Autophagy and Senescence: The Molecular Mechanisms and Implications in Liver Diseases
Abstract
1. Introduction
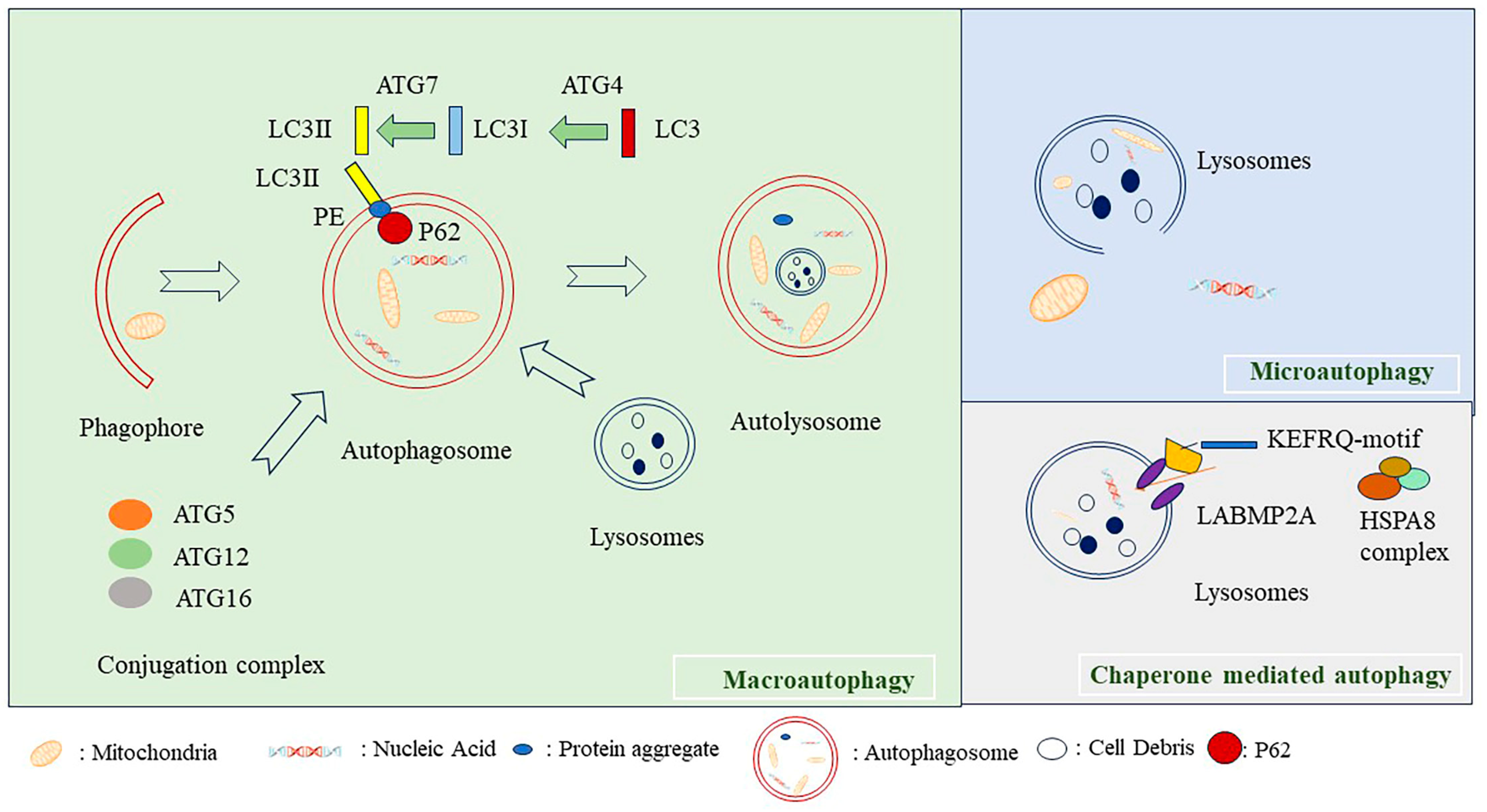
2. Classification of Autophagy
2.1. Macroautophagy
2.2. Microautophagy
2.3. Mhaperon-Mediated Autophagy
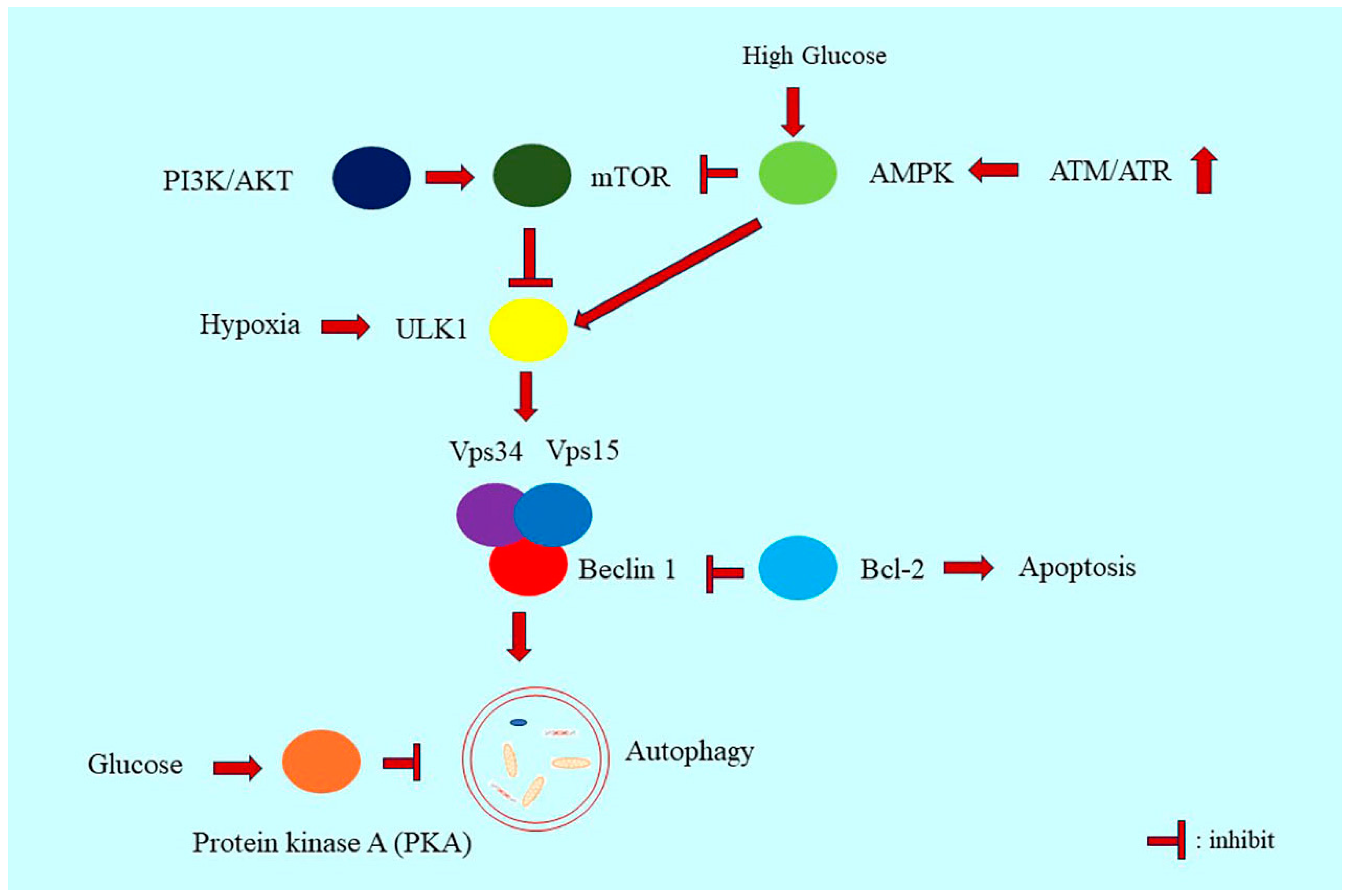
3. Regulatory Mechanisms of Autophagy
3.1. ATG5, P62, and LC3
3.2. PI3K/Akt/mTOR Signaling in Autophagy Regulation
3.3. AMPK Signaling in Autophagy Regulation
3.4. Beclin 1 in Autophagy Regulation
3.5. Extracellular Signal-Regulated Kinase Signaling
4. The Role of Autophagy in Liver Diseases
4.1. The Role of Autophagy in Liver Fibrosis, NAFLD and NASH
4.2. Autophagy and Liver Malignancies
4.3. Autophagy and Other Liver Dysfunction
5. Senescence and Senescence-Associated Biomarkers
6. Classification of Senescence
6.1. Replicative Senescence
6.2. Oncogene and Stress-Induced Senescence
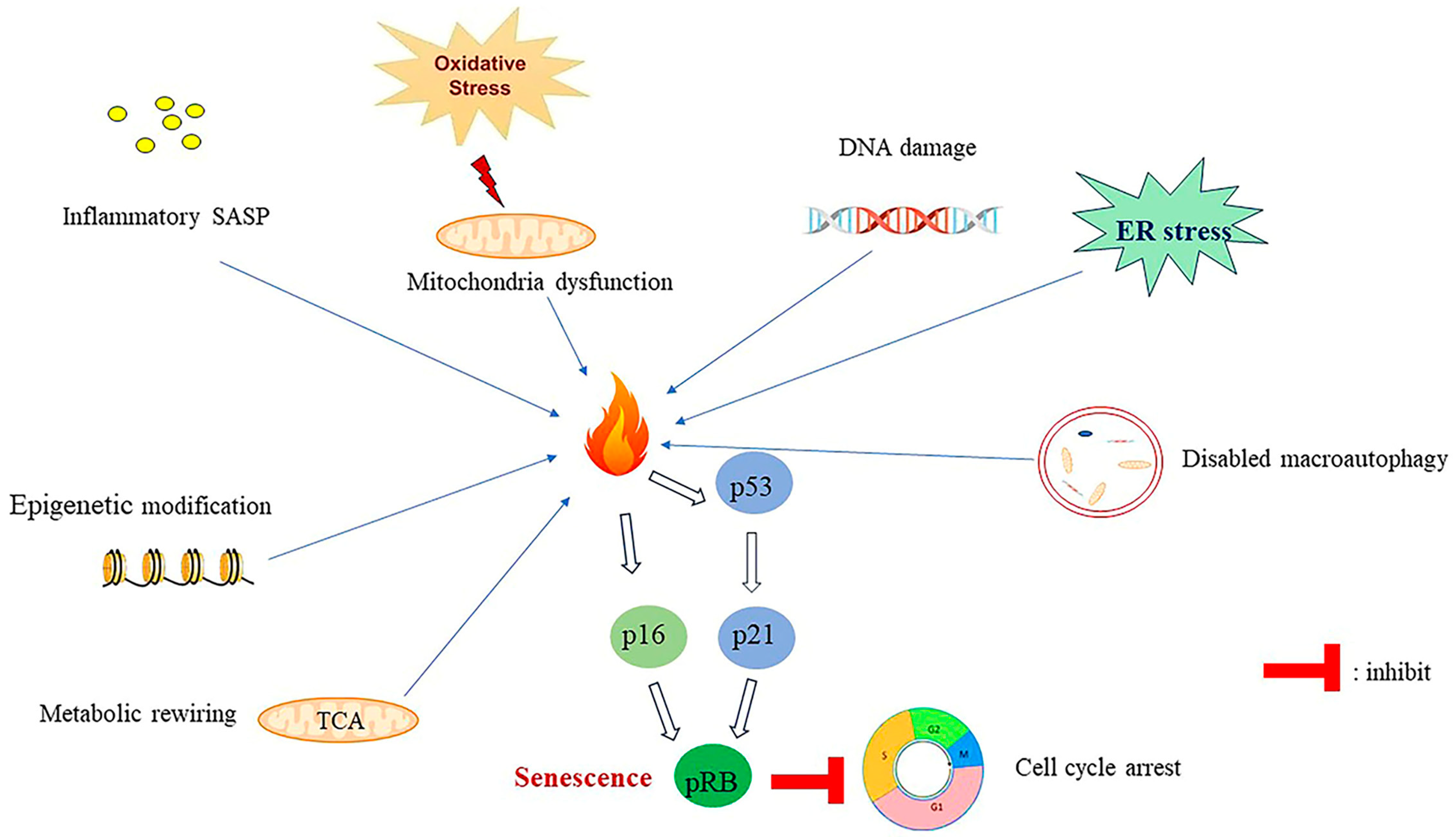
7. Mechanisms of Senescence Induction
7.1. SASP-Induced Senescence
7.2. Epigenetic Regulation-Induced Senescence
7.3. Deregulation of Cellular Metabolism Induce Senescence
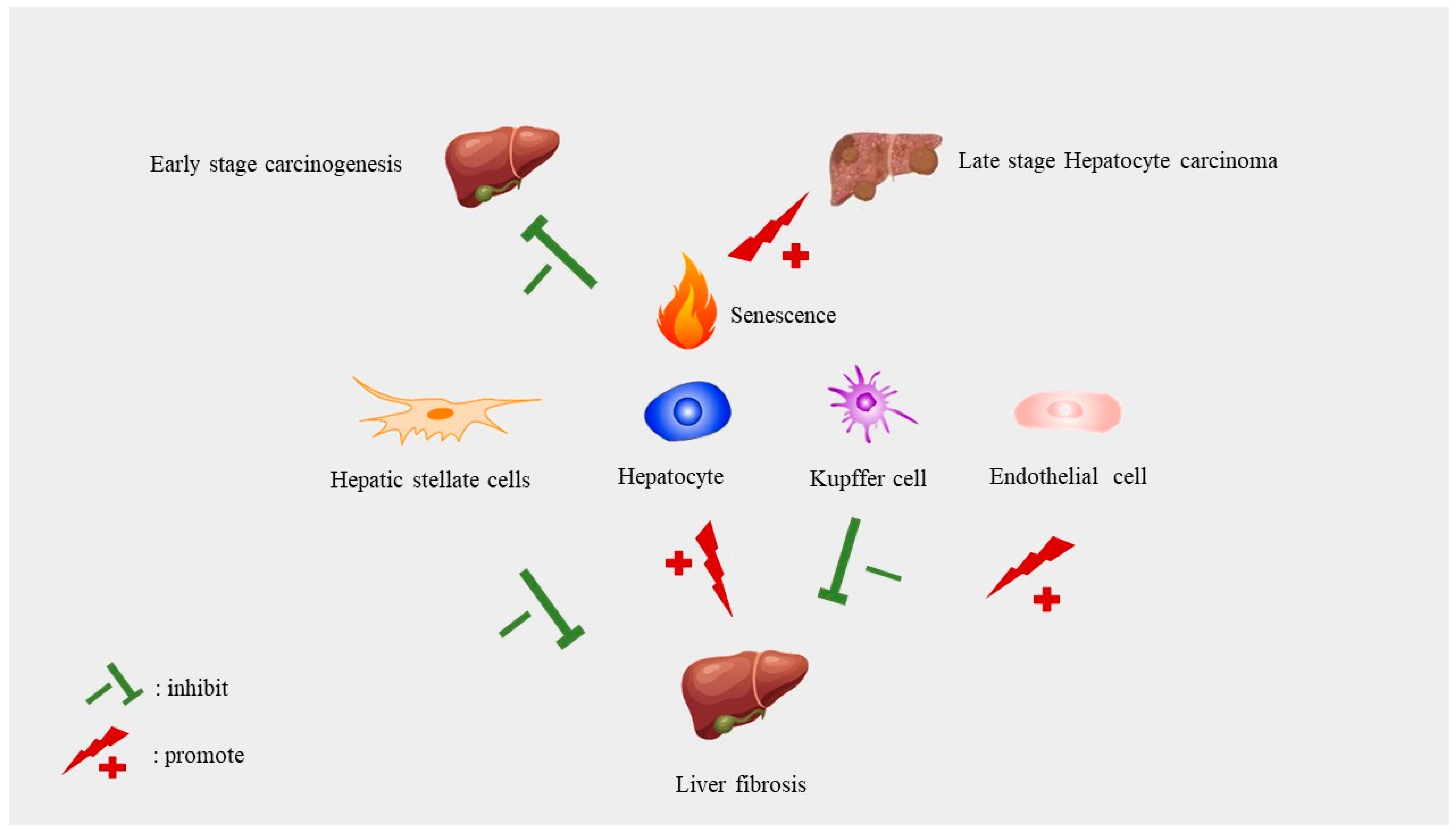
8. Implications of Senescence in Liver Diseases
8.1. The Role of Cellular Senescence in Liver Cancer
8.2. Senescence in Liver Fibrosis and Cirrhosis
8.3. Senescence and NAFLD
9. The Relationship between Autophagy and Senescence
10. Perspectives and Conclusions
Therapeutic Strategies for Liver Diseases Targeting Autophagy Dysregulation or Senescence
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maeso-Díaz, R.; Gracia-Sancho, J. Aging and Chronic Liver Disease. Semin. Liver Dis. 2020, 40, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Tasset, I.; Arias, E.; Pampliega, O.; Wong, E.; Martinez-Vicente, M.; Cuervo, A.M. Autophagy and the hallmarks of aging. Ageing Res. Rev. 2021, 72, 101468. [Google Scholar] [CrossRef]
- D’arcy, M.S. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Aman, Y.; Schmauck-Medina, T.; Hansen, M.; Morimoto, R.I.; Simon, A.K.; Bjedov, I.; Palikaras, K.; Simonsen, A.; Johansen, T.; Tavernarakis, N.; et al. Autophagy in healthy aging and disease. Nat. Aging 2021, 1, 634–650. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Klionsky, D.J.; Shen, H.-M. The emerging mechanisms and functions of microautophagy. Nat. Rev. Mol. Cell Biol. 2023, 24, 186–203. [Google Scholar] [CrossRef] [PubMed]
- Schuck, S. Microautophagy—Distinct molecular mechanisms handle cargoes of many sizes. J. Cell Sci. 2020, 133, jcs246322. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, P.; Bourdenx, M.; Madrigal-Matute, J.; Tiano, S.; Diaz, A.; Bartholdy, B.A.; Will, B.; Cuervo, A.M. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol. 2019, 17, e3000301. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.L.; Terlecky, S.R.; Plant, C.P.; Dice, J.F. A role for a 70 kDa heat shock protein in lysosomal degradation of intracellular protein. Science 1989, 246, 382–385. [Google Scholar] [CrossRef]
- Ashrafi, G.; Schwarz, T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013, 20, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Chao, X.; Williams, J.; Fulte, S.; Li, T.; Yang, L.; Ding, W.X. Autophagy in liver diseases: A review. Mol. Asp. Med. 2021, 82, 100973. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020, 63, 1–10. [Google Scholar] [CrossRef]
- Kharaziha, P.; Panaretakis, T. Dynamics of Atg5-Atg12-Atg16L1 Aggregation and Deaggregation. Methods Enzym. 2017, 587, 247–255. [Google Scholar]
- Islam, M.A.; Sooro, M.A.; Zhang, P. Autophagic regulation of p62 is critical for cancer therapy. Int. J. Mol. Sci. 2018, 19, 1405. [Google Scholar] [CrossRef] [PubMed]
- Mallat, A.; Lodder, J.; Teixeira-Clerc, F.; Moreau, R.; Codogno, P.; Lotersztajn, S. Autophagy: A multifaceted partner in liver fibrosis. Biomed. Res. Int. 2014, 2014, 869390. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.V.; Mills, J.; Lapierre, L.R. Selective Autophagy Receptor p62/SQSTM1, a Pivotal Player in Stress and Aging. Front. Cell Dev. Biol. 2022, 10, 793328. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2503–2518. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef]
- Fujita, N.; Itoh, T.; Omori, H.; Fukuda, M.; Noda, T.; Yoshimori, T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 2008, 19, 2092–2100. [Google Scholar] [CrossRef]
- Vucicevic, L.; Misirkic, M.; Ciric, D.; Martinovic, T.; Jovanovic, M.; Isakovic, A.; Markovic, I.; Saponjic, J.; Foretz, M.; Rabanal-Ruiz, Y.; et al. Transcriptional block of AMPK-induced autophagy promotes glutamate excitotoxicity in nutrient-deprived SH-SY5Y neuroblastoma cells. Cell Mol. Life Sci. 2020, 77, 3383–3399. [Google Scholar] [CrossRef]
- Mauthe, M.; Orhon, I.; Rocchi, C.; Zhou, X.; Luhr, M.; Hijlkema, K.-J.; Coppes, R.P.; Engedal, N.; Mari, M.; Reggiori, F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018, 14, 1435–1455. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and Autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar] [PubMed]
- Czaja, M.J. Functions of Autophagy in Hepatic and Pancreatic Physiology and Disease. Gastroenterology 2011, 140, 1895–1908. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Ji, H.G.; Yan, X.J.; Liu, M.; Ding, Y.H.; Chen, X.H. Dioscin ameliorates doxorubicin-induced heart failure via inhibiting autophagy and apoptosis by controlling the PDK1-mediated Akt/mTOR signaling pathway. Kaohsiung J. Med. Sci. 2023, 39, 1022–1029. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. Feb. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Dossou, A.S.; Basu, A. The Emerging Roles of mTORC1 in Macromanaging Autophagy. Cancers 2019, 11, 1422. [Google Scholar] [CrossRef] [PubMed]
- Puente, C.; Hendrickson, R.C.; Jiang, X. Nutrient-regulated Phosphorylation of ATG13 Inhibits Starvation-induced Autophagy. J. Biol. Chem. 2016, 291, 6026–6035. [Google Scholar] [CrossRef]
- Kimura, T.; Takabatake, Y.; Takahashi, A.; Isaka, Y. Chloroquine in cancer therapy: A double-edged sword of autophagy. Cancer Res. 2013, 73, 3–7. [Google Scholar] [CrossRef]
- Shang, L.; Wang, X. AMPK and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy 2011, 7, 924–926. [Google Scholar] [CrossRef]
- Hardie, D.G. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007, 8, 774–785. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, Q.; Luo, L.; Ning, B.; Fang, Y. β-asarone inhibited cell growth and promoted autophagy via P53/Bcl-2/Bclin-1 and P53/AMPK/mTOR pathways in Human Glioma U251 cells. J. Cell. Physiol. 2018, 233, 2434–2443. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Kroemer, G. Cytoplasmic functions of the tumour suppressor p53. Nature 2009, 458, 1127–1130. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Di, J.; Cao, H.; Bai, J.; Zheng, J. p53-mediated autophagic regulation: A prospective strategy for cancer therapy. Cancer Lett. 2015, 363, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Li, J.; Mao, L.; Wu, J.; Deng, Z.; He, M.; An, S.; Zeng, Z.; Huang, Q.; Chen, Z. p53 Deacetylation Alleviates Sepsis-Induced Acute Kidney Injury by Promoting Autophagy. Front. Immunol. 2021, 12, 685523. [Google Scholar] [CrossRef]
- Zhu, Y.; Jiang, X.; Ding, Z.; Ming, J. Interleukin 7 inhibit autophagy via P53 regulated AMPK/mTOR signaling pathway in non-small cell lung cancer. Sci. Rep. 2022, 12, 11208. [Google Scholar] [CrossRef]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.C.; Tsai, C.Y.; Tsai, M.M.; Yeh, C.T.; Lin, K.H. Molecular functions and clinical impact of thyroid hormone-triggered autophagy in liver-related diseases. J. Biomed. Sci. 2019, 26, 24. [Google Scholar] [CrossRef]
- Maejima, Y.; Isobe, M.; Sadoshima, J. Regulation of autophagy by Beclin 1 in the heart. J. Mol. Cell. Cardiol. 2016, 95, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A. Activation of immunosuppressive network in the aging process. Ageing Res. Rev. 2020, 57, 100998. [Google Scholar] [CrossRef]
- Fernández, Á.F.; Sebti, S.; Wei, Y.; Zou, Z.; Shi, M.; McMillan, K.L.; He, C.; Ting, T.; Liu, Y.; Chiang, W.C.; et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 2018, 558, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Cagnol, S.; Chambard, J.C. ERK and cell death: Mechanisms of ERK-induced cell death—Apoptosis, autophagy and senescence. FEBS J. 2010, 277, 2–21. [Google Scholar] [CrossRef]
- Sivaprasad, U.; Basu, A. Inhibition of ERK attenuates autophagy and potentiates tumour necrosis factor-alpha-induced cell death in MCF-7 cells. J. Cell Mol. Med. 2008, 12, 1265–1271. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Qiu, F.; Tashiro, S.I.; Onodera, S.; Ikejima, T. ERK and JNK mediate TNFa-induced p53 activation in apoptotic and autophagic L929 cell death. Biochem. Biophys. Res. Commun. 2008, 376, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Wang, J.; Yan, X.; Zhang, Q.; Chai, L.; Wang, Q.; Shi, W.; Chen, Y.; Liu, J.; Qu, Z.; et al. ERK/Drp1-dependent mitochondrial fission contributes to HMGB1-induced autophagy in pulmonary arterial hypertension. Cell Prolif. 2021, 54, e13048. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiang, N.; Ping, J.; Xu, L. TGF-β1-induced autophagy activates hepatic stellate cells via the ERK and JNK signaling pathways. Int. J. Mol. Med. 2021, 47, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Venida, A.; Perera, R.M.; Kimmelman, A.C. Selective autophagy of MHC-I promotes immune evasion of pancreatic cancer. Autophagy 2020, 16, 1524–1525. [Google Scholar] [CrossRef] [PubMed]
- Nakai, A.; Yamaguchi, O.; Takeda, T.; Higuchi, Y.; Hikoso, S.; Taniike, M.; Omiya, S.; Mizote, I.; Matsumura, Y.; Asahi, M.; et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007, 13, 619–624. [Google Scholar] [CrossRef]
- Taneike, M.; Yamaguchi, O.; Nakai, A.; Hikoso, S.; Takeda, T.; Mizote, I.; Oka, T.; Tamai, T.; Oyabu, J.; Murakawa, T.; et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 2010, 6, 600–606. [Google Scholar] [CrossRef]
- Kubli, D.A.; Zhang, X.; Lee, Y.; Hanna, R.A.; Quinsay, M.N.; Nguyen, C.K.; Jimenez, R.; Petrosyan, S.; Murphy, A.N.; Gustafsson, A.B. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J. Biol. Chem. 2013, 288, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998, 392, 605–608. [Google Scholar] [CrossRef]
- Franco-Iborra, S.; Plaza-Zabala, A.; Montpeyo, M.; Sebastian, D.; Vila, M.; Martinez-Vicente, M. Mutant HTT (huntingtin) impairs mitophagy in a cellular model of Huntington disease. Autophagy 2021, 17, 672–689. [Google Scholar] [CrossRef]
- Tan, M.; Zhang, Q.B.; Liu, T.H.; Yang, Y.Y.; Zheng, J.X.; Zhou, W.J.; Xiong, Q.; Qing, Y.F. Autophagy dysfunction may be involved in the pathogenesis of ankylosing spondylitis. Exp. Ther. Med. 2020, 20, 3578–3586. [Google Scholar] [CrossRef] [PubMed]
- Zanfardino, P.; Amati, A.; Petracca, E.A.; Santorelli, F.M.; Petruzzella, V. Torin1 restores proliferation rate in Charcot-Marie-Tooth disease type 2A cells harbouring MFN2 (mitofusin 2) mutation. Acta Myol. 2022, 41, 201–206. [Google Scholar]
- Lin, M.Y.; Cheng, X.T.; Tammineni, P.; Xie, Y.; Zhou, B.; Cai, Q.; Sheng, Z.H. Releasing Syntaphilin Removes Stressed Mitochondria from Axons Independent of Mitophagy under Pathophysiological Conditions. Neuron 2017, 94, 595–610.e6. [Google Scholar] [CrossRef] [PubMed]
- Danese, A.; Patergnani, S.; Maresca, A.; Peron, C.; Raimondi, A.; Caporali, L.; Marchi, S.; La Morgia, C.; Del Dotto, V.; Zanna, C.; et al. Pathological mitophagy disrupts mitochondrial homeostasis in Leber’s hereditary optic neuropathy. Cell Rep. 2022, 40, 111124. [Google Scholar] [CrossRef]
- Nah, J.; Zhai, P.; Huang, C.Y.; Fernandez, A.F.; Mareedu, S.; Levine, B.; Sadoshima, J. Upregulation of Rubicon promotes autosis during myocardial ischemia/reperfusion injury. J. Clin. Investig. 2020, 130, 2978–2991. [Google Scholar] [CrossRef] [PubMed]
- Tosco, A.; De Gregorio, F.; Esposito, S.; De Stefano, D.; Sana, I.; Ferrari, E.; Sepe, A.; Salvadori, L.; Buonpensiero, P.; Di Pasqua, A.; et al. A novel treatment of cystic fibrosis acting on-target: Cysteamine plus epigallocatechin gallate for the autophagy-dependent rescue of class II-mutated CFTR. Cell Death Differ. 2016, 23, 1380–1393. [Google Scholar] [CrossRef]
- Sciarretta, S.; Zhai, P.; Shao, D.; Maejima, Y.; Robbins, J.; Volpe, M.; Condorelli, G.; Sadoshima, J. Rheb is a critical regulator of autophagy during myocardial ischemia: Pathophysiological implications in obesity and metabolic syndrome. Circulation 2012, 125, 1134–1146. [Google Scholar] [CrossRef]
- Rudnick, J.A.; Monkkonen, T.; Mar, F.A.; Barnes, J.M.; Starobinets, H.; Goldsmith, J.; Roy, S.; Bustamante Eguiguren, S.; Weaver, V.M.; Debnath, J. Autophagy in stromal fibroblasts promotes tumor desmoplasia and mammary tumorigenesis. Genes Dev. 2021, 35, 963–975. [Google Scholar] [CrossRef]
- Strohecker, A.M.; White, E. Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy 2014, 10, 384–385. [Google Scholar] [CrossRef]
- Huang, F.; Wang, B.R.; Wang, Y.G. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J. Gastroenterol. 2018, 24, 4643–4651. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Blas, D.; Gorostieta-Salas, E.; Castro-Obregón, S. Connecting chaperone-mediated autophagy dysfunction to cellular senescence. Ageing Res. Rev. 2018, 41, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Nishio, N.; Ito, S.; Suzuki, H.; Isobe, K. Autophagic activity in thymus and liver during aging. Age 2012, 34, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.C.; Wang, J.L.; Ren, H.Z.; Shi, X.L. Autophagy plays a double-edged sword role in liver diseases. J. Physiol. Biochem. 2022, 78, 9–17. [Google Scholar] [CrossRef]
- López-Otín, C.; Pietrocola, F.; Roiz-Valle, D.; Galluzzi, L.; Kroemer, G. Meta-hallmarks of aging and cancer. Cell Metab. 2023, 35, 12–35. [Google Scholar] [CrossRef]
- Lavallard, V.J.; Gual, P. Autophagy and non-alcoholic fatty liver disease. Biomed. Res. Int. 2014, 2014, 120179. [Google Scholar] [CrossRef]
- Gonzalez-Rodriguez, A.; Mayoral, R.; Agra, N.; Valdecantos, M.P.; Pardo, V.; Miquilena-Colina, M.E.; Vargas-Castrillon, J.; Lo Iacono, O.; Corazzari, M.; Fimia, G.M.; et al. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014, 5, e1179. [Google Scholar] [CrossRef]
- Ost, A.; Svensson, K.; Ruishalme, I.; Brannmark, C.; Franck, N.; Krook, H.; Sandstrom, P.; Kjolhede, P.; Stralfors, P. Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol. Med. 2010, 16, 235–246. [Google Scholar] [CrossRef]
- Sakurai, Y.; Kubota, N.; Yamauchi, T.; Kadowaki, T. Role of Insulin Resistance in MAFLD. Int. J. Mol. Sci. 2021, 22, 4156. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Udoh, U.S.; Rajan, P.K.; Nakafuku, Y.; Finley, R.; Sanabria, J.R. Cell Autophagy in NASH and NASH-Related Hepatocellular Carcinoma. Int. J. Mol. Sci. 2022, 23, 7734. [Google Scholar] [CrossRef] [PubMed]
- Hammoutene, A.; Biquard, L.; Lasselin, J.; Kheloufi, M.; Tanguy, M.; Vion, A.-C.; Mérian, J.; Colnot, N.; Loyer, X.; Tedgui, A.; et al. A defect in endothelial autophagy occurs in patients with non-alcoholic steatohepatitis and promotes inflammation and fibrosis. J. Hepatol. 2020, 72, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, S.; Matsushita, Y.; Kurosaki, S.; Tange, M.; Fujiwara, N.; Hayata, Y.; Hayakawa, Y.; Suzuki, N.; Hata, M.; Tsuboi, M.; et al. Inhibiting SCAP/SREBP exacerbates liver injury and carcinogenesis in murine nonalcoholic steatohepatitis. J. Clin. Investig. 2022, 132, e151895. [Google Scholar] [CrossRef] [PubMed]
- Brenner, C.; Galluzzi, L.; Kepp, O.; Kroemer, G. Decoding cell death signals in liver inflammation. J. Hepatol. 2013, 59, 583–594. [Google Scholar] [CrossRef]
- Li, J.; Zeng, C.; Zheng, B.; Liu, C.; Tang, M.; Jiang, Y.; Chang, Y.; Song, W.; Wang, Y.; Yang, C. HMGB1-induced autophagy facilitates hepatic stellate cells activation: A new pathway in liver fibrosis. Clin. Sci. 2018, 132, 1645–1667. [Google Scholar] [CrossRef]
- Chen, R.; Hou, W.; Zhang, Q.; Kang, R.; Fan, X.-G.; Tang, D. Emerging Role of High-Mobility Group Box 1 (HMGB1) in Liver Diseases. Mol. Med. 2013, 19, 357–366. [Google Scholar] [CrossRef]
- Khambu, B.; Yan, S.; Huda, N.; Yin, X.-M. Role of High-Mobility Group Box-1 in Liver Pathogenesis. Int. J. Mol. Sci. 2019, 20, 5314. [Google Scholar] [CrossRef]
- Khambu, B.; Huda, N.; Chen, X.; Antoine, D.J.; Li, Y.; Dai, G.; Köhler, U.A.; Zong, W.-X.; Waguri, S.; Werner, S.; et al. HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers. J. Clin. Investig. 2018, 128, 2419–2435. [Google Scholar] [CrossRef]
- Schneider, J.L.; Cuervo, A.M. Liver autophagy: Much more than just taking out the trash. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 187–200. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Ghiassi-Nejad, Z.; Rozenfeld, R.; Gordon, R.; Fiel, M.I.; Yue, Z.; Czaja, M.J.; Friedman, S.L. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 2012, 142, 938–946. [Google Scholar] [CrossRef]
- Wan, Y.; Li, X.; Slevin, E.; Harrison, K.; Li, T.; Zhang, Y.; Klaunig, J.E.; Wu, C.; Shetty, A.K.; Dong, X.C.; et al. Endothelial dysfunction in pathological processes of chronic liver disease during aging. FASEB J. 2022, 36, e22125. [Google Scholar] [CrossRef]
- Ruart, M.; Chavarria, L.; Campreciós, G.; Suárez-Herrera, N.; Montironi, C.; Guixé-Muntet, S.; Bosch, J.; Friedman, S.L.; Garcia-Pagán, J.C.; Hernández-Gea, V. Impaired endothelial autophagy promotes liver fibrosis by aggravating the oxidative stress response during acute liver injury. J. Hepatol. 2019, 70, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Marengo, A.; Rosso, C.; Bugianesi, E. Liver Cancer: Connections with Obesity, Fatty Liver, and Cirrhosis. Annu. Rev. Med. 2016, 67, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Chao, X.; Qian, H.; Wang, S.; Fulte, S.; Ding, W.X. Autophagy and liver cancer. Clin. Mol. Hepatol. 2020, 26, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, L.D.; Narita, M. Autophagy at the intersection of aging, senescence, and cancer. Mol. Oncol. 2022, 16, 3259–3275. [Google Scholar] [CrossRef]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef]
- Mathew, R.; Kongara, S.; Beaudoin, B.; Karp, C.M.; Bray, K.; Degenhardt, K.; Chen, G.; Jin, S.; White, E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes. Dev. 2007, 21, 1367–1381. [Google Scholar] [CrossRef]
- Ding, Z.B.; Shi, Y.H.; Zhou, J.; Qiu, S.J.; Xu, Y.; Dai, Z.; Shi, G.M.; Wang, X.Y.; Ke, A.W.; Wu, B.; et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008, 68, 9167–9175. [Google Scholar] [CrossRef]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef]
- Vempati, R.K.; Malla, R.R. Autophagy-Induced Drug Resistance in Liver Cancer. Crit. Rev. Oncog. 2020, 25, 21–30. [Google Scholar] [CrossRef]
- Zhao, H.; Alam, A.; Soo, A.P.; George, A.J.T.; Ma, D. Ischemia-Reperfusion Injury Reduces Long Term Renal Graft Survival: Mechanism and Beyond. EBioMedicine 2018, 28, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Yan, M.; Li, S.; Song, H.; Liu, L.; Shen, Z.; Cai, J.Z. NOD1 activates autophagy to aggravate hepatic ischemia-reperfusion injury in mice. J. Cell. Biochem. 2019, 120, 10605–10612. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Yiang, G.T.; Liao, W.T.; Tsai, A.P.; Cheng, Y.L.; Cheng, P.W.; Li, C.Y.; Li, C.J. Current Mechanistic Concepts in Ischemia and Reperfusion Injury. Cell. Physiol. Biochem. 2018, 46, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Fang, H.; Wei, W.; Dirsch, O.; Dahmen, U. Ischemic preconditioning attenuates ischemia/reperfusion injury in rat steatotic liver: Role of heme oxygenase-1-mediated autophagy. Crit. Care Med. 2014, 42, e762–e771. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, X.; Yang, J.; Liu, S.; Lv, Y.; Zhang, C.; Dong, W.; Liu, A. GDF11 impairs liver regeneration in mice after partial hepatectomy. Clin. Sci. 2019, 133, 2069–2084. [Google Scholar] [CrossRef]
- Liu, A.; Dong, W.; Peng, J.; Dirsch, O.; Dahmen, U.; Fang, H.; Zhang, C.; Sun, J. Growth differentiation factor 11 worsens hepatocellular injury and liver regeneration after liver ischemia reperfusion injury. FASEB J. 2018, 32, 5186–5198. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Yang, J.; Hu, Q.; Dirsch, O.; Dahmen, U.; Zhang, C.; Gewirtz, D.A.; Fang, H.; Sun, J. Young plasma attenuates age-dependent liver ischemia reperfusion injury. FASEB J. 2019, 33, 3063–3073. [Google Scholar] [CrossRef]
- Huang, H.; Evankovich, J.; Yan, W.; Nace, G.; Zhang, L.; Ross, M.; Liao, X.; Billiar, T.; Xu, J.; Esmon, C.T.; et al. Endogenous histones function as alarmins in sterile inflammatory liver injury through Toll-like receptor 9 in mice. Hepatology 2011, 54, 999–1008. [Google Scholar] [CrossRef]
- Huang, H.; Tohme, S.; Al-Khafaji, A.B.; Tai, S.; Loughran, P.; Chen, L.; Wang, S.; Kim, J.; Billiar, T.; Wang, Y.; et al. Damage-associated molecular pattern-activated neutrophil extracellular trap exacerbates sterile inflammatory liver injury. Hepatology 2015, 62, 600–614. [Google Scholar] [CrossRef]
- Kuilman, T.; Michaloglou, C.; Mooi, W.J.; Peeper, D.S. The essence of senescence. Genes. Dev. 2010, 24, 2463–2479. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Hickson, L.T.; Eirin, A.; Kirkland, J.L.; Lerman, L.O. Cellular senescence: The good, the bad and the unknown. Nat. Rev. Nephrol. 2022, 18, 611–627. [Google Scholar] [CrossRef]
- Kudlova, N.; De Sanctis, J.B.; Hajduch, M. Cellular Senescence: Molecular Targets, Biomarkers, and Senolytic Drugs. Int. J. Mol. Sci. 2022, 23, 4168. [Google Scholar] [CrossRef] [PubMed]
- Narita, M.; Nuñez, S.; Heard, E.; Narita, M.; Lin, A.W.; Hearn, S.A.; Spector, D.L.; Hannon, G.J.; Lowe, S.W. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 2003, 113, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; d’Adda di Fagagna, F. Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2020, 22, 75–95. [Google Scholar] [CrossRef] [PubMed]
- Roger, L.; Tomas, F.; Gire, V. Mechanisms and functions of cellular senescence. Int. J. Mol. Sci. 2021, 22, 13173. [Google Scholar] [CrossRef] [PubMed]
- Legassie, J.D.; Jarstfer, M.B. Telomerase as a DNA-dependent DNA polymerase. Biochemistry 2005, 44, 14191–14201. [Google Scholar] [CrossRef]
- MacNeil, D.E.; Bensoussan, H.J.; Autexier, C. Telomerase Regulation from Beginning to the end. Genes 2016, 7, 64. [Google Scholar] [CrossRef]
- Engin, A.B.; Engin, A. The Connection Between Cell Fate and Telomere. Adv. Exp. Med. Biol. 2021, 1275, 71–100. [Google Scholar]
- Shay, J.W.; Wright, W.E. Hayflick, his limit, and cellular ageing. Nat. Rev. Mol. Cell Biol. 2000, 1, 72–76. [Google Scholar] [CrossRef]
- Birch, J.; Gil, J. Senescence and the SASP: Many therapeutic avenues. Genes Dev. 2020, 34, 1565–1576. [Google Scholar] [CrossRef]
- McHugh, D.; Gil, J. Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 2018, 217, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Borodkina, A.V.; Deryabin, P.I.; Giukova, A.A.; Nikolsky, N.N. “Social Life” of Senescent Cells: What Is SASP and Why Study It? Acta Naturae 2018, 10, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Acosta, J.C.; Banito, A.; Wuestefeld, T.; Georgilis, A.; Janich, P.; Morton, J.P.; Athineos, D.; Kang, T.W.; Lasitschka, F.; Andrulis, M.; et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 2013, 15, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Tchkonia, T.; Zhu, Y.; van Deursen, J.; Campisi, J.; Kirkland, J.L. Cellular senescence and the senescent secretory phenotype: Therapeutic opportunities. J. Clin. Investig. 2013, 123, 966–972. [Google Scholar] [CrossRef] [PubMed]
- Cichowski, K.; Hahn, W.C. Unexpected pieces to the senescence puzzle. Cell 2008, 133, 958–961. [Google Scholar] [CrossRef]
- Wang, K.; Liu, H.; Hu, Q.; Wang, L.; Liu, J.; Zheng, Z.; Zhang, W.; Ren, J.; Zhu, F.; Liu, G.H. Epigenetic regulation of aging: Implications for interventions of aging and diseases. Signal Transduct. Target. Ther. 2022, 7, 374. [Google Scholar] [CrossRef]
- Hao, X.; Wang, C.; Zhang, R. Chromatin basis of the senescence-associated secretory phenotype. Trends Cell Biol. 2022, 32, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, Z.; Shen, W.; Huang, G.; Sedivy, J.M.; Wang, H.; Ju, Z. Inflammation, epigenetics, and metabolism converge to cell senescence and ageing: The regulation and intervention. Signal Transduct. Target. Ther. 2021, 6, 245. [Google Scholar] [CrossRef]
- Dimauro, T.; David, G. Chromatin modifications: The driving force of senescence and aging? Aging 2009, 1, 182–190. [Google Scholar] [CrossRef][Green Version]
- Kida, Y.; Goligorsky, M.S. Sirtuins, Cell Senescence, and Vascular Aging. Can. J. Cardiol. 2016, 32, 634–641. [Google Scholar] [CrossRef]
- Anwar, T.; Khosla, S.; Ramakrishna, G. Increased expression of SIRT2 is a novel marker of cellular senescence and is dependent on wild type p53 status. Cell Cycle 2016, 15, 1883–1897. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Cheng, B.; Su, Y.; Li, M.; Ma, S.; Zhang, Y.; Zhang, A.; Cai, S.; Bao, Q.; Wang, S.; et al. The Potential Role of m6A RNA Methylation in the Aging Process and Aging-Associated Diseases. Front. Genet. 2022, 13, 869950. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene Expression Regulation Mediated through Reversible m6A RNA Methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Krizhanovsky, V.; Yon, M.; Dickins, R.A.; Hearn, S.; Simon, J.; Miething, C.; Yee, H.; Zender, L.; Lowe, S.W. Senescence of activated stellate cells limits liver fibrosis. Cell 2008, 134, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lu, M.; Liu, D.; Shi, Y.; Ren, J.; Wang, S.; Jing, Y.; Zhang, S.; Zhao, Q.; Li, H.; et al. m6A epitranscriptomic regulation of tissue homeostasis during primate aging. Nat. Aging 2023, 3, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, R.; Baboota, R.K.; Gogg, S.; Beguinot, F.; Blüher, M.; Nerstedt, A.; Smith, U. Increased cell senescence in human metabolic disorders. J. Clin. Investig. 2023, 133, e169922. [Google Scholar] [CrossRef]
- Ou, M.Y.; Zhang, H.; Tan, P.C.; Zhou, S.B.; Li, Q.F. Adipose tissue aging: Mechanisms and therapeutic implications. Cell Death Dis. 2022, 13, 300. [Google Scholar] [CrossRef]
- Schaum, N.; Lehallier, B.; Hahn, O.; Pálovics, R.; Hosseinzadeh, S.; Lee, S.E.; Sit, R.; Lee, D.P.; Losada, P.M.; Zardeneta, M.E.; et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nature 2020, 583, 596–602. [Google Scholar] [CrossRef]
- Shmulevich, R.; Krizhanovsky, V. Cell Senescence, DNA Damage, and Metabolism. Antioxid. Redox Signal. 2021, 34, 324–334. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Scaffa, A.M.; Peterson, A.L.; Carr, J.F.; Garcia, D.; Yao, H.; Dennery, P.A. Hyperoxia causes senescence and increases glycolysis in cultured lung epithelial cells. Physiol. Rep. 2021, 9, e14839. [Google Scholar] [CrossRef] [PubMed]
- Moiseeva, O.; Bourdeau, V.; Roux, A.; Deschênes-Simard, X.; Ferbeyre, G. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol. Cell Biol. 2009, 29, 4495–4507. [Google Scholar] [CrossRef] [PubMed]
- Nacarelli, T.; Sell, C. Targeting metabolism in cellular senescence, a role for intervention. Mol. Cell. Endocrinol. 2017, 455, 83–92. [Google Scholar] [CrossRef]
- Moiseeva, O.; Guillon, J.; Ferbeyre, G. Senescence: A program in the road to cell elimination and cancer. Semin. Cancer Biol. 2022, 81, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.C. Pleiotropy, natural selection and the evolution of senescence. Evolution 1957, 11, 398–411. [Google Scholar] [CrossRef]
- Kang, T.W.; Yevsa, T.; Woller, N.; Hoenicke, L.; Wuestefeld, T.; Dauch, D.; Hohmeyer, A.; Gereke, M.; Rudalska, R.; Potapova, A.; et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 2011, 479, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Mudbhary, R.; Hoshida, Y.; Chernyavskaya, Y.; Jacob, V.; Villanueva, A.; Fiel, M.I.; Chen, X.; Kojima, K.; Thung, S.; Bronson, R.T.; et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 2014, 25, 196–209. [Google Scholar] [CrossRef]
- Xue, W.; Zender, L.; Miething, C.; Dickins, R.A.; Hernando, E.; Krizhanovsky, V.; Cordon-Cardo, C.; Lowe, S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007, 445, 656–660. [Google Scholar] [CrossRef]
- Li, F.; Huangyang, P.; Burrows, M.; Guo, K.; Riscal, R.; Godfrey, J.; Lee, K.E.; Lin, N.; Lee, P.; Blair, I.A.; et al. FBP1 loss disrupts liver metabolism and promotes tumorigenesis through a hepatic stellate cell senescence secretome. Nat. Cell Biol. 2020, 22, 728–739. [Google Scholar] [CrossRef]
- Krtolica, A.; Parrinello, S.; Lockett, S.; Desprez, P.Y.; Campisi, J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: A link between cancer and aging. Proc. Natl. Acad. Sci. USA 2001, 98, 12072–12077. [Google Scholar] [CrossRef]
- Ruhland, M.K.; Loza, A.J.; Capietto, A.H.; Luo, X.; Knolhoff, B.L.; Flanagan, K.C.; Belt, B.A.; Alspach, E.; Leahy, K.; Luo, J.; et al. Stromal senescence establishes an immunosuppressive microenvironment that drives tumorigenesis. Nat. Commun. 2016, 7, 11762. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Hornsby, P.J. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 2007, 67, 3117–3126. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef]
- Laberge, R.M.; Awad, P.; Campisi, J.; Desprez, P.Y. Epithelial-mesenchymal transition induced by senescent fibroblasts. Cancer Microenviron. 2012, 5, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Efimova, E.V.; Hamzeh, K.W.; Darga, T.E.; Mauceri, H.J.; Fu, Y.X.; Kron, S.J.; Weichselbaum, R.R. Radiation-inducible immunotherapy for cancer: Senescent tumor cells as a cancer vaccine. Mol. Ther. 2012, 20, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Udomsinprasert, W.; Sobhonslidsuk, A.; Jittikoon, J.; Honsawek, S.; Chaikledkaew, U. Cellular senescence in liver fibrosis: Implications for age-related chronic liver diseases. Expert Opin. Ther. Targets 2021, 25, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Caligiuri, A.; Gentilini, A.; Pastore, M.; Gitto, S.; Marra, F. Cellular and Molecular Mechanisms Underlying Liver Fibrosis Regression. Cells 2021, 10, 2759. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Kostallari, E.; Ibrahim, S.H.; Iwakiri, Y. The evolving role of liver sinusoidal endothelial cells in liver health and disease. Hepatology 2023, 78, 649–669. [Google Scholar] [CrossRef]
- Maeso-Díaz, R.; Ortega-Ribera, M.; Fernández-Iglesias, A.; Hide, D.; Muñoz, L.; Hessheimer, A.J.; Vila, S.; Francés, R.; Fondevila, C.; Albillos, A.; et al. Effects of aging on liver microcirculatory function and sinusoi dal phenotype. Aging Cell 2018, 17, e12829. [Google Scholar] [CrossRef]
- Delire, B.; Lebrun, V.; Selvais, C.; Henriet, P.; Bertrand, A.; Horsmans, Y.; Leclercq, I.A. Aging enhances liver fibrotic response in mice through hampering extracellular matrix remodeling. Aging 2016, 9, 98–113. [Google Scholar] [CrossRef]
- Yang, L.; Kwon, J.; Popov, Y.; Gajdos, G.B.; Ordog, T.; Brekken, R.A.; Mukhopadhyay, D.; Schuppan, D.; Bi, Y.; Simonetto, D.; et al. Vascular endothelial growth factor promotes fibrosis resolution and repair in mice. Gastroenterology 2014, 146, 1339–1350.e1. [Google Scholar] [CrossRef] [PubMed]
- Wijayasiri, P.; Astbury, S.; Kaye, P.; Oakley, F.; Alexander, G.J.; Kendall, T.J.; Aravinthan, A.D. Role of Hepatocyte Senescence in the Activation of Hepatic Stellate Cells and Liver Fibrosis Progression. Cells 2022, 11, 2221. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Jiang, X.; Dong, F.; Zhang, F.; Ji, X.; Xue, M.; Yang, F.; Chen, J.; Hu, X.; Bao, Z. Lipid accumulation-induced hepatocyte senescence regulates the activation of hepatic stellate cells through the Nrf2-antioxidant response element pathway. Exp. Cell Res. 2021, 405, 112689. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, S.U.; Satyanarayana, A.; Tsahuridu, M.; Tillmann, H.L.; Zender, L.; Klempnauer, J.; Flemming, P.; Franco, S.; Blasco, M.A.; Manns, M.P.; et al. Hepatocyte telomere shortening and senescence are general markers of human liver cirrhosis. FASEB J. 2002, 16, 935–942. [Google Scholar] [CrossRef]
- Calzadilla Bertot, L.; Adams, L.A. The Natural Course of Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2016, 17, 774. [Google Scholar] [CrossRef]
- Papatheodoridi, A.M.; Chrysavgis, L.; Koutsilieris, M.; Chatzigeorgiou, A. The Role of Senescence in the Development of Nonalcoholic Fatty Liver Disease and Progression to Nonalcoholic Steatohepatitis. Hepatology 2020, 71, 363–374. [Google Scholar] [CrossRef]
- Ogrodnik, M.; Miwa, S.; Tchkonia, T.; Tiniakos, D.; Wilson, C.L.; Lahat, A.; Day, C.P.; Burt, A.; Palmer, A.; Anstee, Q.M.; et al. Cellular senescence drives age-dependent hepatic steatosis. Nat. Commun. 2017, 8, 15691. [Google Scholar] [CrossRef]
- Park, H.; Ishigami, A.; Shima, T.; Mizuno, M.; Maruyama, N.; Yamaguchi, K.; Mitsuyoshi, H.; Minami, M.; Yasui, K.; Itoh, Y.; et al. Hepatic senescence marker protein-30 is involved in the progression of nonalcoholic fatty liver disease. J. Gastroenterol. 2010, 45, 426–434. [Google Scholar] [CrossRef]
- Engelmann, C.; Tacke, F. The Potential Role of Cellular Senescence in Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2022, 23, 652. [Google Scholar] [CrossRef]
- Novo, E.; Cappon, A.; Villano, G.; Quarta, S.; Cannito, S.; Bocca, C.; Turato, C.; Guido, M.; Maggiora, M.; Protopapa, F.; et al. SerpinB3 as a Pro-Inflammatory Mediator in the Progression of Experimental Non-Alcoholic Fatty Liver Disease. Front. Immunol. 2022, 13, 910526. [Google Scholar] [CrossRef]
- Dabravolski, S.A.; Bezsonov, E.E.; Orekhov, A.N. The role of mitochondria dysfunction and hepatic senescence in NAFLD development and progression. Biomed. Pharmacother. 2021, 142, 112041. [Google Scholar] [CrossRef] [PubMed]
- Asrih, M.; Jornayvaz, F.R. Metabolic syndrome and nonalcoholic fatty liver disease: Is insulin resistance the link? Mol. Cell. Endocrinol. 2015, 418, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, M.; Ammerpohl, O.; von Schönfels, W.; Kolarova, J.; Bens, S.; Itzel, T.; Teufel, A.; Herrmann, A.; Brosch, M.; Hinrichsen, H.; et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013, 18, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Sim, B.C.; Kang, Y.E.; You, S.K.; Lee, S.E.; Nga, H.T.; Lee, H.Y.; Nguyen, T.L.; Moon, J.S.; Tian, J.; Jang, H.J.; et al. Hepatic T-cell senescence and exhaustion are implicated in the progression of fatty liver disease in patients with type 2 diabetes and mouse model with nonalcoholic steatohepatitis. Cell Death Dis. 2023, 14, 618. [Google Scholar] [CrossRef] [PubMed]
- Barbé-Tuana, F.; Funchal, G.; Schmitz, C.R.R.; Maurmann, R.M.; Bauer, M.E. The interplay between immunosenescence and age-related diseases. Semin. Immunopathol. 2020, 42, 545–557. [Google Scholar] [CrossRef]
- Bektas, A.; Schurman, S.H.; Sen, R.; Ferrucci, L. Human T cell immunosenescence and inflammation in aging. J. Leukoc. Biol. 2017, 102, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Bonafè, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254. [Google Scholar] [CrossRef]
- Gupta, N.A.; Kolachala, V.L.; Jiang, R.; Abramowsky, C.; Shenoi, A.; Kosters, A.; Pavuluri, H.; Anania, F.; Kirk, A.D. Mitigation of autophagy ameliorates hepatocellular damage following ischemia-reperfusion injury in murine steatotic liver. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G1088–G1099. [Google Scholar] [CrossRef]
- Huda, N.; Khambu, B.; Liu, G.; Nakatsumi, H.; Yan, S.; Chen, X.; Ma, M.; Dong, Z.; Nakayama, K.I.; Yin, X.-M. Senescence Connects Autophagy Deficiency to Inflammation and Tumor Progression in the Liver. Cell. Mol. Gastroenterol. Hepatol. 2022, 14, 333–355. [Google Scholar] [CrossRef]
- Zhang, C.; Cuervo, A.M. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 2008, 14, 959–965. [Google Scholar] [CrossRef]
- Rajendran, P.; Alzahrani, A.M.; Hanieh, H.N.; Kumar, S.A.; Ben Ammar, R.; Rengarajan, T.; Alhoot, M.A. Autophagy and senescence: A new insight in selected human diseases. J. Cell Physiol. 2019, 234, 21485–21492. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Su, Y.; Duan, C.; Wang, S.; He, W.; Zhang, Y.; An, X.; He, M. Emerging role of aging in the progression of NAFLD to HCC. Ageing Res. Rev. 2023, 84, 101833. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Lin, Y.C. Autophagy Dysregulation in Metabolic Associated Fatty Liver Disease: A New Therapeutic Target. Int. J. Mol. Sci. 2022, 23, 10055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zeng, Y.; Zhao, L.; Xu, Q.; Miao, D.; Yu, F. Targeting Hepatic Stellate Cell Death to Reverse Hepatic Fibrosis. Curr. Drug Targets 2023, 24, 568–583. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, C.; Ziani, S.; Nelson, L.J.; Avila, M.A.; Nevzorova, Y.A.; Cubero, F.J. Fibrotic Events in the Progression of Cholestatic Liver Disease. Cells 2021, 10, 1107. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Elledge, S.J. How autophagy both activates and inhibits cellular senescence. Autophagy 2016, 12, 898–899. [Google Scholar] [CrossRef] [PubMed]
- Abdellatif, M.; Rainer, P.P.; Sedej, S.; Kroemer, G. Hallmarks of cardiovascular ageing. Nat. Rev. Cardiol. 2023, 20, 754–777. [Google Scholar] [CrossRef]
- Sung, J.Y.; Kim, S.G.; Kim, J.R.; Choi, H.C. SIRT1 suppresses cellular senescence and inflammatory cytokine release in human dermal fibroblasts by promoting the deacetylation of NF-κB and activating autophagy. Exp. Gerontol. 2021, 150, 111394. [Google Scholar] [CrossRef]
- Lee, I.H.; Cao, L.; Mostoslavsky, R.; Lombard, D.B.; Liu, J.; Bruns, N.E.; Tsokos, M.; Alt, F.W.; Finkel, T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA 2008, 105, 3374–3379. [Google Scholar] [CrossRef]
- Xu, C.; Wang, L.; Fozouni, P.; Evjen, G.; Chandra, V.; Jiang, J.; Lu, C.; Nicastri, M.; Bretz, C.; Winkler, J.D.; et al. SIRT1 is downregulated by autophagy in senescence and ageing. Nat. Cell Biol. 2020, 22, 1170–1179. [Google Scholar] [CrossRef]
- Chun, S.K.; Lee, S.; Flores-Toro, J.; U, R.Y.; Yang, M.J.; Go, K.L.; Biel, T.G.; Miney, C.E.; Pierre Louis, S.; Law, B.K.; et al. Loss of sirtuin 1 and mitofusin 2 contributes to enhanced ischemia/reperfusion injury in aged livers. Aging Cell 2018, 17, e12761. [Google Scholar] [CrossRef] [PubMed]
- Dou, Z.; Xu, C.; Donahue, G.; Shimi, T.; Pan, J.A.; Zhu, J.; Ivanov, A.; Capell, B.C.; Drake, A.M.; Shah, P.P.; et al. Autophagy mediates degradation of nuclear lamina. Nature 2015, 527, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Toshima, T.; Shirabe, K.; Fukuhara, T.; Ikegami, T.; Yoshizumi, T.; Soejima, Y.; Ikeda, T.; Okano, S.; Maehara, Y. Suppression of autophagy during liver regeneration impairs energy charge and hepatocyte senescence in mice. Hepatology 2014, 60, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, F.; Galluzzi, L.; Bravo-San Pedro, J.M.; Madeo, F.; Kroemer, G. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 2015, 21, 805–821. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, Y.; Dunfield, J.; Roderique, T.; Ni, H.M. Liver-adipose tissue crosstalk in alcohol-associated liver disease: The role of mTOR. Liver Res. 2022, 6, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Basu, A. The interplay between apoptosis and cellular senescence: Bcl-2 family proteins as targets for cancer therapy. Pharmacol. Ther. 2022, 230, 107943. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, J.L.; Tchkonia, T. Cellular Senescence: A Translational Perspective. eBioMedicine 2017, 21, 21–28. [Google Scholar] [CrossRef]
- Hang, L.; Pitcher, L.E.; Prahalad, V.; Niedernhofer, L.J.; Robbins, P.D. Targeting cellular senescence with senotherapeutics: Senolytics and senomorphics. FEBS J. 2023, 290, 1362–1383. [Google Scholar]
- Yang, M.; Luo, S.; Chen, W.; Zhao, L.; Wang, X. Chaperone-Mediated Autophagy: A Potential Target for Metabolic Diseases. Curr. Med. Chem. 2023, 30, 1887–1899. [Google Scholar] [CrossRef]
- Kaushik, S.; Kiffin, R.; Cuervo, A.M. Chaperone-mediated autophagy and aging: A novel regulatory role of lipids revealed. Autophagy 2007, 3, 387–389. [Google Scholar] [CrossRef][Green Version]
- Cuervo, A.M.; Dice, J.F. Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 2000, 275, 31505–31513. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, Y.; Yang, X.; Dong, W.; Yang, J.; Hu, Q.; Zhang, C.; Fang, H.; Liu, A. Growth differentiation factor 11 accelerates liver senescence through the inhibition of autophagy. Aging Cell 2022, 21, e13532. [Google Scholar] [CrossRef] [PubMed]
- Byun, S.; Seok, S.; Kim, Y.C.; Zhang, Y.; Yau, P.; Iwamori, N.; Xu, H.E.; Ma, J.; Kemper, B.; Kemper, J.K. Fasting-induced FGF21 signaling activates hepatic autophagy and lipid degradation via JMJD3 histone demethylase. Nat. Commun. 2020, 11, 807. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Xia, A.; Meng, F.; Chunyu, J.; Sun, X.; Ren, G.; Yu, D.; Jiang, X.; Tang, L.; Xiao, W.; et al. FGF21 alleviates chronic inflammatory injury in the aging process through modulating polarization of macrophages. Int. Immunopharmacol. 2021, 96, 107634. [Google Scholar] [CrossRef] [PubMed]
- Lauri, A.; Pompilio, G.; Capogrossi, M.C. The mitochondrial genome in aging and senescence. Ageing Res. Rev. 2014, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.S.; Gubbi, S.; Barzilai, N. Benefits of Metformin in Attenuating the Hallmarks of Aging. Cell Metab. 2020, 32, 15–30. [Google Scholar] [CrossRef]
- Bharath, L.P.; Agrawal, M.; McCambridge, G.; Nicholas, D.A.; Hasturk, H.; Liu, J.; Jiang, K.; Liu, R.; Guo, Z.; Deeney, J.; et al. Metformin Enhances Autophagy and Normalizes Mitochondrial Function to Alleviate Aging-Associated Inflammation. Cell Metab. 2020, 32, 44–55.e6. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.Y.K.; Chiu, D.K.C.; Yuen, V.W.H.; Law, C.T.; Wong, B.P.Y.; Thu, K.L.; Wong, C.C.L.; Cescon, D.W.; Soria-Bretones, I.; Cheu, J.W.S.; et al. CFI-402257, a TTK inhibitor, effectively suppresses hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2022, 119, e2119514119. [Google Scholar] [CrossRef]
- Lv, Z.; Guo, Y. Metformin and Its Benefits for Various Diseases. Front. Endocrinol. 2020, 11, 191. [Google Scholar] [CrossRef]
- Hunt, N.J.; Lockwood, G.P.; Kang, S.W.S.; Pulpitel, T.; Clark, X.; Mao, H.; McCourt, P.A.G.; Cooney, G.J.; Wali, J.A.; Le Couteur, F.H.; et al. The Effects of Metformin on Age-Related Changes in the Liver Sinusoidal Endothelial Cell. J. Gerontol. Ser. A 2020, 75, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Zhang, X.; Zhai, R.; Liang, H.; Song, G.; Yuan, Y.; Xu, Y.; Yan, Y.; Qiu, L.; Sun, T. Metformin attenuated sepsis-associated liver injury and inflammatory response in aged mice. Bioengineered 2022, 13, 4598–4609. [Google Scholar] [CrossRef] [PubMed]
- Van Deursen, J.M. Senolytic therapies for healthy longevity. Science 2019, 364, 636–637. [Google Scholar] [CrossRef] [PubMed]
- Thadathil, N.; Selvarani, R.; Mohammed, S.; Nicklas, E.H.; Tran, A.L.; Kamal, M.; Luo, W.; Brown, J.L.; Lawrence, M.M.; Borowik, A.K.; et al. Senolytic treatment reduces cell senescence and necroptosis in Sod1 knockout mice that is associated with reduced inflammation and hepatocellular carcinoma. Aging Cell 2022, 21, e13676. [Google Scholar] [CrossRef]
| Cell Type | Disease Type | Function | Reference |
|---|---|---|---|
| Hepatocyte | Fibrosis | Protect the hepatocyte from fibrosis | [64,79] |
| LSECs | Fibrosis | Limit the development of fibrosis | [81,82] |
| HSCs | Fibrosis | Accelerate the development of fibrosis | [80] |
| LSEC | NASH | Prevent the development of NASH | [72] |
| Hepatocyte | Early stage of liver cancer | Tumor-Preventing | [84] |
| Hepatocyte | Late stage of liver cancer | Tumor-Promoting | [89] |
| Cell Type | Disease Type | Function | Reference |
|---|---|---|---|
| Hepatocyte | Fibrosis | Promoting fibrosis | [146] |
| LSECs | Fibrosis | Accelerating the development of fibrosis | [148] |
| HSCs | Fibrosis | Reducing the development of fibrosis | [124] |
| Hepatocyte | Cirrhosis | Contributing to liver cirrhosis | [154] |
| Hepatocyte | Early stage of liver cancer | Tumor-Preventing | [134] |
| Hepatocyte | Late stage of liver cancer | Tumor-Promoting | [65] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Lin, Y.; Liang, G.; Xiao, N.; Zhang, H.; Yang, X.; Yang, J.; Liu, A. Autophagy and Senescence: The Molecular Mechanisms and Implications in Liver Diseases. Int. J. Mol. Sci. 2023, 24, 16880. https://doi.org/10.3390/ijms242316880
Li Q, Lin Y, Liang G, Xiao N, Zhang H, Yang X, Yang J, Liu A. Autophagy and Senescence: The Molecular Mechanisms and Implications in Liver Diseases. International Journal of Molecular Sciences. 2023; 24(23):16880. https://doi.org/10.3390/ijms242316880
Chicago/Turabian StyleLi, Qiao, Yan Lin, Guangyu Liang, Nanyin Xiao, Heng Zhang, Xiao Yang, Jiankun Yang, and Anding Liu. 2023. "Autophagy and Senescence: The Molecular Mechanisms and Implications in Liver Diseases" International Journal of Molecular Sciences 24, no. 23: 16880. https://doi.org/10.3390/ijms242316880
APA StyleLi, Q., Lin, Y., Liang, G., Xiao, N., Zhang, H., Yang, X., Yang, J., & Liu, A. (2023). Autophagy and Senescence: The Molecular Mechanisms and Implications in Liver Diseases. International Journal of Molecular Sciences, 24(23), 16880. https://doi.org/10.3390/ijms242316880






