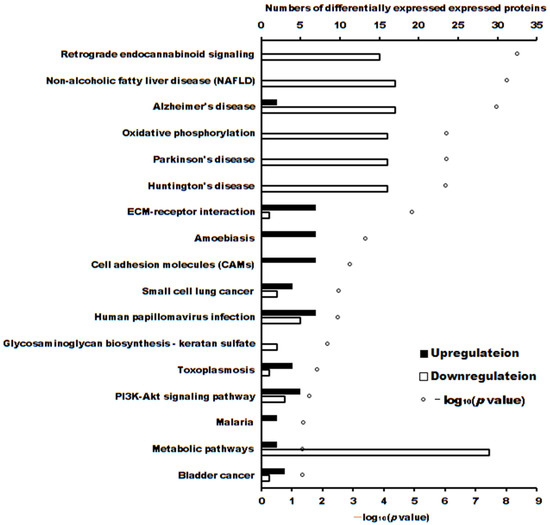
Figure 7.
The number of differentially expressed proteins enriched in Kyoto Encyclopedia of Genes and Genomes (p < 0.05).
These DEPs played roles in different pathways. The significantly enriched pathways (p < 0.05) of upregulated and downregulated proteins were separately analyzed, as shown in Table 3.

Table 3.
The significantly enriched pathways (adjusted p < 0.05) of differentially expressed proteins.
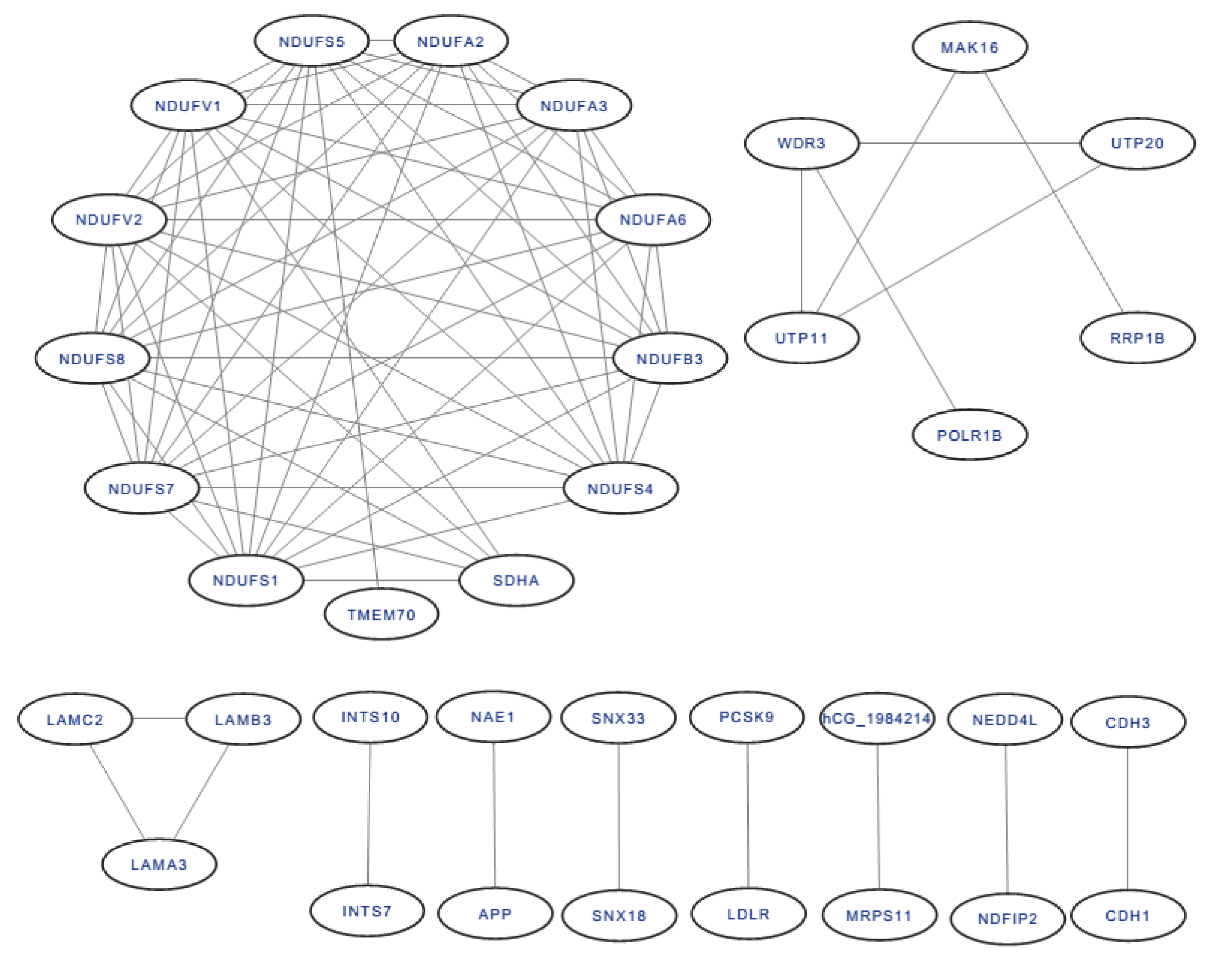
All of the DEP interactions are shown in Figure 8. Among the interacting proteins, NADH dehydrogenase [ubiquinone] flavoprotein 2 (mitochondrial), mitochondrial NADH-ubiquinone oxidoreductase 75 kDa subunit, NADH dehydrogenase [ubiquinone] flavoprotein 1 (mitochondrial), NADH dehydrogenase (ubiquinone) Fe–S protein 5, 15 kDa (NADH-coenzyme Q reductase), NADH dehydrogenase [ubiquinone] iron–sulfur protein 8 (mitochondrial), and NADH dehydrogenase [ubiquinone] iron–sulfur protein 7 (mitochondrial) had the most protein interactions, with 11 DEPs.

Figure 8.
The protein-protein interaction of DEPs.
2.4. Relative Gene Expression
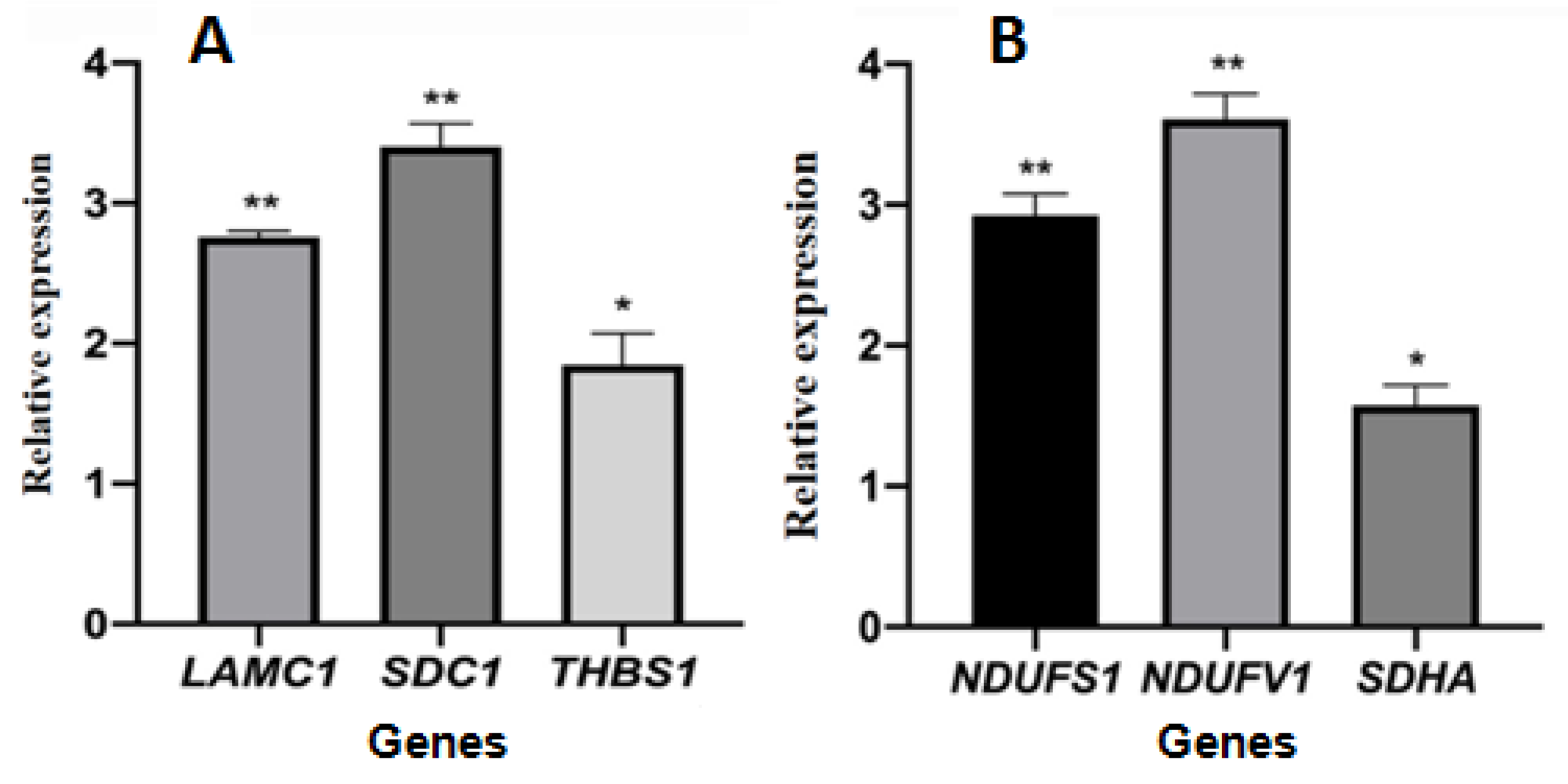
The cycle threshold (ct) data of selected genes is shown in Supplementary Table S1. The relative gene expression levels of selected genes encoding differentially expressed proteins are shown in Figure 9. The expression levels of the three genes LAMC1, SDC1, and THBS1 involved in the ECM-receptor interaction pathway were significantly upregulated in the treated group compared with the control group, and the gene expression was consistent with the expression of proteins. The expression levels of three genes, NDUFS1, NDUFV1, and SDHA, involved in the oxidative phosphorylation pathway, were significantly upregulated and inconsistent with the expression of proteins.
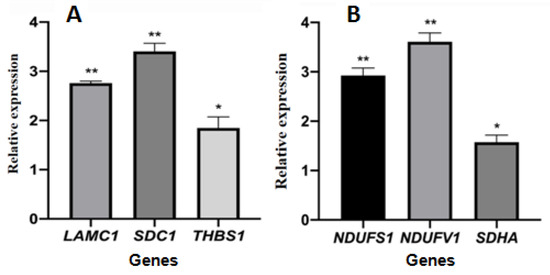
Figure 9.
The relative gene expression of selected genes encoding differentially expressed proteins. (A): genes coded proteins enriched in ECM-receptor interaction pathway; (B): genes coded proteins enriched in oxidative phosphorylation pathway. The symbols * and ** indicates significant differences compared with the solvent control group, p < 0.05 or p < 0.01, respectively.
2.5. The Effect of EEP on A431 Cell Xenograft Tumors in Nude Mice
The tumor volumes of nude mice in the control group, solvent group, 50 mg/kg propolis group, and 100 mg/kg propolis group after 12 days of gavage are shown in Table 4. There was a significant difference in the 100 mg/kg propolis group compared with the control group (p < 0.05), which indicated that the 100 mg/kg propolis group had in vivo inhibitory effects on A431 cell tumors.

Table 4.
Volume of A431 cell xenograft tumors in nude mice (mm3, n = 5).
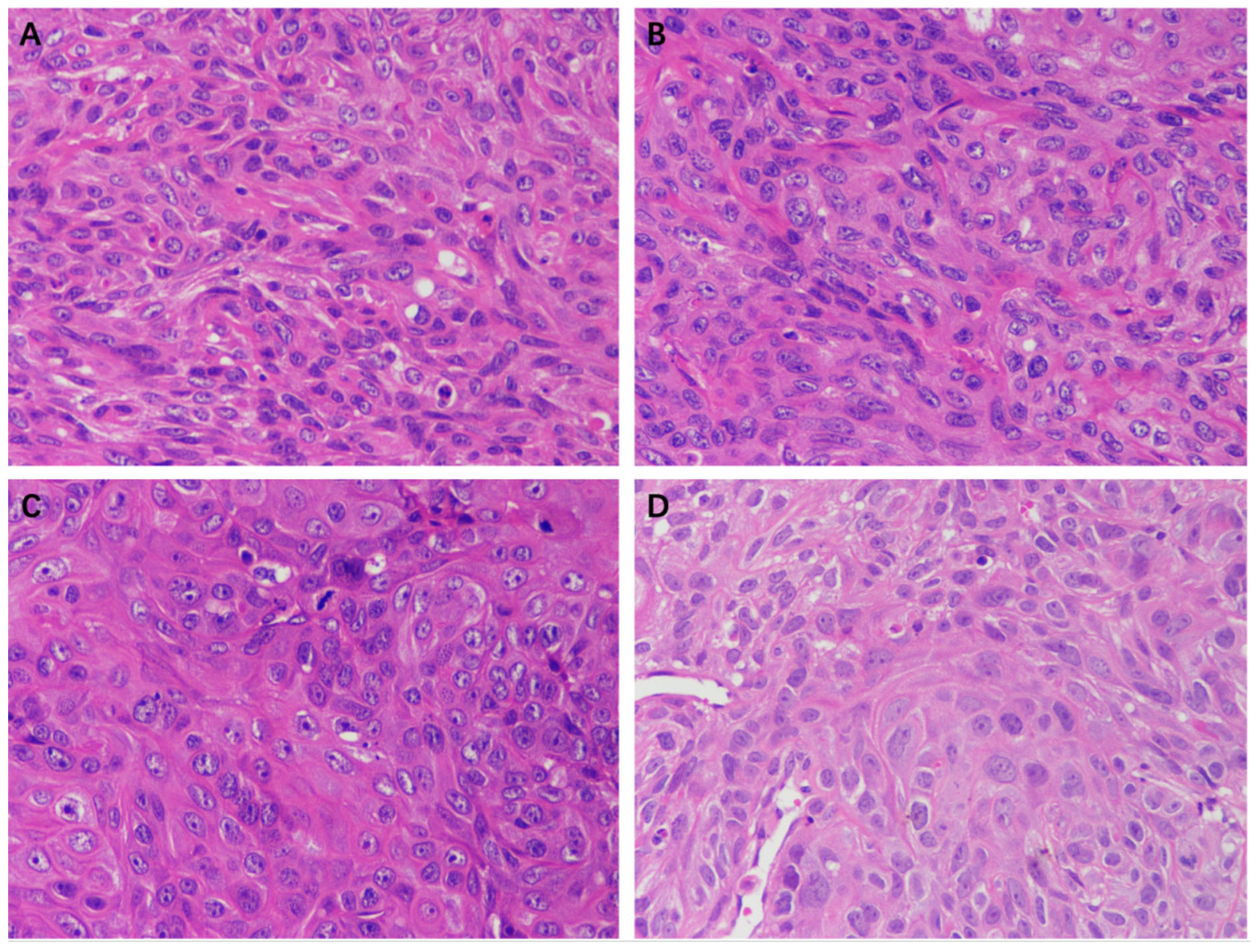
The HE staining results of the tumor tissue of the EEP, solvent control, and control groups are shown in Figure 10. A large number of tumor cells were observed in each group except in the 100 mg/kg EEP group. The morphology and size of cells varied and exhibited atypia, with enlarged nucleoli and unclear cell spacing. There was a small amount of cell necrosis in the control group, solvent control group, and 50 mg/kg group, while a large amount of cell necrosis was observed in the 100 mg/kg EEP group.
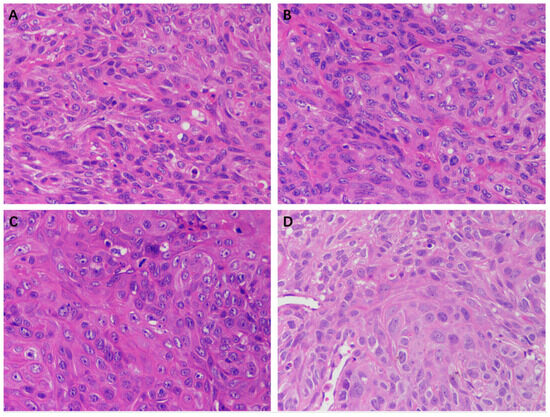
Figure 10.
HE staining of tumors in nude mice of different groups (400×). (A) Control group, (B) Solvent control group, (C) 50 mg/kg EEP group, and (D) 100 mg/kg EEP.
3. Discussion
Propolis exhibits antitumor activity against different cell lines. Brazilian red propolis (from Brejo Grande, Brazil) can inhibit the growth of cancer cells, and after 24 h of treatment, the IC50 values against Hep-2 cells and HeLa cells were 63.48 ± 3.30 μg/mL and 81.40 ± 6.40 μg/mL, respectively [16]. EEP (from Ardabil, Iran) has dose-dependent toxic effects on both KB and A431 cancer cells. The IC50 values of EEP in the KB cell line and A431 cell line were 40 ± 8.9 μg/mL and 98 μg/mL, respectively, after 48 h of incubation [17]. The IC50 values of EEPs (from Podlasie, Masovia, and West Pomerania; Poland) against tongue cancer cells treated for 24 h were approximately 88 µg/mL, 110 µg/mL, and 150 µg/mL, respectively [12]. The IC50 values of EEPs range from 26.33 to 143.09 μg/mL against the human colon cancer cell line HCT-16 [18]. The IC50 values of EEP (from Phayao, Chiang Mai, and Nan, Thailand) against A549 cells were 106 ± 0.004 µg/mL, 199 ± 0.009 µg/mL, and 87 ± 0.012 µg/mL, respectively, and for HeLa cells were 81 ± 0.006 µg/mL, 116 ± 0.023 µg, and 54 ± 0.005 µg/mL, respectively [19]. The IC50 of EEP (from Hebei Province, China) against the 5 × l05/mL DLBCL SU-DHL-2 cell line for 24 h was 5.729 μg/mL [9]. In this study, the IC50 of EEP (same as [9]) against A431 cells for 48 h was 39.17 μg/mL (Figure 3). These different median lethal doses against tumor cell lines may be related to the type of cancer cells, concentration of cancer cells, incubation duration, botanical origin of propolis, extraction process of propolis, and storage of propolis.
The cytotoxicity mechanism of propolis against A431 tumor cells was different. Proteins play important roles in the proliferation of cells. Label-free proteomics is commonly used to explore DEPs in cells subjected to different treatments [20,21,22]. In this manuscript, there were 103 upregulated and 90 downregulated DEPs between treatment and control cells. GO enrichment and KEGG enrichment analysis (as shown in Figure 6 and Figure 7) showed that the main upregulated proteins enriched in the ECM-receptor interaction and cell adhesion molecule (CAM) pathways inhibits the ability of A431 cells to metastasize and invade. The downregulated proteins were mainly enriched in adenosine triphosphate (ATP) production by downregulating the main proteins of the retrograde endocannabinoid signaling and oxidative phosphorylation pathways, thereby reducing adenosine triphosphate (ATP) production and inhibiting the proliferation of A431 cells.
The most significantly enriched pathway of the upregulated DEPs was the ECM-receptor interaction pathway. The interaction between tumor cells and extracellular matrix (ECM) components, such as laminin, fibronectin, and collagen, plays a crucial role in tumor invasion and metastasis. The genes LAMC1, SDC1, and THBS1, which are involved in the ECM-receptor interaction pathway, were upregulated. Similar results were also found in gastric cancer, in which the upregulated genes were COL1A2 and COL6A3 [23] or COL6A3, COL3A1, and COL1A1 [24]. There were seven upregulated proteins involved in the ECM-receptor interaction pathway, which is associated with an enhanced migratory/invasive capacity of ovarian cancer cells [25] and non-small cell lung cancer tumors [26,27]. Most of the upregulated DEPs, except Thrombospondin 1, Agrin, and Syndecan-1, involved in the ECM-receptor interaction pathway were also enriched in the amoebiasis pathway. Differentially expressed genes related to cervical cancer were enriched in amoebiasis and other pathways [28,29]. Similar differentially expressed genes were also found in colorectal cancer cells with cetuximab insensitivity [30]. The differentially expressed proteins were involved in amoebiasis pathways of non-small cell lung cancer [26] and gastric cancer tumors of the patients [27].
Another important pathway of the upregulated DEPs was the amoebiasis pathway. The amoebiasis pathway was significantly enriched and identified as one of the important processes or signaling pathways of melanoma metastasis [31,32], the pathogenesis of nasopharyngeal carcinoma [33], carcinogenesis and pathogenesis of cervical cancer [28], breast cancer [34], and gastric cancer [35] by bioinformatics analysis based on the Gene Expression Omnibus database. Our result is in accordance with these previous scientific studies.
The third pathway of the upregulated DEPs was the cell adhesion molecule (CAM) pathway. CAMs, having four main groups including cadherins, integrins, selectins, and immunoglobulins, are primarily glycoproteins on cell surface membranes and can promote homeostasis between cells and between cells and the extracellular matrix. With higher levels of neural cell adhesion molecule expression, neuroblastoma cells have more intense homophilic tumor binding [36]. Knockdown of E-cadherin and cell adhesion molecule 1-related genes decreased cell growth, migration, and cell-to-cell adhesion of BAP1-mutant uveal melanoma cells [37]. High expression of prostaglandin F2 receptor inhibitor (PTGFRN), a type I (single pass) transmembrane Ig superfamily of CAM, could protect cells from apoptosis, thereby promoting growth and migration in glioblastoma cells [38]. It was also shown that the CAM pathway is one of the key processes or signaling pathways of melanoma metastasis and the pathogenesis of nasopharyngeal carcinoma [31,32,33]. CAMs may be the stress response to adverse factors on cancer cells.
The most significantly downregulated pathway was the nonalcoholic fatty liver disease (NAFLD) pathway. NAFLD affected the cell cycle and p53 pathways. SNORA71A knockdown in HT-29 cells led to significant inhibition of cell migration and invasion ability, which targeted LBP to participate in NAFLD in colorectal cancer cells [39]. Corosolic acid inhibited NAFLD-related hepatocellular carcinoma progression by downregulating the NAFLD pathway [40]. Levodopa downregulates oxidative phosphorylation (OXPHO), NAFLD, and Parkinson’s disease-related pathways to inhibit esophageal squamous cell carcinoma cells [41]. Similar results were also obtained in this experiment, in which the OXPHO and Parkinson’s disease-related pathways were also suppressed. OXPHO mainly occurs in the inner mitochondrial membrane of eukaryotic cells or the cytoplasm of prokaryotic organisms [42]. Many tumors require energy from the mitochondria for biosynthesis to synthesize ATP through OXPHO [43]. The average contribution of OXPHOS to ATP generation in normal cells is 80% and 83% in cancer cells [44]. Triple-negative breast cancer (TNBC) cells can be inhibited by a decrease in OXPHOS caused by OXPHOS inhibitors, which causes ATP deficiency that cannot be fully compensated by other mechanisms [45]. Oxidative phosphorylation also acts on other cancer cells, such as liver cancer [46], rectal cancer [47], and pancreatic cancer cells [48]. OXPHO is now widely used as a therapeutic target in cancer [49].
Another suppressed pathway was retrograde endocannabinoid signaling. This pathway was also suppressed in retinoblastoma [50], glioblastoma [51], and glioma patients [52]. The metabolites were also enriched in the retrograde endocannabinoid signaling pathway in breast cancer cells treated with Faecalibacterium prausnitzii [53]. This pathway was also mainly enriched in DEPs and DEGs related to nonfunctional pituitary adenoma [54]. The DEPs between the tumor and adjacent healthy tissue of patients with diffuse gastric cancer and those of patients with advanced gastric cancer were enriched in this pathway [55]. Other suppressed pathways were Alzheimer’s disease-related pathways, Huntington’s disease-related pathways, metabolic pathways, and glycosaminoglycan biosynthesis—keratan sulfate. The proteins involved in these pathways were downregulated and inhibited the growth of A431 cells.
The expression levels of genes subjected to RT–PCR were not completely consistent with the protein expression levels. Some studies have pointed out that transcription levels alone are not sufficient to predict protein levels in many cases [56,57] because the protein concentration is affected by both transcription and translation. Cancer-related genetic changes can affect proteins involved in nearly all levels of transcriptional control [58].
This experiment showed that EEP inhibited tumor growth in nude mice. It was also found that the growth of gastric pyloric tumors and colorectal cancer was inhibited by a diet containing propolis [59,60]. Propolis has immunological enhancement activity [61], which can enhance the immune system of mice and then nonspecificly inhibit tumor growth.
The limitation of this study is that metabonomics, cancer stem cell, molecular docking, or other methods can be employed to explore more accurately the regulation of the EPP antitumor approach against the A431 cells for new drug development. More research about the cell cycle of A431 cells inhibited by EEP and the active components, such as caffeic acid, dihydro cinnamic acid, and p-coumaric acid, or others, who play the main antitumor effect can be determined in the future.
4. Materials and Methods
4.1. Propolis Samples and Its Chemical Components Determination
The crude poplar propolis sample and extraction procedure of ethanol extract of propolis (EEP) were the same as in our previous report [9].
The total flavonoid content determination of EEP was performed using the spectrophotometer method according to the national standards for propolis in China (GB/T 24283-2018)[62]. Rutin was used as a standard substance to determine the absorbance of the samples at 510 nm.
The chemical components of EEP were determined using a UHPLC-MS/MS system (Vanquish UHPLC system (Thermo Fisher Scientific Inc., Germering, Germany) coupled with an Orbitrap Q ExactiveTMHF-X mass spectrometer (ThermoFisher, Germering, Germany)) by Untargeted Metabolomics mothed and performed by Novogene Co., Ltd. (Beijing, China). Simply, The EEP samples (1 mL) were resuspended with prechilled 80% methanol by well vortex and then incubated on ice for 5 min and centrifuged at 15,000× g, 4 °C for 15 min. The supernatant was diluted to 53% methanol final concentration by LC-MS grade water. The solution was transferred to an Eppendorf tube and subsequently centrifuged at 15,000× g, 4 °C for 15 min. The elution conditions and processes were the same as in reference [63]. Q ExactiveTM HF-X mass spectrometer was operated in positive/negative polarity mode with a spray voltage of 3.5 kV, capillary temperature of 320 °C, sheath gas flow rate of 35 psi, and aux gas flow rate of 10 L/min, S-lens RF level of 60, Aux gas heater temperature of 350 °C.
4.2. Antitumor Bioassay
The human skin squamous cell carcinoma A431 cell line (purchased from Wuhan Purosai Life Sciences Co., Ltd., Wuhan, China) was cultured in a special complete culture medium (Wuhan Purosai Life Sciences Co., Ltd.) in a 5% CO2 humidified incubator at 37 °C (C150, Binder, Tuttlingen, German).
EEP (0.1 g) was dissolved in 0.5 mL DMSO and diluted with a complete culture medium at a concentration of 1 mg/mL. Propolis solution was diluted with a complete medium to 100, 75, 50, and 25 µg/mL. DMSO (0.05%, v/v; equal to DMSO in the 100 µg/mL propolis group) was added to the complete medium as a solvent control.
The A431 cells were irrigated with PBS buffer (pH 7.2–7.4, Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) twice and then digested with 1 mL 0.25% trypsin solution (HyClone, Thermo Scientific, Waltham, UT, USA). The digested solution was centrifuged at 137× g for 5 min. The sediment was suspended in 2 mL of complete culture medium. The concentration was determined via 0.4% trypan blue staining (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). At a beginning concentration of 5 × 104 cells/mL, 100 µL cell suspension was added in 96-well plates with 6 repetitions per treatment. After 48 h of cell culture with EEP 100, 75, 50, and 25 µg/mL, and 0.05% DMSO, the viability of cells was determined by a CCK8 kit (DOJINDO, Kumamoto, Japan) at 450 nm using a microplate reader (1510, Thermo Fisher Waltham, MA, USA). The IC50 of EEP on A431 cells for 48 h was calculated using GraphPad Prism 8.4.3 for Windows (GraphPad Software, Inc., La Jolla, CA, USA). 5-Fluorouracil (HPLC grade; purchased from Sigma-Aldrich Co., St. Louis, MO, USA) at 0, 4, 8, 16, 32, and 64 µg/mL were employed to determine the IC50 value against A431 cells.
A431 cells in the logarithmic growth phase were digested with trypsin and seeded in 6-well plates at 1.5 × 105 cells/well. After 24 h of cell culture, the cells were treated with control, and IC50 EEP solution for 48 h, and then the morphological changes of the cells were observed using an inverted microscope (TS-100f, Nikon, Tokyo, Japan).
4.3. Differentially Expressed Proteins in A431 Cells Treated with Propolis
The A431 cells were treated as described for the morphology observation experiment, which was also treated with control and IC50 EEP solution. After these cells were treated with propolis or non-propolis for 48 h, the culture medium was removed. Then, the cells were irrigated twice with precooled PBS buffer, digested, and irrigated twice again. Cells were collected in a centrifuge tube (1.5 mL) after centrifugation. Therefore, these cells were frozen in liquid nitrogen and further stored in a refrigerator at −80 °C (Haier Biomedical, Qingdao, China).
The extraction and concentration determination of total proteins and spectra of proteins were performed as described in our previous report [9].
4.4. Detection of Relative Gene Expression
According to the proteomics results, the genes coded NDUFS1, NDUFV1, and SDHA proteins involved in oxidative phosphorylation and LAMC1, SDC1, and THBS1 ECM-receptor interaction were selected to determine the gene expression levels between the control and IC50 EEP groups using RT–PCR assay. The primers were designed through NCBI’s free online primer design platform, which is given in Supplementary Table S2. The internal reference gene was β-actin.
4.5. Xenograft Tumor Nude Mice
BALB/C male nude mice of SPF grade, 5 weeks old, weighing 18–20 g (purchased from Shanghai Jihui Experimental Animal Breeding Co., Ltd., Shanghai, China) were experimental animals. The A431 cell suspension (1 × 107 cell/mL) (0.1 mL) was injected subcutaneously into the axilla of the right forelimb of nude mice. The small nodules at the inoculation site mean the heterologous tumor model in nude mice was successful. After 1 week, the tumor sizes were 4–7 mm3. Then, 20 nude mice were randomly divided into 4 groups: the control, solvent, 50 mg/kg EEP, and 100 mg/kg EEP groups (according to [64]). They were intragastrically administered 0.2 mL PBS buffer solution, 10% PEG-400 solution, 50 mg/kg, and 100 mg/kg. The tumor volumes of each mouse were measured every 2 days.
These nude mice were sacrificed after the final treatment. The tumor tissue was immediately peeled off with scissors and tweezers, washed with normal saline, and fixed in 4% paraformaldehyde (Biosharp, Labgic Technology Co., Ltd., Beijing, China) for paraffin section preparation and hematoxylin-eosin staining (HE, Wuhan Servicebio Technology Co., Ltd., Wuhan, China).
4.6. Data Analysis
All experiments were performed in triplicate. All these data are expressed as the mean ± standard error. One-way ANOVA was used to analyze the significance of differences (p < 0.01: extremely statistically significant differences between treatment and control groups, p < 0.05: statistically significant differences). The relative gene expression was represented by the ratio of gene expression in propolis-treated cells to that of control cells. Differences in tumor volumes among groups were analyzed using repeated-measures ANOVA using Stat View 5.0.1 (SAS Institute Inc. 1992–1998, Cary, NC, USA).
The spectra obtained from label-free proteomics by LC-MS/MS were analyzed as described in our previous report [9].
The raw data files generated from Untargeted Metabolomics by UHPLC-MS/MS were processed using the Compound Discoverer 3.1 (CD3.1, ThermoFisher) to perform peak alignment, peak picking, and quantitation for each metabolite, whose main parameters were set as follows: retention time tolerance, 0.2 min; actual mass tolerance, 5 ppm; signal intensity tolerance,30%; signal/noise ratio, 3; and minimum intensity.
5. Conclusions
A431 cancer cells can be inhibited by poplar propolis via the main pathways enriched DEPs were ECM-receptor interaction, amoebiasis, cell adhesion molecules (CAMs), nonalcoholic fatty liver disease (NAFLD) related pathway, retrograde endocannabinoid signaling, and other pathways. The inhibition effect was also found in a xenograft tumor for nude mice. Poplar propolis has the potential to be a new treatment strategy for CSCC patients.
Supplementary Materials
The supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms242316753/s1.
Author Contributions
Conceptualization, C.Z. and W.Y.; methodology, W.Y.; data curation, C.Z., Y.T. and X.L.; formal analysis, C.Z., A.Y. and W.T.; writing—original draft preparation, Y.T. and C.Z.; writing—review and editing, W.Y.; supervision, W.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The animal study protocol was approved by the Ethics Committee of Fujian Agriculture and Forestry University (protocol code PZCASFAFU23074).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within this article and supplementary materials.
Acknowledgments
Thanks for the help from Xin Lin during the performance of the protocol.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tufaro, A.P.; Chuang, J.C.M.; Prasad, N.; Chuang, A.; Chuang, T.C.; Fischer, A.C. Molecular markers in cutaneous squamous cell carcinoma. Int. J. Surg. Oncol. 2011, 12, 231475. [Google Scholar] [CrossRef] [PubMed]
- Cleavenger, J.; Johnson, S.M. Non melanoma skin cancer review. J. Ark. Med. Soc. 2014, 110, 230–234. [Google Scholar] [PubMed]
- Lomas, A.; Leonardi-Bee, J.; Bath-Hextall, F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br. J. Dermatol. 2012, 166, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Halim, A.S.; Ramasenderan, N. High-risk cutaneous squamous cell carcinoma (CSCC): Challenges and emerging therapies. Asian J. Surg. 2023, 46, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Altabbal, S.; Athamnah, K.; Rahma, A.; Wali, A.F.; Eid, A.H.; Iratni, R.; Al Dhaheri, Y. Propolis: A detailed insight of its anticancer molecular mechanisms. Pharmaceuticals 2023, 16, 450. [Google Scholar] [CrossRef]
- Bobiş, O. Plants: Sources of diversity in propolis properties. Plants 2022, 11, 2298. [Google Scholar] [CrossRef]
- Forma, E.; Bryś, M. Anticancer activity of propolis and its compounds. Nutrients 2021, 13, 2594. [Google Scholar] [CrossRef]
- Pasupuleti, V.R.; Sammugam, L.; Ramesh, N.; Gan, S.H. Honey, propolis, and royal jelly: A comprehensive review of their biological actions and health benefits. Oxid. Med. Cell. Longev. 2017, 2017, 1259510. [Google Scholar] [CrossRef]
- Liu, X.; Tian, Y.; Yang, A.; Zhang, C.; Miao, X.; Yang, W. Antitumor effects of poplar propolis on DLBCL SU-DHL-2 cells. Foods 2023, 12, 283. [Google Scholar] [CrossRef]
- Fu, Y.K.; Wang, B.J.; Tseng, J.C.; Huang, S.H.; Lin, C.Y.; Kuo, Y.Y.; Hour, T.C.; Chuu, C.P. Combination treatment of docetaxel with caffeic acid phenethyl ester suppresses the survival and the proliferation of docetaxel-resistant prostate cancer cells via induction of apoptosis and metabolism interference. J. Biomed. Sci. 2022, 29, 16. [Google Scholar] [CrossRef] [PubMed]
- Gui, C.; Zhang, C.; Xiong, X.; Huang, J.; Xi, J.; Gong, L.; Huang, B.; Zhang, X. Total flavone extract from Ampelopsis megalophylla induces apoptosis in the MCF-7 cell line. Int. J. Oncol. 2021, 58, 409–418. [Google Scholar] [CrossRef]
- Wezgowiec, J.; Wieczynska, A.; Wieckiewicz, W.; Kulbacka, J.; Saczko, J.; Pachura, N.; Wieckiewicz, M.; Gancarz, R.; Wilk, K.A. Polish propolis—Chemical composition and biological effects in tongue cancer cells and macrophages. Molecules 2020, 25, 2426. [Google Scholar] [CrossRef]
- Frión-Herrera, Y.; Díaz-García, A.; Ruiz-Fuentes, J.; Rodríguez-Sánchez, H.; Sforcin, J.M. The cytotoxic effects of propolis on breast cancer cells involve PI3K/Akt and ERK1/2 pathways, mitochondrial membrane potential, and reactive oxygen species generation. Inflammopharmacology 2018, 27, 1081–1089. [Google Scholar] [CrossRef]
- Misir, S.; Aliyazicioglu, Y.; Demir, S.; Turan, I.; Hepokur, C. Effect of Turkish Propolis on miRNA Expression, Cell Cycle, and Apoptosis in Human Breast Cancer (MCF-7) Cells. Nutr. Cancer 2019, 72, 133–145. [Google Scholar] [CrossRef]
- Nör, F.; Nör, C.; Bento, L.W.; Zhang, Z.; Bretz, W.A.; Nör, J.E. Propolis reduces the stemness of head and neck squamous cell carcinoma. Arch. Oral Biol. 2021, 125, 105087. [Google Scholar] [CrossRef] [PubMed]
- da Silva Frozza, C.O.; Garcia, C.S.C.; Gambato, G.; de Souza, M.D.O.; Salvador, M.; Moura, S.; Padilha, F.F.; Seixas, F.K.; Collares, T.; Borsuk, S.; et al. Chemical characterization, antioxidant and cytotoxic activities of Brazilian red propolis. Food Chem. Toxicol. 2013, 52, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Asgharpour, F.; Moghadamnia, A.A.; Zabihi, E.; Namvar, A.E.; Gholinia, H.; Motallebnejad, M.; Nouri, H.R. Iranian propolis efficiently inhibits growth of oral streptococci and cancer cell lines. BMC Complement. Altern. Med. 2019, 19, 266. [Google Scholar] [CrossRef] [PubMed]
- Žižić, J.B.; Vuković, N.L.; Jadranin, M.B.; Andelković, B.D.; Tešević, V.V.; Kacaniova, M.M.; Sukdolak, S.B.; Marković, S.D. Chemical composition, cytotoxic and antioxidative activities of ethanolic extracts of propolis on HCT-116 cell line. J. Sci. Food Agric. 2013, 93, 3001–3009. [Google Scholar] [CrossRef] [PubMed]
- Khacha-Ananda, S.; Tragoolpua, K.; Chantawannakul, P.; Tragoolpua, Y. Propolis extracts from the northern region of Thailand suppress cancer cell growth through induction of apoptosis pathways. Investig. New Drugs 2016, 34, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, S.; Saieva, L.; Cristaldi, M.; Monteleone, F.; Fontana, S.; Alessandro, R. Label-free quantitative proteomic profiling of colon cancer cells identifies acetyl-CoA carboxylase alpha as antitumor target of Citrus limon-derived nanovesicles. J. Proteom. 2018, 173, 1–11. [Google Scholar] [CrossRef]
- Yin, S.; Mai, Z.; Liu, C.; Xu, L.; Xia, C. Label-free-based quantitative proteomic analysis of the inhibition of cisplatin-resistant ovarian cancer cell proliferation by cucurbitacin B. Phytomedicine 2023, 111, 154669. [Google Scholar] [CrossRef]
- Li, M.; Liao, H.X.; Bando, K.; Nawa, Y.; Fujita, S.; Fujita, K. Label-free monitoring of drug-induced cytotoxicity and its molecular fingerprint by live-cell Raman and autofluorescence imaging. Anal. Chem. 2022, 94, 10019–10026. [Google Scholar] [CrossRef] [PubMed]
- Yasui, W.; Oue, N.; Ito, R.; Kuraoka, K.; Nakayama, H. Search for new biomarkers of gastric cancer through serial analysis of gene expression and its clinical implications. Cancer Sci. 2004, 95, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Chen, F. Identification of significant pathways in gastric cancer based on protein-protein interaction networks and cluster analysis. Genet. Mol. Biol. 2012, 35, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Sodek, K.L.; Evangelou, A.I.; Ignatchenko, A.; Agochiya, M.; Brown, T.J.; Ringuette, M.J.; Jurisica, I.; Kislinger, T. Identification of pathways associated with invasive behavior by ovarian cancer cells using multidimensional protein identification technology (MudPIT). Mol. BioSyst. 2008, 4, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yang, J.; Zhao, G.; Shen, Z.; Yang, K.; Tian, L.; Zhou, Q.; Chen, Y.; Huang, Y. Dysregulation of ferroptosis may involve in the development of non-small-cell lung cancer in Xuanwei area. J. Cell. Mol. Med. 2021, 25, 2872–2884. [Google Scholar] [CrossRef] [PubMed]
- Dhondrup, R.; Zhang, X.; Feng, X.; Lobsang, D.; Hua, Q.; Liu, J.; Cuo, Y.; Zhuoma, S.; Duojie, G.; Caidan, S.D.; et al. Proteomic analysis reveals molecular differences in the development of gastric cancer. Evid.-Based Complement. Altern. Med. 2022, 2022, 8266544. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.J.; Xue, J.M.; Li, J.; Wan, L.H.; Zhu, Y.X. Identification of key genes and pathways of diagnosis and prognosis in cervical cancer by bioinformatics analysis. Mol. Genet. Genomic Med. 2020, 8, e1200. [Google Scholar] [CrossRef]
- Wu, K.; Yi, Y.; Liu, F.; Wu, W.; Chen, Y.; Zhang, W. Identification of key pathways and genes in the progression of cervical cancer using bioinformatics analysis. Oncol. Lett. 2018, 16, 1003–1009. [Google Scholar] [CrossRef]
- Yu, C.; Hong, H.; Lu, J.; Zhao, X.; Hu, W.; Zhang, S.; Zong, Y.; Mao, Z.; Li, J.; Wang, M.; et al. Prediction of target genes and pathways associated with cetuximab insensitivity in colorectal cancer. Technol. Cancer Res. Treat. 2018, 17, 1–13. [Google Scholar] [CrossRef]
- Chen, J.; Wu, F.; Shi, Y.; Yang, D.; Xu, M.; Lai, Y.; Liu, Y. Identification of key candidate genes involved in melanoma metastasis. Mol. Med. Rep. 2019, 20, 903–914. [Google Scholar] [CrossRef]
- Xie, R.; Li, B.; Jia, L.; Li, Y.; Zhang, Y.; Tu, Y.; He, Z. Identification of core genes and pathways in melanoma metastasis via bioinformatics analysis. Int. J. Mol. Sci. 2022, 23, 794. [Google Scholar] [CrossRef]
- Han, B.; Yang, X.; Zhang, P.; Yuan, J.; Dong, Y.; Hosseini, D.K.; Zhou, T.; Sun, H.; He, Z.; Tu, Y.; et al. DNA methylation biomarkers for nasopharyngeal carcinoma. PLoS ONE 2020, 15, e0230524. [Google Scholar] [CrossRef] [PubMed]
- Sivakumaran, N.; Samarakoon, S.R.; Adhikari, A.; Ediriweera, M.K.; Tennekoon, K.H.; Malavige, N.; Thabrew, I.; Shrestha, R.L.S. Cytotoxic and apoptotic effects of govaniadine isolated from corydalis govaniana wall. roots on human breast cancer (MCF-7) Cells. BioMed Res. Int. 2018, 2018, 3171348. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Kong, S.; Zhao, W. In silico analyses for potential key genes associated with gastric cancer. PeerJ 2018, 6, e6092. [Google Scholar] [CrossRef]
- Blaheta, R.; Daher, F.; Michaelis, M.; Hasenberg, C.W.; Weich, E.M.; Jonas, D.; Kotchetkov, R.; Doerr, H.W.; Cinatl, J., Jr. Chemoresistance induces enhanced adhesion and transendothelial penetration of neuroblastoma cells by down-regulating NCAM surface expression. BMC Cancer 2006, 6, 294. [Google Scholar] [CrossRef] [PubMed]
- Baqai, U.; Purwin, T.J.; Bechtel, N.; Chua, V.; Han, A.; Hartsough, E.J.; Kuznetsoff, J.N.; Harbour, J.W.; Aplin, A.E. Multi-omics profiling shows BAP1 loss is associated with upregulated cell adhesion molecules in uveal melanoma. Mol. Cancer Res. 2022, 20, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Mala, U.; Baral, T.K.; Somasundaram, K. Integrative analysis of cell adhesion molecules in glioblastoma identified prostaglandin F2 receptor inhibitor (PTGFRN) as an essential gene. BMC Cancer 2022, 22, 642. [Google Scholar] [CrossRef]
- Zhang, Z.; Tao, Y.; Hua, Q.; Cai, J.; Ye, X.; Li, H. SNORA71A promotes colorectal cancer cell proliferation, migration, and invasion. BioMed Res. Int. 2020, 2020, 8284576. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, H.; An, Y.; Shen, K.; Yu, L. Biological effects of corosolic acid as an anti-inflammatory, anti-metabolic syndrome and anti-neoplasic natural compound. Oncol. Lett. 2021, 21, 84. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; He, X.; Jia, X.; Zhang, X.; Lu, B.; Zhao, J.; Lu, J.; Chen, L.; Dong, Z.; et al. Proteomics reveal the inhibitory mechanism of levodopa against esophageal squamous cell carcinoma. Front. Pharmacol. 2020, 11, 568459. [Google Scholar] [CrossRef] [PubMed]
- Jin, E.H.; Sung, J.K.; Lee, S.I.; Hong, J.H. Mitochondrial NADH dehydrogenase subunit 3 (mtnd3) polymorphisms are associated with gastric cancer susceptibility. Int. J. Med. Sci. 2018, 15, 1329–1333. [Google Scholar] [CrossRef] [PubMed]
- Solaini, G.; Sgarbi, G.; Baracca, A. Oxidative phosphorylation in cancer cells. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 534–542. [Google Scholar] [CrossRef]
- Sica, V.; Bravo-San Pedro, J.M.; Stoll, G.; Kroemer, G. Oxidative phosphorylation as a potential therapeutic target for cancer therapy. Int. J. Cancer 2020, 146, 10–17. [Google Scholar] [CrossRef]
- Evans, K.W.; Yuca, E.; Scott, S.S.; Zhao, M.; Arango, N.P.; Pico, C.X.C.; Saridogan, T.; Shariati, M.; Class, C.A.; Bristow, C.A.; et al. Oxidative phosphorylation is a metabolic vulnerability in chemotherapy-resistant triple-negative breast cancer. Cancer Res. 2021, 81, 5572–5581. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.L.; Li, F.; Yeo, J.Z.; Yong, K.J.; Bassal, M.A.; Ng, G.H.; Lee, M.Y.; Leong, C.Y.; Tan, H.K.; Wu, C.; et al. New high-throughput screening identifies compounds that reduce viability specifically in liver cancer cells that express high levels of SALL4 by inhibiting oxidative phosphorylation. BMC Gastroenterol. 2019, 157, 1615–1629.e17. [Google Scholar] [CrossRef]
- Wu, Z.; Zuo, M.; Zeng, L.; Cui, K.; Liu, B.; Yan, C.; Chen, L.; Dong, J.; Shangguan, F.; Hu, W.; et al. OMA1 reprograms metabolism under hypoxia to promote colorectal cancer development. EMBO Rep. 2021, 22, e50827. [Google Scholar] [CrossRef]
- Xue, D.; Xu, Y.; Kyani, A.; Roy, J.; Dai, L.; Sun, D.; Neamati, N. Multiparameter optimization of oxidative phosphorylation inhibitors for the treatment of pancreatic cancer. J. Med. Chem. 2022, 65, 3404–3419. [Google Scholar] [CrossRef]
- Sullivan, L.B.; Luengo, A.; Danai, L.V.; Bush, L.N.; Diehl, F.F.; Hosios, A.M.; Lau, A.N.; Elmiligy, S.; Malstrom, S.; Lewis, C.A.; et al. Aspartate is an endogenous metabolic limitation for tumour growth. Nat. Cell Biol. 2018, 20, 782–788. [Google Scholar] [CrossRef]
- Gómez-Romero, L.; Alvarez-Suarez, D.E.; Hernández-Lemus, E.; Ponce-Castañeda, M.V.; Tovar, H. The regulatory landscape of retinoblastoma: A pathway analysis perspective. R. Soc. Open Sci. 2022, 9, 220031. [Google Scholar] [CrossRef]
- Dong, C.; Fan, W.; Fang, S. PBK as a potential biomarker associated with prognosis of glioblastoma. J. Mol. Neurosci. 2020, 70, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Geng, R.; Li, N.; Xu, Y.; Liu, J.; Yuan, F.; Sun, Q.; Liu, B.; Chen, Q. Identification of core biomarkers associated with outcome in glioma: Evidence from bioinformatics analysis. Dis. Markers 2018, 2018, 3215958. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Sun, L.; Liu, Y.; Ren, H.; Shen, Y.; Bi, F.; Zhang, T.; Wang, X. Alter between gut bacteria and blood metabolites and the anti-tumor effects of Faecalibacterium prausnitzii in breast cancer. BMC Microbiol. 2020, 20, 82. [Google Scholar] [CrossRef]
- Cheng, T.; Wang, Y.; Lu, M.; Zhan, X.; Zhou, T.; Li, B.; Zhan, X. Quantitative analysis of proteome in non-functional pituitary adenomas: Clinical relevance and potential benefits for the patients. Front. Endocrinol. 2019, 10, 854. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, Y.; Wu, Y.; Zhang, Z.; Tong, M.; Yu, R. Application of personalized differential expression analysis in human cancer proteome. bioRxiv 2021. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, J.; Wang, X.; Zhu, J.; Liu, Q.; Shi, Z.; Chambers, M.C.; Zimmerman, L.J.; Shaddox, K.F.; Kim, S.; et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014, 513, 382–387. [Google Scholar] [CrossRef]
- Bradner, J.E.; Hnisz, D.; Young, R.A. Transcriptional addiction in cancer. Cell 2017, 168, 629–643. [Google Scholar] [CrossRef]
- Cho, Y.; Gutierrez, L.; Bordonaro, M.; Russo, D.; Anzelmi, F.; Hooven, J.T.; Cerra, C.; Lazarova, D.L. Effects of propolis and gamma-cyclodextrin on intestinal neoplasia in normal weight and obese mice. Cancer Med. 2016, 5, 2448–2458. [Google Scholar] [CrossRef]
- Desamero, M.J.; Kakuta, S.; Tang, Y.; Chambers, J.K.; Uchida, K.; Estacio, M.A.; Cervancia, C.; Kominami, Y.; Ushio, H.; Nakayama, J.; et al. Tumor-suppressing potential of stingless bee propolis in in vitro and in vivo models of differentiated-type gastric adenocarcinoma. Sci. Rep. 2019, 9, 19635. [Google Scholar] [CrossRef]
- Tao, Y.; Wang, D.; Hu, Y.; Huang, Y.; Yu, Y.; Wang, D. The immunological enhancement activity of propolis flavonoids liposome in vitro and in vivo. J. Evid.-Based Complement. Altern. Med. 2014, 2014, 483513. [Google Scholar] [CrossRef]
- GB/T 24283-2018; Propolis. National Standards of People’s Republic of China: Beijing, China, 2018.
- Cao, Y.; Ding, W.; Liu, C. Unraveling the metabolite signature of endophytic Bacillus velezensis strain showing defense response towards Fusarium oxysporum. Agronomy 2021, 11, 683. [Google Scholar] [CrossRef]
- Oršolić, N. A review of propolis antitumor action in vivo and in vitro. JAAS 2010, 2, 1–20. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).