Comparative Analysis of Transposable Elements in Strawberry Genomes of Different Ploidy Levels
Abstract
:1. Introduction
2. Results
2.1. Construction of a Pan-Genome TE Library with Ten Fragaria Species
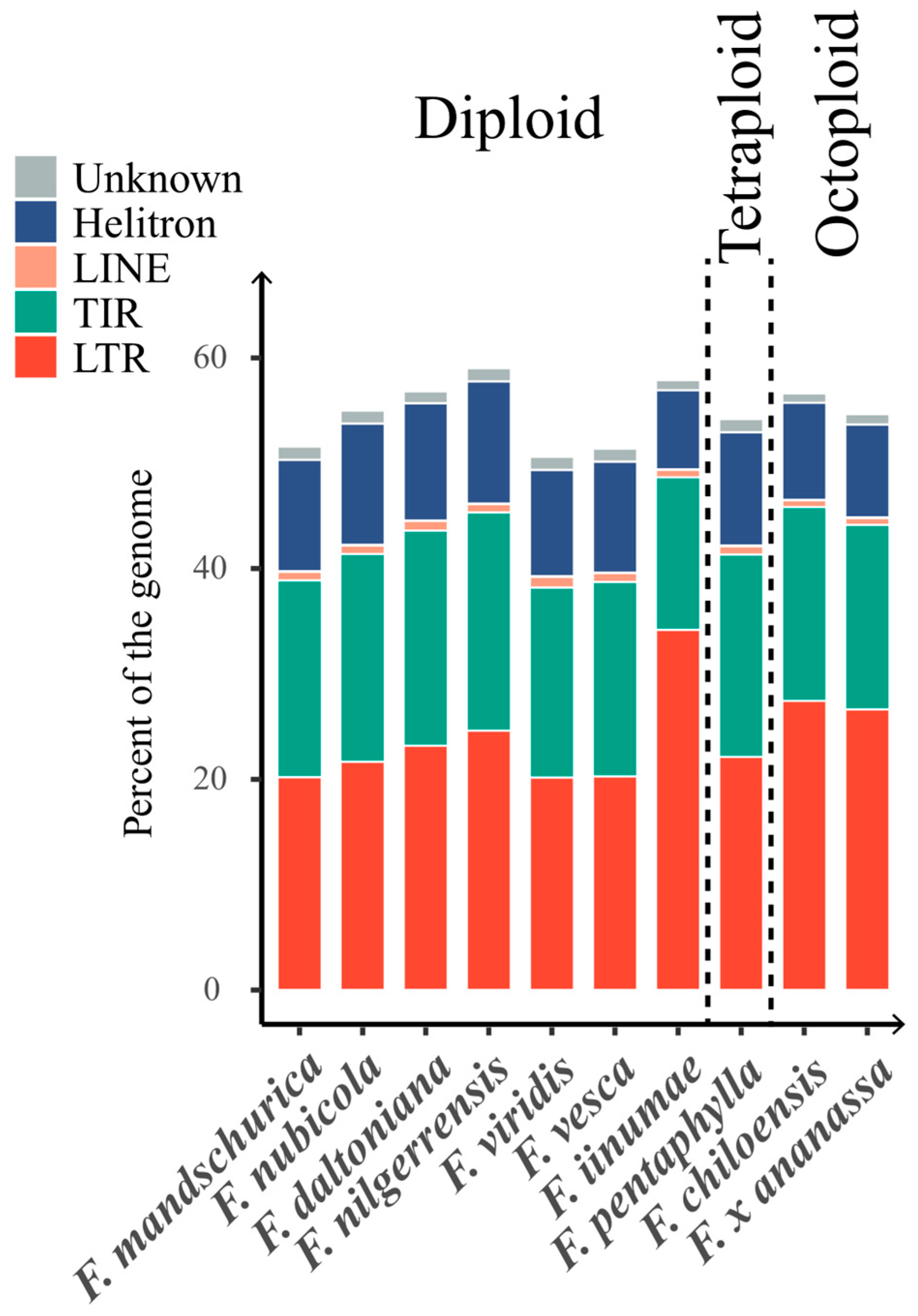
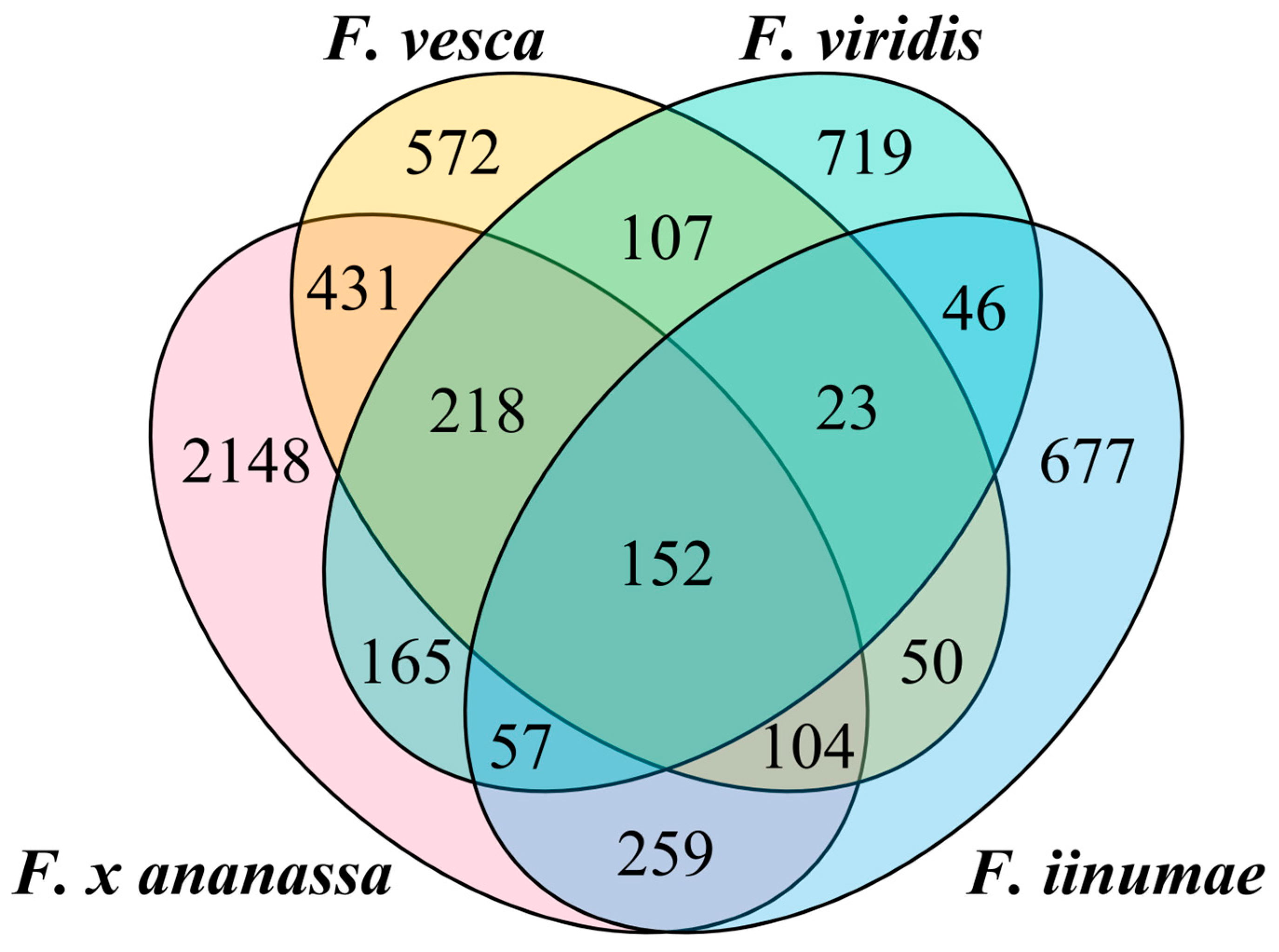
2.1.1. Pan-Genome TE Content across Genus Fragaria
2.1.2. Diversity of TE Content in the Genome of Strawberries
2.2. The Evolution History of TEs in the Fragaria Genus
2.2.1. Amplification of TE Subfamilies
2.2.2. TE Contents Are Strongly Correlated with Genome Sizes of Diploid Strawberries
2.2.3. Removal Rate of LTR-RTs and the Strawberry Genome Size
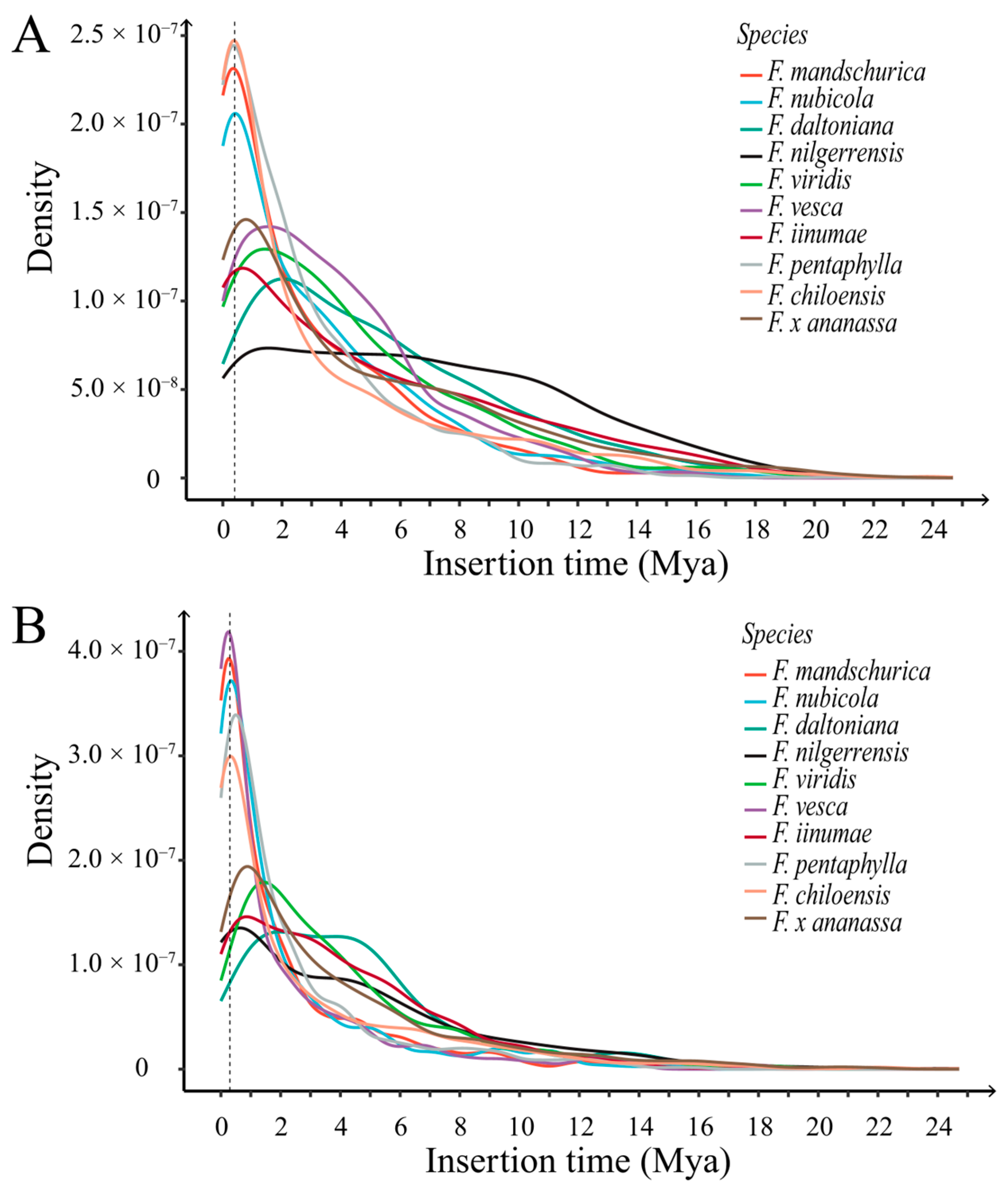
2.2.4. The Transposition Bursts of LTR-RTs in Fragaria Genomes
2.2.5. Recent Transposition Burst of LTR Retrotransposons
2.3. Evolution History of TEs in Cultivated Strawberry
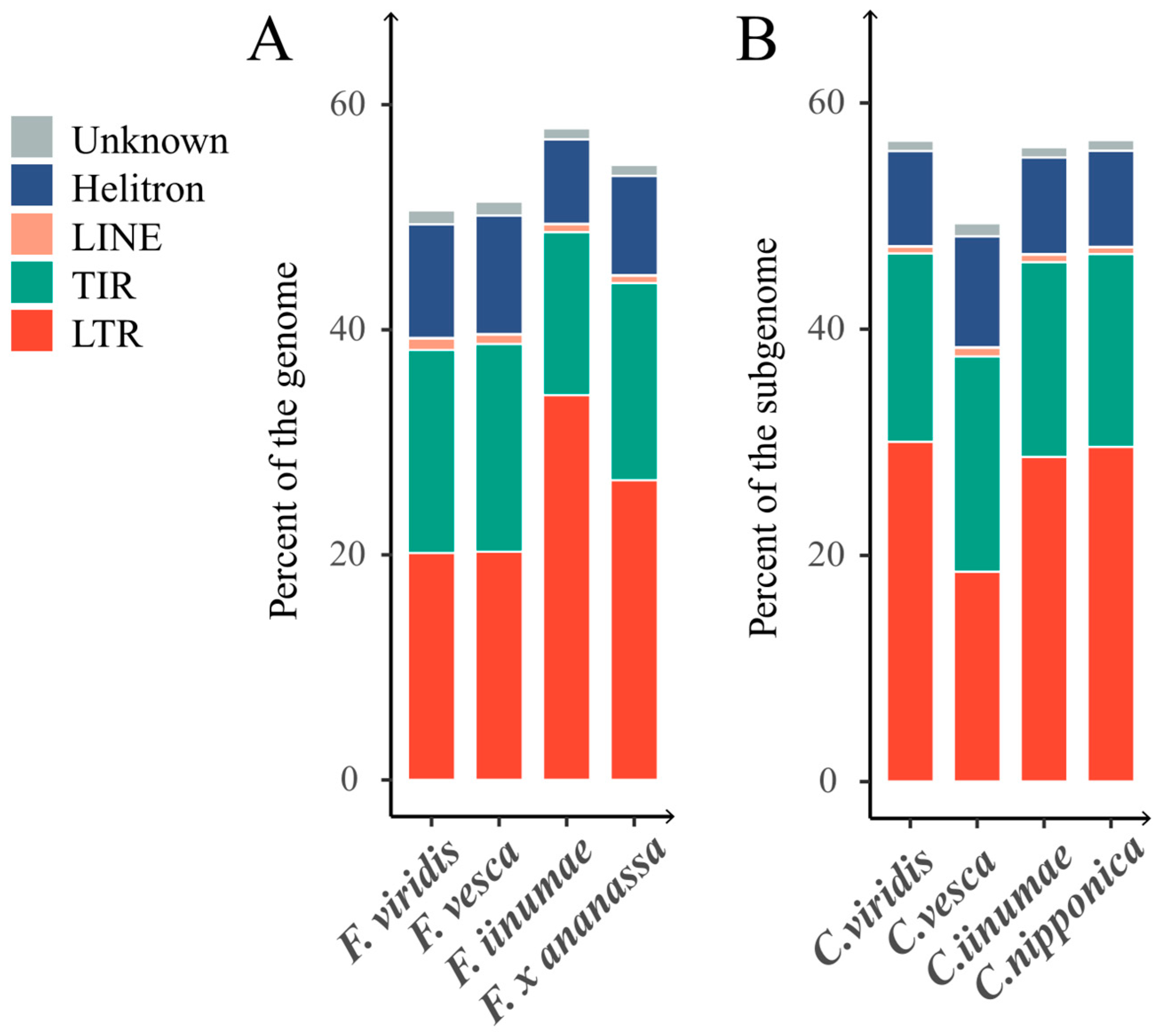
2.3.1. Comparative Analysis of the TE Contribution between Cultivated Strawberry Subgenomes and Their Diploid Ancestor Genomes
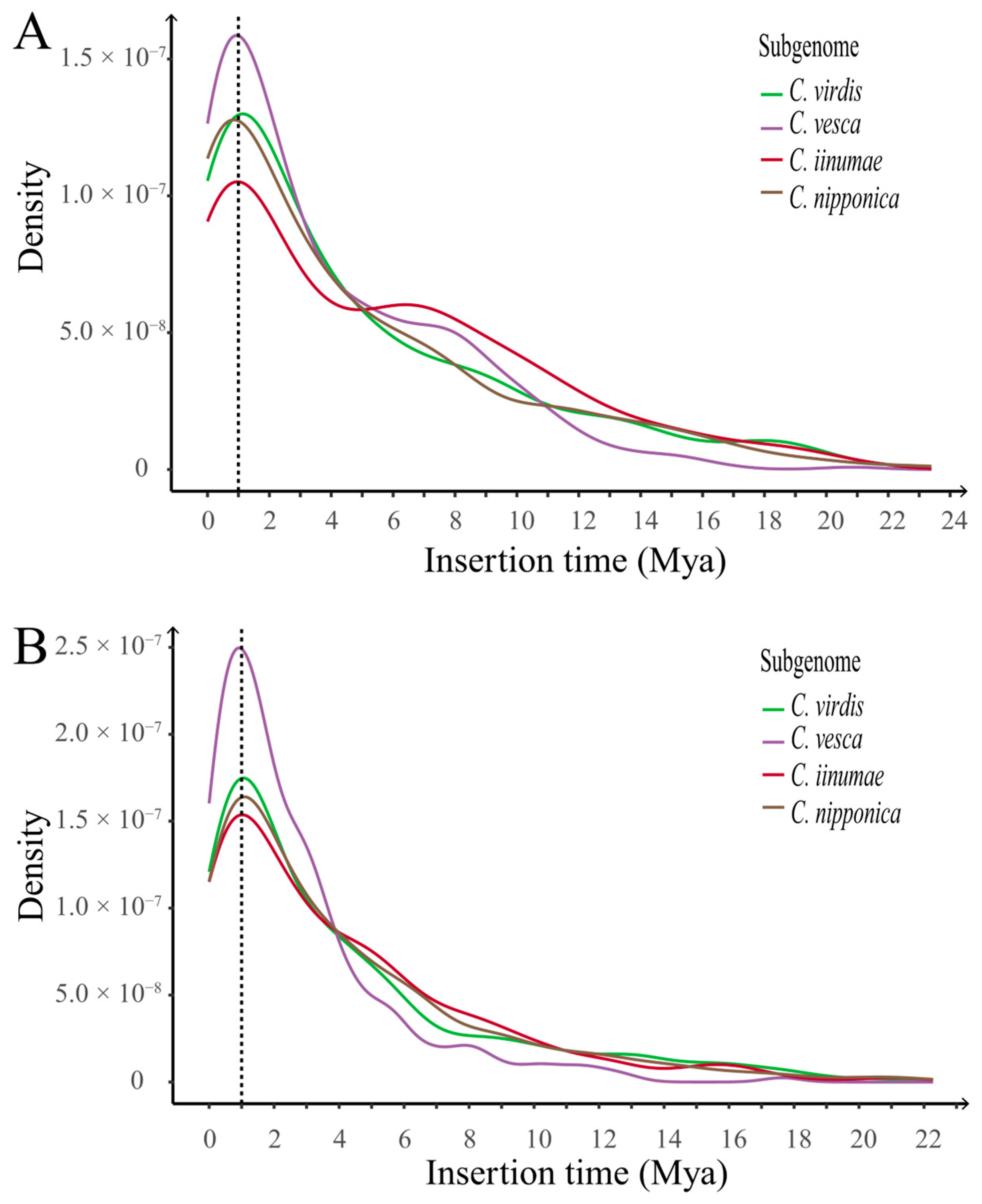
2.3.2. Evolution History of LTR-RTs in Cultivated Strawberry
2.3.3. Shared TEs in Cultivated Strawberry Genome and the Ancestor Genomes
3. Discussion
3.1. The Contributions of TE Amplification and DNA Removal to Genome Size
3.2. The Role of TEs in Strawberry Genome Evolution
3.3. New Perspective on the Diploid Ancestors of Cultivated Strawberry
4. Materials and Methods
4.1. Genome Datasets
4.2. Construction of the Pan-Genome TE Library for Fragaria Species
4.3. Genome Masking
4.4. Kimura Distance-Based Propagation Activity Analysis
4.5. Estimate the Insertion Time of LTR Retrotransposon
4.6. Estimate the Half-Life Rate of LTR Retrotransposon
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvak, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Wang, H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Baucom, R.S.; Estill, J.C.; Chaparro, C.; Upshaw, N.; Jogi, A.; Deragon, J.-M.; Westerman, R.P.; SanMiguel, P.J.; Bennetzen, J.L. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 2009, 5, e1000732. [Google Scholar] [CrossRef]
- Wells, J.N.; Feschotte, C. A field guide to eukaryotic transposable elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Bennetzen, J.L. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 2005, 15, 621–627. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, J.; Han, X.; Li, J.; Gao, Y.; Richards, C.M.; Zhang, C.; Tian, Y.; Liu, G.; Gul, H.; et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019, 10, 1494. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gao, Y.; Jia, H.; Liu, L.; Zhang, D.; Zhang, Z. Comparative transcriptomics uncovers alternative splicing changes and signatures of selection from maize improvement. BMC Genom. 2015, 16, 363. [Google Scholar] [CrossRef]
- McCann, J.; Macas, J.; Novak, P.; Stuessy, T.F.; Villasenor, J.L.; Weiss-Schneeweiss, H. Differential genome size and repetitive DNA evolution in diploid species of Melampodium sect. Melampodium (Asteraceae). Front. Plant Sci. 2020, 11, 362. [Google Scholar] [CrossRef]
- Zeng, X.; Xu, T.; Ling, Z.; Wang, Y.; Li, X.; Xu, S.; Xu, Q.; Zha, S.; Qimei, W.; Basang, Y.; et al. An improved high-quality genome assembly and annotation of Tibetan hulless barley. Sci. Data 2020, 7, 139. [Google Scholar] [CrossRef]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef]
- Rohilla, M.; Mazumder, A.; Saha, D.; Pal, T.; Begam, S.; Mondal, T.K. Genome-wide identification and development of miniature inverted-repeat transposable elements and intron length polymorphic markers in tea plant (Camellia sinensis). Sci. Rep. 2022, 12, 16233. [Google Scholar] [CrossRef]
- Castanera, R.; Morales-Diaz, N.; Gupta, S.; Purugganan, M.; Casacuberta, J.M. Transposons are important contributors to gene expression variability under selection in rice populations. eLife 2023, 12, RP86324. [Google Scholar] [CrossRef]
- Yan, H.; Haak, D.C.; Li, S.; Huang, L.; Bombarely, A. Exploring transposable element-based markers to identify allelic variations underlying agronomic traits in rice. Plant Commun. 2022, 3, 100270. [Google Scholar] [CrossRef]
- Han, Y.; Qin, S.; Wessler, S.R. Comparison of class 2 transposable elements at superfamily resolution reveals conserved and distinct features in cereal grass genomes. BMC Genom. 2013, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; Zhang, J.; Feng, Y.; Chen, Q.; Liu, Z.S.; Liu, C.L.; He, W.; Wang, H.; Yang, S.F.; et al. Comparative analysis of transposable elements and the identification of candidate centromeric elements in the Prunus subgenus Cerasus and Its relatives. Genes 2022, 13, 641. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; She, H.B.; Yang, L.L.; Lan, L.N.; Zhang, X.Y.; Wang, L.Y.; Zhang, Y.L.; Li, N.; Deng, C.L.; Qian, W.; et al. Impact of LTR-retrotransposons on genome structure, evolution, and function in Curcurbitaceae species. Int. J. Mol. Sci. 2022, 23, 10158. [Google Scholar] [CrossRef] [PubMed]
- Nouroz, F.; Noreen, S.; Heslop-Harrison, J.S. Evolutionary genomics of miniature inverted-repeat transposable elements (MITEs) in Brassica. Mol. Genet. Genom. 2015, 290, 2297–2312. [Google Scholar] [CrossRef] [PubMed]
- Liston, A.; Cronn, R.; Ashman, T.L. Fragaria: A genus with deep historical roots and ripe for evolutionary and ecological insights. Am. J. Bot. 2014, 101, 1686–1699. [Google Scholar] [CrossRef] [PubMed]
- Tennessen, J.A.; Govindarajulu, R.; Ashman, T.L.; Liston, A. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol. 2014, 6, 3295–3313. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- Edger, P.P.; VanBuren, R.; Colle, M.; Poorten, T.J.; Wai, C.M.; Niederhuth, C.E.; Alger, E.I.; Ou, S.; Acharya, C.B.; Wang, J.; et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 2018, 7, gix124. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; McKain, M.R.; Yocca, A.E.; Knapp, S.J.; Qiao, Q.; Zhang, T. Reply to: Revisiting the origin of octoploid strawberry. Nat. Genet. 2020, 52, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Liston, A.; Wei, N.; Tennessen, J.A.; Li, J.; Dong, M.; Ashman, T.L. Revisiting the origin of octoploid strawberry. Nat. Genet. 2020, 52, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.L.; Govindarajulu, R.; Ashman, T.-L. Bioclimatic evaluation of geographical range in Fragaria (Rosaceae) consequences of variation in breeding system, ploidy and species age. Bot. J. Linn. Soc. 2014, 176, 99–114. [Google Scholar] [CrossRef]
- Qiao, Q.; Edger, P.P.; Xue, L.; Qiong, L.; Lu, J.; Zhang, Y.; Cao, Q.; Yocca, A.E.; Platts, A.E.; Knapp, S.J.; et al. Evolutionary history and pan-genome dynamics of strawberry (Fragaria spp.). Proc. Natl. Acad. Sci. USA 2021, 118, e2105431118. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Xue, L.; Luo, G.; Zhang, T.; Lei, J. Investigation and taxonomy of wild Fragaria resources in Tibet, China. Genet. Resour. Crop Evol. 2017, 65, 405–415. [Google Scholar] [CrossRef]
- Luo, G.; Xue, L.; Guo, R.; Ding, Y.; Xu, W.; Lei, J. Creating interspecific hybrids with improved cold resistance in Fragaria. Sci. Hortic. 2018, 234, 1–9. [Google Scholar] [CrossRef]
- Zhang, J.; Lei, Y.; Wang, B.; Li, S.; Yu, S.; Wang, Y.; Li, H.; Liu, Y.; Ma, Y.; Dai, H.; et al. The high-quality genome of diploid strawberry (Fragaria nilgerrensis) provides new insights into anthocyanin accumulation. Plant Biotechnol. J. 2020, 18, 1908–1924. [Google Scholar] [CrossRef]
- Sangiacomo, M.A.; Sullivan, J.A. Introgression of wild species into the cultivated strawberry using synthetic octoploids. Theor. Appl. Genet. 1994, 88, 349–354. [Google Scholar] [CrossRef]
- Noguchi, Y.; Mochizuki, T.; Sone, K. Breeding of a new aromatic strawberry by interspecific hybridization Fragaria × ananassa × F. nilgerrensis. Engei Gakkai Zasshi 2002, 71, 208–213. [Google Scholar] [CrossRef]
- Castillejo, C.; Waurich, V.; Wagner, H.; Ramos, R.; Oiza, N.; Muñoz, P.; Triviño, J.C.; Caruana, J.; Liu, Z.; Cobo, N.; et al. Allelic variation of MYB10 is the major force controlling natural variation in skin and flesh color in strawberry (Fragaria spp.) fruit. Plant Cell 2020, 32, 3723–3749. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, C.; He, B.; Wan, Z.; Zhao, Y.; Hu, F.; Lv, Y. Transcriptome profiling of transposon-derived long non-coding RNAs response to hormone in strawberry fruit development. Front. Plant Sci. 2022, 13, 915569. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Wang, J.; Harris, A.J.; Folta, K.M.; Zhao, M.; Kang, M. Tracing the diploid ancestry of the cultivated octoploid strawberry. Mol. Biol. Evol. 2021, 38, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Cauret, C.M.S.; Mortimer, S.M.E.; Roberti, M.C.; Ashman, T.L.; Liston, A. Chromosome-scale assembly with a phased sex-determining region resolves features of early Z and W chromosome differentiation in a wild octoploid strawberry. G3 (Bethesda) 2022, 12, jkac139. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Shirasawa, K.; Kosugi, S.; Tashiro, K.; Nakayama, S.; Yamada, M.; Kohara, M.; Watanabe, A.; Kishida, Y.; Fujishiro, T.; et al. Dissection of the octoploid strawberry genome by deep sequencing of the genomes of Fragaria species. DNA Res. 2013, 21, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Su, W.; Liao, Y.; Chougule, K.; Agda, J.R.A.; Hellinga, A.J.; Lugo, C.S.B.; Elliott, T.A.; Ware, D.; Peterson, T.; et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019, 20, 275. [Google Scholar] [CrossRef] [PubMed]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Wicker, T.; Keller, B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 2007, 17, 1072–1081. [Google Scholar] [CrossRef]
- Zhang, R.G.; Li, G.Y.; Wang, X.L.; Dainat, J.; Wang, Z.X.; Ou, S.; Ma, Y. TEsorter: An accurate and fast method to classify LTR-retrotransposons in plant genomes. Hortic. Res. 2022, 9, uhac017. [Google Scholar] [CrossRef]
- Vicient, C.M.; Casacuberta, J.M. Impact of transposable elements on polyploid plant genomes. Ann. Bot. 2017, 120, 195–207. [Google Scholar] [CrossRef]
- Castro, M.R.J.; Goubert, C.; Monteiro, F.A.; Vieira, C.; Carareto, C.M.A. Homology-free detection of transposable elements unveils their dynamics in three ecologically distinct Rhodnius species. Genes 2020, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Laurie, J.D.; Ali, S.; Linning, R.; Mannhaupt, G.; Wong, P.; Güldener, U.; Münsterkötter, M.; Moore, R.; Kahmann, R.; Bakkeren, G.; et al. Genome comparison of barley and maize smut fungi reveals targeted Loss of RNA silencing components and species-specific presence of transposable elements. Plant Cell 2012, 24, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Armisen, D.; Gibbs, R.A.; Hering, L.; Khila, A.; Mayer, G.; Richards, S.; Niehuis, O.; Misof, B. Diversity and evolution of the transposable element repertoire in arthropods with particular reference to insects. BMC Ecol. Evol. 2019, 19, 11. [Google Scholar] [CrossRef] [PubMed]
- Chalopin, D.; Naville, M.; Plard, F.; Galiana, D.; Volff, J.N. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol. 2015, 7, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Suntsova, M.V.; Buzdin, A.A. Differences between human and chimpanzee genomes and their implications in gene expression, protein functions and biochemical properties of the two species. BMC Genom. 2020, 21, 535. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Ammiraju, J.S.; Zuccolo, A.; Yu, Y.; Song, X.; Piegu, B.; Chevalier, F.; Walling, J.G.; Ma, J.; Talag, J.; Brar, D.S.; et al. Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J. 2007, 52, 342–351. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, L.; Dou, L.; Yue, B.; Xing, J.; Li, J. Transposable elements shape the genome diversity and the evolution of noctuidae species. Genes 2023, 14, 1244. [Google Scholar] [CrossRef]
- Wong, W.Y.; Simakov, O.; Bridge, D.M.; Cartwright, P.; Bellantuono, A.J.; Kuhn, A.; Holstein, T.W.; David, C.N.; Steele, R.E.; Martinez, D.E. Expansion of a single transposable element family is associated with genome-size increase and radiation in the genus Hydra. Proc. Natl. Acad. Sci. USA 2019, 116, 22915–22917. [Google Scholar] [CrossRef]
- Kapusta, A.; Suh, A. Evolution of bird genomes-a transposon’s-eye view. Ann. N. Y. Acad. Sci. 2017, 1389, 164–185. [Google Scholar] [CrossRef]
- Sun, X.; Jiao, C.; Schwaninger, H.; Chao, C.T.; Ma, Y.; Duan, N.; Khan, A.; Ban, S.; Xu, K.; Cheng, L.; et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Alioto, T.; Alexiou, K.G.; Bardil, A.; Barteri, F.; Castanera, R.; Cruz, F.; Dhingra, A.; Duval, H.; Fernández i Martí, Á.; Frias, L.; et al. Transposons played a major role in the diversification between the closely related almond and peach genomes: Results from the almond genome sequence. Plant J. 2019, 101, 455–472. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zhang, H.H.; Wang, X. Machine learning for big data analytics in plants. Trends Plant Sci. 2014, 19, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Cortes, A.J.; Restrepo-Montoya, M.; Bedoya-Canas, L.E. Modern strategies to assess and breed forest tree adaptation to changing climate. Front. Plant Sci. 2020, 11, 583323. [Google Scholar] [CrossRef]
- Cortes, A.J.; Lopez-Hernandez, F. Harnessing crop wild diversity for climate change adaptation. Genes 2021, 12, 783. [Google Scholar] [CrossRef]
- Tong, H.; Nikoloski, Z. Machine learning approaches for crop improvement: Leveraging phenotypic and genotypic big data. J. Plant Physiol. 2021, 257, 153354. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the genome database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef]
- Ou, S.; Jiang, N. LTR_retriever: A highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018, 176, 1410–1422. [Google Scholar] [CrossRef]
- Su, W.; Gu, X.; Peterson, T. TIR-Learner, a new ensemble method for TIR transposable element annotation, orovides evidence for abundant new transposable elements in the maize genome. Mol. Plant 2019, 12, 447–460. [Google Scholar] [CrossRef]
- Xiong, W.; He, L.; Lai, J.; Dooner, H.K.; Du, C. HelitronScanner uncovers a large overlooked cache of Helitron transposons in many plant genomes. Proc. Natl. Acad. Sci. USA 2014, 111, 10263–10268. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Collins, T.; Qiu, Y.; Seetharam, A.S.; Menard, C.C.; Manchanda, N.; Gent, J.I.; Schatz, M.C.; Anderson, S.N.; Hufford, M.B.; et al. Differences in activity and stability drive transposable element variation in tropical and temperate maize. bioRxiv 2022. [Google Scholar] [CrossRef]
- Yan, H.; Bombarely, A.; Li, S. DeepTE: A computational method for de novo classification of transposons with convolutional neural network. Bioinformatics 2020, 36, 4269–4275. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]




| Type of TE | Number of Sequences | |
|---|---|---|
| Class I | 8931 | |
| LTR | 8539 | |
| Copia | 3002 | |
| Gypsy | 5255 | |
| Unknown | 282 | |
| NonLTR | 392 | |
| LINE | 320 | |
| SINE | 46 | |
| DIRS | 3 | |
| PLE | 23 | |
| Class II | 16,154 | |
| TIR | 13,945 | |
| CACTA | 3928 | |
| Mutator | 5007 | |
| P | 2 | |
| PIF-Harbinger | 1408 | |
| Tc1-Mariner | 517 | |
| hAT | 3083 | |
| Helitron | 2209 | |
| Unknown | 1141 | |
| Total | 26,226 | |
| Subgenome | Total | F. viridis | F. vesca | F. iinumae |
|---|---|---|---|---|
| Camarosa viridis | 1639 | 85 | 127 | 157 |
| Camarosa vesca | 2136 | 120 | 516 | 73 |
| Camarosa iinumae | 1855 | 82 | 139 | 243 |
| Camarosa nipponica | 1824 | 80 | 151 | 154 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, K.; Xiao, J.; Lyu, S.; Liu, R. Comparative Analysis of Transposable Elements in Strawberry Genomes of Different Ploidy Levels. Int. J. Mol. Sci. 2023, 24, 16935. https://doi.org/10.3390/ijms242316935
Lyu K, Xiao J, Lyu S, Liu R. Comparative Analysis of Transposable Elements in Strawberry Genomes of Different Ploidy Levels. International Journal of Molecular Sciences. 2023; 24(23):16935. https://doi.org/10.3390/ijms242316935
Chicago/Turabian StyleLyu, Keliang, Jiajing Xiao, Shiheng Lyu, and Renyi Liu. 2023. "Comparative Analysis of Transposable Elements in Strawberry Genomes of Different Ploidy Levels" International Journal of Molecular Sciences 24, no. 23: 16935. https://doi.org/10.3390/ijms242316935
APA StyleLyu, K., Xiao, J., Lyu, S., & Liu, R. (2023). Comparative Analysis of Transposable Elements in Strawberry Genomes of Different Ploidy Levels. International Journal of Molecular Sciences, 24(23), 16935. https://doi.org/10.3390/ijms242316935







