Chromosomal Instability in Gastric Cancer: Role in Tumor Development, Progression, and Therapy
Abstract
:1. Introduction
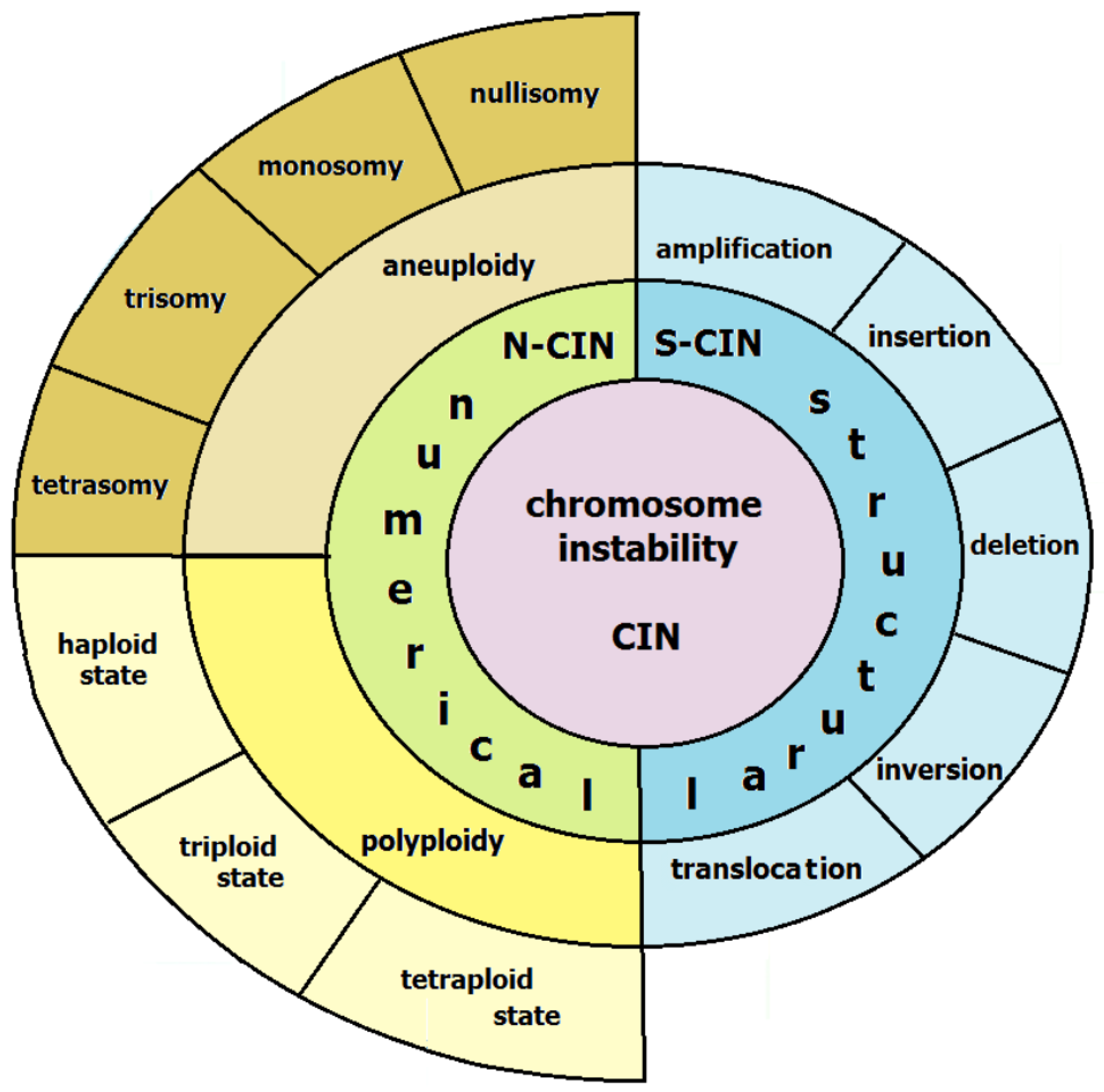
2. Types and Mechanisms of Chromosomal Instability in CIN Subtype of GC
3. Chromosomal Rearrangements Characterizing Chromosomal Instability in Gastric Cancer
4. Driver Genes in the CIN Subtypes of GC and Their Associations with Its Tumorigenesis and Therapy
4.1. Genes Regulating the Cell Cycle and the Response to DNA Damage
4.2. Genes Regulating Cell Division
4.3. Genes Regulating EGFR Signaling
5. The Link between Mutations in Driver Genes and Chromasomal Instability
6. Therapeutic Approaches for Treatment of the CIN Subtype of GC
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Japanese Gastric Cancer Association. Japanese Gastric Cancer Treatment Guidelines 2018 (5th Edition). Gastric Cancer 2021, 24, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Grabsch, H.I.; Tan, P. Gastric Cancer Pathology and Underlying Molecular Mechanisms. Dig. Surg. 2013, 30, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; El Hajj, N.; Sittler, S.; Lammert, N.; Barnes, R.; Meloni-Ehrig, A. Gastric Cancer: Classification, Histology and Application of Molecular Pathology. J. Gastrointest. Oncol. 2012, 3, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhao, Y.; Song, W.-M.; Zhang, B. Molecular Classification and Prediction in Gastric Cancer. Comput. Struct. Biotechnol. J. 2015, 13, 448–458. [Google Scholar] [CrossRef]
- McCracken, K.W.; Aihara, E.; Martin, B.; Crawford, C.M.; Broda, T.; Treguier, J.; Zhang, X.; Shannon, J.M.; Montrose, M.H.; Wells, J.M. Wnt/β-Catenin Promotes Gastric Fundus Specification in Mice and Humans. Nature 2017, 541, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shen, H.; Kapesa, L.; Zeng, S. Lauren Classification and Individualized Chemotherapy in Gastric Cancer. Oncol. Lett. 2016, 11, 2959–2964. [Google Scholar] [CrossRef]
- Correa, P.; Piazuelo, M.B. Helicobacter Pylori Infection and Gastric Adenocarcinoma. US Gastroenterol. Hepatol. Rev. 2011, 7, 59–64. [Google Scholar]
- Kolb, J.M.; Ozbek, U.; Harpaz, N.; Holcombe, R.F.; Ang, C. Effect of Helicobacter Pylori Infection on Outcomes in Resected Gastric and Gastroesophageal Junction Cancer. J. Gastrointest. Oncol. 2017, 8, 583–588. [Google Scholar] [CrossRef]
- Miyahara, R.; Niwa, Y.; Matsuura, T.; Maeda, O.; Ando, T.; Ohmiya, N.; Itoh, A.; Hirooka, Y.; Goto, H. Prevalence and Prognosis of Gastric Cancer Detected by Screening in a Large Japanese Population: Data from a Single Institute over 30 Years. J. Gastroenterol. Hepatol. 2007, 22, 1435–1442. [Google Scholar] [CrossRef]
- van der Kaaij, R.T.; Koemans, W.J.; van Putten, M.; Snaebjornsson, P.; Luijten, J.C.H.B.M.; van Dieren, J.M.; Cats, A.; Lemmens, V.E.P.P.; Verhoeven, R.H.A.; van Sandick, J.W. A Population-Based Study on Intestinal and Diffuse Type Adenocarcinoma of the Oesophagus and Stomach in the Netherlands between 1989 and 2015. Eur. J. Cancer 2020, 130, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Henson, D.E.; Dittus, C.; Younes, M.; Nguyen, H.; Albores-Saavedra, J. Differential Trends in the Intestinal and Diffuse Types of Gastric Carcinoma in the United States, 1973–2000: Increase in the Signet Ring Cell Type. Arch. Pathol. Lab. Med. 2004, 128, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Rexin, P.; Qin, Z.; Changbo, C.; Guanghui, C.; Luyao, W.; Voelker, H.-U.; Stauch, G. Different Incidence of Early-Onset Gastric Carcinoma Depending on Ethnicity: Preliminary Results of a Hospital in Liangshan. Sci. World J. 2020, 2020, 6845413. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Strand, M.S.; Lockhart, A.C.; Fields, R.C. Genetics of Gastric Cancer. Surg. Clin. N. Am. 2017, 97, 345–370. [Google Scholar] [CrossRef] [PubMed]
- Geigl, J.B.; Obenauf, A.C.; Schwarzbraun, T.; Speicher, M.R. Defining “Chromosomal Instability”. Trends Genet. 2008, 24, 64–69. [Google Scholar] [CrossRef]
- Loeb, L.A. A Mutator Phenotype in Cancer. Cancer Res. 2001, 61, 3230–3239. [Google Scholar] [PubMed]
- Kawakami, H.; Zaanan, A.; Sinicrope, F.A. Microsatellite Instability Testing and Its Role in the Management of Colorectal Cancer. Curr. Treat. Options Oncol. 2015, 16, 30. [Google Scholar] [CrossRef]
- Carter, S.L.; Cibulskis, K.; Helman, E.; McKenna, A.; Shen, H.; Zack, T.; Laird, P.W.; Onofrio, R.C.; Winckler, W.; Weir, B.A.; et al. Absolute Quantification of Somatic DNA Alterations in Human Cancer. Nat. Biotechnol. 2012, 30, 413–421. [Google Scholar] [CrossRef]
- Bielski, C.M.; Zehir, A.; Penson, A.V.; Donoghue, M.T.A.; Chatila, W.; Armenia, J.; Chang, M.T.; Schram, A.M.; Jonsson, P.; Bandlamudi, C.; et al. Genome Doubling Shapes the Evolution and Prognosis of Advanced Cancers. Nat. Genet. 2018, 50, 1189–1195. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Cantley, L.C. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 2018, 174, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Maleki, S.S.; Röcken, C. Chromosomal Instability in Gastric Cancer Biology. Neoplasia 2017, 19, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Sansregret, L.; Vanhaesebroeck, B.; Swanton, C. Determinants and Clinical Implications of Chromosomal Instability in Cancer. Nat. Rev. Clin. Oncol. 2018, 15, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.J.; Cleveland, D.W. Boveri Revisited: Chromosomal Instability, Aneuploidy and Tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009, 10, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Cowan, G.; Mead, A.J.; Filippi, S.; Bohn, G.; Chaidos, A.; Tunstall, O.; Chan, J.K.Y.; Choolani, M.; Bennett, P.; et al. Perturbation of Fetal Liver Hematopoietic Stem and Progenitor Cell Development by Trisomy 21. Proc. Natl. Acad. Sci. USA 2012, 109, 17579–17584. [Google Scholar] [CrossRef]
- Castellanos, G.; Valbuena, D.S.; Pérez, E.; Villegas, V.E.; Rondón-Lagos, M. Chromosomal Instability as Enabling Feature and Central Hallmark of Breast Cancer. Breast Cancer 2023, 15, 189–211. [Google Scholar] [CrossRef]
- Wilhelm, T.; Said, M.; Naim, V. DNA Replication Stress and Chromosomal Instability: Dangerous Liaisons. Genes 2020, 11, 642. [Google Scholar] [CrossRef]
- Gregan, J.; Polakova, S.; Zhang, L.; Tolić-Nørrelykke, I.M.; Cimini, D. Merotelic Kinetochore Attachment: Causes and Effects. Trends Cell Biol. 2011, 21, 374–381. [Google Scholar] [CrossRef]
- Ma, H.; He, Z.; Chen, J.; Zhang, X.; Song, P. Identifying of Biomarkers Associated with Gastric Cancer Based on 11 Topological Analysis Methods of CytoHubba. Sci. Rep. 2021, 11, 1331. [Google Scholar] [CrossRef]
- Mazouzi, A.; Velimezi, G.; Loizou, J.I. DNA Replication Stress: Causes, Resolution and Disease. Exp. Cell Res. 2014, 329, 85–93. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Linstra, R.; van Vugt, M.A.T.M. Genomic Instability, Inflammatory Signaling and Response to Cancer Immunotherapy. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188661. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-E.; Yeh, D.-W.; Pan, Y.-R.; Huang, W.-K.; Chen, M.-H.; Chang, J.W.-C.; Chen, J.-S.; Wang, Y.-C.; Yeh, C.-N. Chromosomal Instability May Not Be a Predictor for Immune Checkpoint Inhibitors from a Comprehensive Bioinformatics Analysis. Life 2020, 10, 276. [Google Scholar] [CrossRef] [PubMed]
- Sunakawa, Y.; Lenz, H.J. Molecular classification of gastric adenocarcinoma: Translating new insights from the cancer genome atlas research network. Curr. Treat Options Oncol. 2015, 16, 17. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Fang, J.-Y.; Xu, J. Gastric Cancer and Gene Copy Number Variation: Emerging Cancer Drivers for Targeted Therapy. Oncogene 2016, 35, 1475–1482. [Google Scholar] [CrossRef]
- Ezaki, T.; Yanagisawa, A.; Ohta, K.; Aiso, S.; Watanabe, M.; Hibi, T.; Kato, Y.; Nakajima, T.; Ariyama, T.; Inazawa, J.; et al. Deletion Mapping on Chromosome 1p in Well-Differentiated Gastric Cancer. Br. J. Cancer 1996, 73, 424–428. [Google Scholar] [CrossRef]
- Blanchet, A.; Bourgmayer, A.; Kurtz, J.-E.; Mellitzer, G.; Gaiddon, C. Isoforms of the P53 Family and Gastric Cancer: A Ménage à Trois for an Unfinished Affair. Cancers 2021, 13, 916. [Google Scholar] [CrossRef]
- Bibi, F.; Ali, I.; Naseer, M.I.; Ali Mohamoud, H.S.; Yasir, M.; Alvi, S.A.; Jiman-Fatani, A.A.; Sawan, A.; Azhar, E.I. Detection of Genetic Alterations in Gastric Cancer Patients from Saudi Arabia Using Comparative Genomic Hybridization (CGH). PLoS ONE 2018, 13, e0202576. [Google Scholar] [CrossRef]
- Barone, G.; Staples, C.J.; Ganesh, A.; Patterson, K.W.; Bryne, D.P.; Myers, K.N.; Patil, A.A.; Eyers, C.E.; Maslen, S.; Skehel, J.M.; et al. Human CDK18 Promotes Replication Stress Signaling and Genome Stability. Nucleic Acids Res. 2016, 44, 8772–8785. [Google Scholar] [CrossRef]
- Kim, Y.-I.; Pecha, R.L.; Keihanian, T.; Mercado, M.; Pena-Munoz, S.V.; Lang, K.; Van Buren, G.; Dhingra, S.; Othman, M.O. MUC1 Expressions and Its Prognostic Values in US Gastric Cancer Patients. Cancers 2023, 15, 998. [Google Scholar] [CrossRef]
- Wistuba, I.I.; Maitra, A.; Carrasco, R.; Tang, M.; Troncoso, P.; Minna, J.D.; Gazdar, A.F. High Resolution Chromosome 3p, 8p, 9q and 22q Allelotyping Analysis in the Pathogenesis of Gallbladder Carcinoma. Br. J. Cancer 2002, 87, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of β-Catenin-Tcf Signaling in Colon Cancer by Mutations in β-Catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef] [PubMed]
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High Frequency of Mutations of the PIK3CA Gene in Human Cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Yamada, Y.; Taniguchi, H.; Fukahori, M.; Sasaki, Y.; Shoji, H.; Honma, Y.; Iwasa, S.; Takashima, A.; Kato, K.; et al. Clinicopathological Features and Prognostic Roles of KRAS, BRAF, PIK3CA and NRAS Mutations in Advanced Gastric Cancer. BMC Res. Notes 2014, 7, 271. [Google Scholar] [CrossRef]
- Buffart, T.E.; Carvalho, B.; van Grieken, N.C.T.; van Wieringen, W.N.; Tijssen, M.; Kranenbarg, E.M.-K.; Verheul, H.M.W.; Grabsch, H.I.; Ylstra, B.; van de Velde, C.J.H.; et al. Losses of Chromosome 5q and 14q Are Associated with Favorable Clinical Outcome of Patients with Gastric Cancer. Oncologist 2012, 17, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Xiong, Y.; Li, J.; Liu, L.; Zhang, W.; Zhang, C.; Wan, J. APC Gene Deletions in Gastric Adenocarcinomas in a Chinese Population: A Correlation with Tumour Progression. Clin. Transl. Oncol. 2012, 14, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Graziano, F.; Galluccio, N.; Lorenzini, P.; Ruzzo, A.; Canestrari, E.; D’Emidio, S.; Catalano, V.; Sisti, V.; Ligorio, C.; Andreoni, F.; et al. Genetic Activation of the MET Pathway and Prognosis of Patients with High-Risk, Radically Resected Gastric Cancer. J. Clin. Oncol. 2011, 29, 4789–4795. [Google Scholar] [CrossRef]
- Hong, L.; Han, Y.; Yang, J.; Zhang, H.; Jin, Y.; Brain, L.; Li, M.; Zhao, Q. Prognostic Value of Epidermal Growth Factor Receptor in Patients with Gastric Cancer: A Meta-Analysis. Gene 2013, 529, 69–72. [Google Scholar] [CrossRef]
- Rossi, E.; Villanacci, V.; Danesino, C.; Donato, F.; Nascimbeni, R.; Bassotti, G. Epidermal Growth Factor Receptor Overexpression/Amplification in Adenocarcinomas Arising in the Gastrointestinal Tract. Rev. Esp. Enferm. Dig. 2011, 103, 632–639. [Google Scholar] [CrossRef]
- Higaki, E.; Kuwata, T.; Nagatsuma, A.K.; Nishida, Y.; Kinoshita, T.; Aizawa, M.; Nitta, H.; Nagino, M.; Ochiai, A. Gene Copy Number Gain of EGFR Is a Poor Prognostic Biomarker in Gastric Cancer: Evaluation of 855 Patients with Bright-Field Dual in Situ Hybridization (DISH) Method. Gastric. Cancer 2016, 19, 63–73. [Google Scholar] [CrossRef]
- Buffart, T.E.; Carvalho, B.; Mons, T.; Reis, R.M.; Moutinho, C.; Silva, P.; van Grieken, N.C.T.; Vieth, M.; Stolte, M.; van de Velde, C.J.H.; et al. DNA Copy Number Profiles of Gastric Cancer Precursor Lesions. BMC Genom. 2007, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Tu, R.; Liu, H.; Qing, G. Regulation of Cancer Cell Metabolism: Oncogenic MYC in the Driver’s Seat. Signal Transduct. Target. Ther. 2020, 5, 124. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Choi, I.J.; Kim, C.G.; Kim, H.S.; Oshima, A.; Yamada, Y.; Arao, T.; Nishio, K.; Michalowski, A.; Green, J.E. Three-Gene Predictor of Clinical Outcome for Gastric Cancer Patients Treated with Chemotherapy. Pharmacogenom. J. 2012, 12, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Arao, T.; Togashi, Y.; Kato, H.; Fujita, Y.; De Velasco, M.A.; Kimura, H.; Matsumoto, K.; Tanaka, K.; Okamoto, I.; et al. The OCT4 Pseudogene POU5F1B Is Amplified and Promotes an Aggressive Phenotype in Gastric Cancer. Oncogene 2015, 34, 199–208. [Google Scholar] [CrossRef] [PubMed]
- de Manzoni, G.; Tomezzoli, A.; Di Leo, A.; Moore, P.S.; Talamini, G.; Scarpa, A. Clinical Significance of Mutator Phenotype and Chromosome 17p and 18q Allelic Loss in Gastric Cancer. Br. J. Surg. 2001, 88, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Liu, Z.; Chang, X.; Gao, Y.; Han, H.; Liu, X.; Cai, H.; Fu, Q.; Liu, L.; Yin, K. Clinical Significance of Chromosomal Integrity in Gastric Cancers. Int. J. Biol. Markers 2022, 37, 296–305. [Google Scholar] [CrossRef] [PubMed]
- de Mello, R.A.; Marques, A.M.; Araújo, A. HER2 Therapies and Gastric Cancer: A Step Forward. World J. Gastroenterol. 2013, 19, 6165–6169. [Google Scholar] [CrossRef]
- Sheng, W.Q.; Huang, D.; Ying, J.M.; Lu, N.; Wu, H.M.; Liu, Y.H.; Liu, J.P.; Bu, H.; Zhou, X.Y.; Du, X. HER2 Status in Gastric Cancers: A Retrospective Analysis from Four Chinese Representative Clinical Centers and Assessment of Its Prognostic Significance. Ann. Oncol. 2013, 24, 2360–2364. [Google Scholar] [CrossRef]
- Shim, J.H.; Yoon, J.H.; Choi, S.S.; Ashktorab, H.; Smoot, D.T.; Song, K.Y.; Nam, S.W.; Lee, J.Y.; Park, C.H.; Park, W.S. The Effect of Helicobacter Pylori CagA on the HER-2 Copy Number and Expression in Gastric Cancer. Gene 2014, 546, 288–296. [Google Scholar] [CrossRef]
- Inoue, T.; Uchino, S.; Shiraishi, N.; Adachi, Y.; Kitano, S. Loss of Heterozygosity on Chromosome 18q in Cohesive-Type Gastric Cancer Is Associated with Tumor Progression and Poor Prognosis. Clin. Cancer Res. 1998, 4, 973–977. [Google Scholar]
- Snijders, A.M.; Mao, J.-H. Multi-Omics Approach to Infer Cancer Therapeutic Targets on Chromosome 20q across Tumor Types. Adv. Mod. Oncol. Res. 2016, 2, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Ptashkin, R.N.; Pagan, C.; Yaeger, R.; Middha, S.; Shia, J.; O’Rourke, K.P.; Berger, M.F.; Wang, L.; Cimera, R.; Wang, J.; et al. Chromosome 20q Amplification Defines a Subtype of Microsatellite Stable, Left-Sided Colon Cancers with Wild-Type RAS/RAF and Better Overall Survival. Mol. Cancer Res. 2017, 15, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Xu, Y.; Liu, M.; Shen, X.; Mao, Y.; Li, Y.; Zhang, K.; Yu, S.; Fan, H. Upregulation of LINC00659 Expression Predicts a Poor Prognosis and Promotes Migration and Invasion of Gastric Cancer Cells. Oncol. Lett. 2021, 22, 557. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.-C.; Yeh, C.-N.; Cheng, C.-T.; Jung, S.-M.; Huang, S.-C.; Chang, T.-W.; Jan, Y.-Y.; Tzeng, C.-H.; Chao, T.-C.; Chen, Y.-Y.; et al. Integrating Bioinformatics and Clinicopathological Research of Gastrointestinal Stromal Tumors: Identification of Aurora Kinase A as a Poor Risk Marker. Ann. Surg. Oncol. 2012, 19, 3491–3499. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Xiong, Y.; Li, J.; Liu, L.; Li, M.; Zhang, C.; Zhang, W.; Wan, J. Copy-Number Increase of AURKA in Gastric Cancers in a Chinese Population: A Correlation with Tumor Progression. Med. Oncol. 2011, 28, 1017–1022. [Google Scholar] [CrossRef]
- Wang, H.-L.; Zhou, P.-Y.; Liu, P.; Zhang, Y. Abnormal FHIT Protein Expression May Be Correlated with Poor Prognosis in Gastric Cancer: A Meta-Analysis. Tumour Biol. 2014, 35, 6815–6821. [Google Scholar] [CrossRef] [PubMed]
- Tabach, Y.; Kogan-Sakin, I.; Buganim, Y.; Solomon, H.; Goldfinger, N.; Hovland, R.; Ke, X.-S.; Oyan, A.M.; Kalland, K.-H.; Rotter, V.; et al. Amplification of the 20q Chromosomal Arm Occurs Early in Tumorigenic Transformation and May Initiate Cancer. PLoS ONE 2011, 6, e14632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, X.; Yu, G.; Liu, L.; Wang, J.; Chen, X.; Bian, Y.; Ji, Y.; Zhou, X.; Chen, Y.; et al. UBE2C Is a Potential Biomarker of Intestinal-Type Gastric Cancer with Chromosomal Instability. Front. Pharmacol. 2018, 9, 847. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, P.; Yang, S.; Yang, Y.; Zhang, Q.; Zhang, W.; Xiao, H.; Gao, H.; Zhang, Q. Identification of Genes with a Correlation between Copy Number and Expression in Gastric Cancer. BMC Med. Genom. 2012, 5, 14. [Google Scholar] [CrossRef]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular Analysis of Gastric Cancer Identifies Subtypes Associated with Distinct Clinical Outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting P53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Soussi, T.; Wiman, K.G. TP53: An Oncogene in Disguise. Cell Death Differ. 2015, 22, 1239–1249. [Google Scholar] [CrossRef]
- Frum, R.A.; Grossman, S.R. Mechanisms of Mutant P53 Stabilization in Cancer. Subcell. Biochem. 2014, 85, 187–197. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, L.; Jiang, J.; Zhang, Y.; Wang, X.; Zhang, Q.; Wang, Y.; Liu, C.; Li, F. CDK1 and CCNB1 as Potential Diagnostic Markers of Rhabdomyosarcoma: Validation Following Bioinformatics Analysis. BMC Med. Genom. 2019, 12, 198. [Google Scholar] [CrossRef]
- Li, B.; Zhu, H.-B.; Song, G.-D.; Cheng, J.-H.; Li, C.-Z.; Zhang, Y.-Z.; Zhao, P. Regulating the CCNB1 Gene Can Affect Cell Proliferation and Apoptosis in Pituitary Adenomas and Activate Epithelial-to-Mesenchymal Transition. Oncol. Lett. 2019, 18, 4651–4658. [Google Scholar] [CrossRef]
- Izadi, S.; Nikkhoo, A.; Hojjat-Farsangi, M.; Namdar, A.; Azizi, G.; Mohammadi, H.; Yousefi, M.; Jadidi-Niaragh, F. CDK1 in Breast Cancer: Implications for Theranostic Potential. Anticancer Agents Med. Chem. 2020, 20, 758–767. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, H.; Zou, Q.; Wu, J. Analysis of Cyclin-Dependent Kinase 1 as an Independent Prognostic Factor for Gastric Cancer Based on Statistical Methods. Front. Cell Dev. Biol. 2020, 8, 620164. [Google Scholar] [CrossRef]
- Sofi, S.; Mehraj, U.; Qayoom, H.; Aisha, S.; Asdaq, S.M.B.; Almilaibary, A.; Mir, M.A. Cyclin-Dependent Kinases in Breast Cancer: Expression Pattern and Therapeutic Implications. Med. Oncol. 2022, 39, 106. [Google Scholar] [CrossRef]
- Yasukawa, M.; Ando, Y.; Yamashita, T.; Matsuda, Y.; Shoji, S.; Morioka, M.S.; Kawaji, H.; Shiozawa, K.; Machitani, M.; Abe, T.; et al. CDK1 Dependent Phosphorylation of HTERT Contributes to Cancer Progression. Nat. Commun. 2020, 11, 1557. [Google Scholar] [CrossRef]
- Huang, X.; Huang, Q.; Chen, S.; Zhang, J.; Lin, K.; Zhang, X. Efficacy of Laparoscopic Adenomyomectomy Using Double-Flap Method for Diffuse Uterine Adenomyosis. BMC Womens Health 2015, 15, 24. [Google Scholar] [CrossRef]
- Huang, S.; Ye, H.; Guo, W.; Dong, X.; Wu, N.; Zhang, X.; Huang, Z. CDK4/6 Inhibitor Suppresses Gastric Cancer with CDKN2A Mutation. Int. J. Clin. Exp. Med. 2015, 8, 11692–11700. [Google Scholar] [PubMed]
- Zhang, M.; Zhang, L.; Hei, R.; Li, X.; Cai, H.; Wu, X.; Zheng, Q.; Cai, C. CDK Inhibitors in Cancer Therapy, an Overview of Recent Development. Am. J. Cancer Res. 2021, 11, 1913–1935. [Google Scholar] [PubMed]
- Sofi, S.; Mehraj, U.; Qayoom, H.; Aisha, S.; Almilaibary, A.; Alkhanani, M.; Mir, M.A. Targeting Cyclin-Dependent Kinase 1 (CDK1) in Cancer: Molecular Docking and Dynamic Simulations of Potential CDK1 Inhibitors. Med. Oncol. 2022, 39, 133. [Google Scholar] [CrossRef] [PubMed]
- Giet, R.; Prigent, C. Aurora/Ipl1p-Related Kinases, a New Oncogenic Family of Mitotic Serine-Threonine Kinases. J. Cell Sci. 1999, 112, 3591–3601. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, J.R.; Plowman, G.D. The Aurora/Ipl1p Kinase Family: Regulators of Chromosome Segregation and Cytokinesis. Trends Cell Biol. 1999, 9, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Huang, C.; Liu, K.; Li, X.; Dong, Z. Targeting AURKA in Cancer: Molecular Mechanisms and Opportunities for Cancer Therapy. Mol. Cancer 2021, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Crosio, C.; Fimia, G.M.; Loury, R.; Kimura, M.; Okano, Y.; Zhou, H.; Sen, S.; Allis, C.D.; Sassone-Corsi, P. Mitotic Phosphorylation of Histone H3: Spatio-Temporal Regulation by Mammalian Aurora Kinases. Mol. Cell Biol. 2002, 22, 874–885. [Google Scholar] [CrossRef] [PubMed]
- LeRoy, P.J.; Hunter, J.J.; Hoar, K.M.; Burke, K.E.; Shinde, V.; Ruan, J.; Bowman, D.; Galvin, K.; Ecsedy, J.A. Localization of Human TACC3 to Mitotic Spindles Is Mediated by Phosphorylation on Ser558 by Aurora A: A Novel Pharmacodynamic Method for Measuring Aurora A Activity. Cancer Res. 2007, 67, 5362–5370. [Google Scholar] [CrossRef]
- Venoux, M.; Basbous, J.; Berthenet, C.; Prigent, C.; Fernandez, A.; Lamb, N.J.; Rouquier, S. ASAP Is a Novel Substrate of the Oncogenic Mitotic Kinase Aurora-A: Phosphorylation on Ser625 Is Essential to Spindle Formation and Mitosis. Hum. Mol. Genet. 2008, 17, 215–224. [Google Scholar] [CrossRef]
- Chou, E.-J.; Hung, L.-Y.; Tang, C.-J.C.; Hsu, W.-B.; Wu, H.-Y.; Liao, P.-C.; Tang, T.K. Phosphorylation of CPAP by Aurora-A Maintains Spindle Pole Integrity during Mitosis. Cell Rep. 2016, 14, 2975–2987. [Google Scholar] [CrossRef]
- Fu, J.; Bian, M.; Xin, G.; Deng, Z.; Luo, J.; Guo, X.; Chen, H.; Wang, Y.; Jiang, Q.; Zhang, C. TPX2 Phosphorylation Maintains Metaphase Spindle Length by Regulating Microtubule Flux. J. Cell Biol. 2015, 210, 373–383. [Google Scholar] [CrossRef]
- Macůrek, L.; Lindqvist, A.; Lim, D.; Lampson, M.A.; Klompmaker, R.; Freire, R.; Clouin, C.; Taylor, S.S.; Yaffe, M.B.; Medema, R.H. Polo-like Kinase-1 Is Activated by Aurora A to Promote Checkpoint Recovery. Nature 2008, 455, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Cazales, M.; Quaranta, M.; Froment, C.; Trabut, V.; Dozier, C.; Mirey, G.; Bouché, J.-P.; Theis-Febvre, N.; Schmitt, E.; et al. Phosphorylation of CDC25B by Aurora-A at the Centrosome Contributes to the G2-M Transition. J. Cell Sci. 2004, 117, 2523–2531. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Belkhiri, A.; El-Rifai, W. The Aurora Kinase A Regulates GSK-3β in Gastric Cancer Cells. Oncogene 2009, 28, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Sasai, K.; Kawai, H.; Yuan, Z.-M.; Bondaruk, J.; Suzuki, F.; Fujii, S.; Arlinghaus, R.B.; Czerniak, B.A.; Sen, S. Phosphorylation by Aurora Kinase A Induces Mdm2-Mediated Destabilization and Inhibition of P53. Nat. Genet. 2004, 36, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Tang, A.; Gao, K.; Chu, L.; Zhang, R.; Yang, J.; Zheng, J. Aurora Kinases: Novel Therapy Targets in Cancers. Oncotarget 2017, 8, 23937–23954. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, Y.; Zhang, C.; Zhang, Q. Role of Aurora Kinase B in Regulating Resistance to Paclitaxel in Breast Cancer Cells. Hum. Cell 2022, 35, 678–693. [Google Scholar] [CrossRef]
- Nie, M.; Wang, Y.; Yu, Z.; Li, X.; Deng, Y.; Wang, Y.; Yang, D.; Li, Q.; Zeng, X.; Ju, J.; et al. AURKB Promotes Gastric Cancer Progression via Activation of CCND1 Expression. Aging 2020, 12, 1304–1321. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, Z.; Wang, G.-H.; Zhou, Y.-M.; Deng, J.-P.; Feng, Y.; Chen, J.-Q.; Tian, L. AURKB Promotes the Metastasis of Gastric Cancer, Possibly by Inducing EMT. Cancer Manag. Res. 2020, 12, 6947–6958. [Google Scholar] [CrossRef]
- Lei, Z.-N.; Teng, Q.-X.; Tian, Q.; Chen, W.; Xie, Y.; Wu, K.; Zeng, Q.; Zeng, L.; Pan, Y.; Chen, Z.-S.; et al. Signaling Pathways and Therapeutic Interventions in Gastric Cancer. Signal Transduct. Target. Ther. 2022, 7, 358. [Google Scholar] [CrossRef]
- Kanayama, K.; Imai, H.; Usugi, E.; Shiraishi, T.; Hirokawa, Y.S.; Watanabe, M. Association of HER2 Gene Amplification and Tumor Progression in Early Gastric Cancer. Virchows Arch. 2018, 473, 559–565. [Google Scholar] [CrossRef]
- Neve, R.M.; Lane, H.A.; Hynes, N.E. The Role of Overexpressed HER2 in Transformation. Ann. Oncol. 2001, 12 (Suppl. S1), S9–S13. [Google Scholar] [CrossRef]
- Dang, H.-Z.; Yu, Y.; Jiao, S.-C. Prognosis of HER2 Over-Expressing Gastric Cancer Patients with Liver Metastasis. World J. Gastroenterol. 2012, 18, 2402–2407. [Google Scholar] [CrossRef]
- Shitara, K.; Bang, Y.-J.; Iwasa, S.; Sugimoto, N.; Ryu, M.-H.; Sakai, D.; Chung, H.-C.; Kawakami, H.; Yabusaki, H.; Lee, J.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Positive Gastric Cancer. N. Engl. J. Med. 2020, 382, 2419–2430. [Google Scholar] [CrossRef]
- Smith, P.; Bradley, T.; Gavarró, L.M.; Goranova, T.; Ennis, D.P.; Mirza, H.B.; De Silva, D.; Piskorz, A.M.; Sauer, C.M.; Al-Khalidi, S.; et al. The Copy Number and Mutational Landscape of Recurrent Ovarian High-Grade Serous Carcinoma. Nat. Commun. 2023, 14, 4387. [Google Scholar] [CrossRef]
- Drews, R.M.; Hernando, B.; Tarabichi, M.; Haase, K.; Lesluyes, T.; Smith, P.S.; Morrill Gavarró, L.; Couturier, D.-L.; Liu, L.; Schneider, M.; et al. A Pan-Cancer Compendium of Chromosomal Instability. Nature 2022, 606, 976–983. [Google Scholar] [CrossRef]
- Cai, H.; Jing, C.; Chang, X.; Ding, D.; Han, T.; Yang, J.; Lu, Z.; Hu, X.; Liu, Z.; Wang, J.; et al. Mutational Landscape of Gastric Cancer and Clinical Application of Genomic Profiling Based on Target Next-Generation Sequencing. J. Transl. Med. 2019, 17, 189. [Google Scholar] [CrossRef]
- Hassin, O.; Oren, M. Drugging P53 in Cancer: One Protein, Many Targets. Nat. Rev. Drug Discov. 2023, 22, 127–144. [Google Scholar] [CrossRef]
- Furukawa, H.; Makino, T.; Yamasaki, M.; Tanaka, K.; Miyazaki, Y.; Takahashi, T.; Kurokawa, Y.; Nakajima, K.; Takiguchi, S.; Mori, M.; et al. PRIMA-1 Induces P53-Mediated Apoptosis by Upregulating Noxa in Esophageal Squamous Cell Carcinoma with TP53 Missense Mutation. Cancer Sci. 2018, 109, 412–421. [Google Scholar] [CrossRef]
- Tan, A.C.; Chan, D.L.; Faisal, W.; Pavlakis, N. New Drug Developments in Metastatic Gastric Cancer. Ther. Adv. Gastroenterol. 2018, 11, 1756284818808072. [Google Scholar] [CrossRef]
- Díaz Del Arco, C.; Estrada Muñoz, L.; Molina Roldán, E.; Cerón Nieto, M.Á.; Ortega Medina, L.; García Gómez de Las Heras, S.; Fernández Aceñero, M.J. Immunohistochemical Classification of Gastric Cancer Based on New Molecular Biomarkers: A Potential Predictor of Survival. Virchows Arch. 2018, 473, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.-H.; Jeng, Y.-M.; Chen, K.-H.; Lee, C.-H.; Yuan, C.-T.; Liau, J.-Y. An Integrative Morphomolecular Classification System of Gastric Carcinoma with Distinct Clinical Outcomes. Am. J. Surg. Pathol. 2020, 44, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Nam, K.H.; Ahn, S.-H.; Park, D.J.; Kim, H.-H.; Kim, S.H.; Chang, H.; Lee, J.-O.; Kim, Y.J.; Lee, H.S.; et al. Prognostic Implications of Immunosuppressive Protein Expression in Tumors as Well as Immune Cell Infiltration within the Tumor Microenvironment in Gastric Cancer. Gastric. Cancer 2016, 19, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-K.; Boku, N.; Satoh, T.; Ryu, M.-H.; Chao, Y.; Kato, K.; Chung, H.C.; Chen, J.-S.; Muro, K.; Kang, W.K.; et al. Nivolumab in Patients with Advanced Gastric or Gastro-Oesophageal Junction Cancer Refractory to, or Intolerant of, at Least Two Previous Chemotherapy Regimens (ONO-4538-12, ATTRACTION-2): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2017, 390, 2461–2471. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.N.S.; Saito, Y.; Yoshikawa, T.; Oshima, T.; Hayden, J.D.; Oosting, J.; Earle, S.; Hewitt, L.C.; Slaney, H.L.; Wright, A.; et al. Increasing Frequency of Gene Copy Number Aberrations Is Associated with Immunosuppression and Predicts Poor Prognosis in Gastric Adenocarcinoma. Br. J. Surg. 2022, 109, 291–297. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, Y.; Chen, R.; Tian, Z.; Zhai, Y.; Janz, S.; Gu, C.; Yang, Y. Chromosomal Instability and Acquired Drug Resistance in Multiple Myeloma. Oncotarget 2017, 8, 78234–78244. [Google Scholar] [CrossRef]
- Kohlruss, M.; Krenauer, M.; Grosser, B.; Pfarr, N.; Jesinghaus, M.; Slotta-Huspenina, J.; Novotny, A.; Hapfelmeier, A.; Schmidt, T.; Steiger, K.; et al. Diverse “Just-Right” Levels of Chromosomal Instability and Their Clinical Implications in Neoadjuvant Treated Gastric Cancer. Br. J. Cancer 2021, 125, 1621–1631. [Google Scholar] [CrossRef]
- Nakamura, Y.; Shitara, K.; Lee, J. The Right Treatment of the Right Patient: Integrating Genetic Profiling into Clinical Decision Making in Advanced Gastric Cancer in Asia. Am. Soc. Clin. Oncol. Educ. Book 2021, 41, e166–e173. [Google Scholar] [CrossRef]


| Gene | Location | Function | Role in Pathogenesis | Refs. |
|---|---|---|---|---|
| TP73 | 1p36.32 | cell cycle regulation, induction of apoptosis | mutations are associated with GC progression | [37] |
| PRDM16 | 1p36.3 | regulation of transcription | high copy gain is associated with late cancer stages and poor prognosis | [38] |
| CDK18 | 1q32.1 | regulation of cell cycle and proliferation | gene inactivation promotes GC progression | [39] |
| MUC1 | 1q21-24 | regulation of apoptosis, protection of gastric epithelial cells from pathogens that initiate inflammation and carcinogenesis | high copy gain is associated with deeper invasion and metastasis | [40] |
| RASSF1A | 3p21.3 | regulation of cell cycle; inhibits epithelial-mesenchymal transition | gene inactivation is associated with metastasis and poor overall survival | [41] |
| FHIT | 3p14.2 | regulation of apoptosis | associated with tumor progression and poor prognosis | [66] |
| RAP2B | 3q25.2 | regulation of proliferation, migration, invasion and apoptosis after DNA damage | gene inactivation promotes GC progression | [38] |
| PIK3CA | 3q26.3 | regulation of proliferation | gene amplification is associated with poor prognosis | [43,44] |
| APC | 5q22.2 | regulation of the mitotic spindle to facilitate proper chromosome segregation | deletion is associated with tumor progression and poor prognosis | [42] |
| EGFR | 7p12 | regulation of cellular growth, proliferation, and differentiation | copy number gain is associated with higher risk of invasion and metastasis | [48,50] |
| MET | 7q21 | regulation of cell motility | gene amplification is associated with late stages and poor prognosis | [47] |
| MYC | 8q24.21 | regulation of cell proliferation and differentiation | promotes invasion and tumor progression | [52] |
| POU5F1B | 8q24 | regulation of angiogenesis, cell proliferation and apoptosis | copy number gain is associated with aggressive phenotype | [54] |
| TP53 | 17p13.1 | regulation of cell cycle, DNA repair and apoptosis | mutations in gene are associated with longer overall survival | [55] |
| HER2 | 17q12 | regulation of cell growth and proliferation | amplification of gene promotes GC | [57] |
| UBE2C | 20q13.12 | regulation of proliferation and cell cycle | associated with poor outcome | [67,68] |
| AURKA | 20q13 | regulation of DNA repair, cell migration and invasion | amplification of gene is associated with poor patient survival | [64,65] |
| C20orf20 | 20q13.33 | regulation of DNA methylation and chromatin remodeling | associated with high tumor aggressiveness | [61,69] |
| LINC00659 | 20q13.33 | regulation of migration and invasion | associated with the stage of the disease and metastasis to the lymph nodes | [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nemtsova, M.V.; Kuznetsova, E.B.; Bure, I.V. Chromosomal Instability in Gastric Cancer: Role in Tumor Development, Progression, and Therapy. Int. J. Mol. Sci. 2023, 24, 16961. https://doi.org/10.3390/ijms242316961
Nemtsova MV, Kuznetsova EB, Bure IV. Chromosomal Instability in Gastric Cancer: Role in Tumor Development, Progression, and Therapy. International Journal of Molecular Sciences. 2023; 24(23):16961. https://doi.org/10.3390/ijms242316961
Chicago/Turabian StyleNemtsova, Marina V., Ekaterina B. Kuznetsova, and Irina V. Bure. 2023. "Chromosomal Instability in Gastric Cancer: Role in Tumor Development, Progression, and Therapy" International Journal of Molecular Sciences 24, no. 23: 16961. https://doi.org/10.3390/ijms242316961





