Predictive Roles of ADAM17 in Patient Survival and Immune Cell Infiltration in Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Results
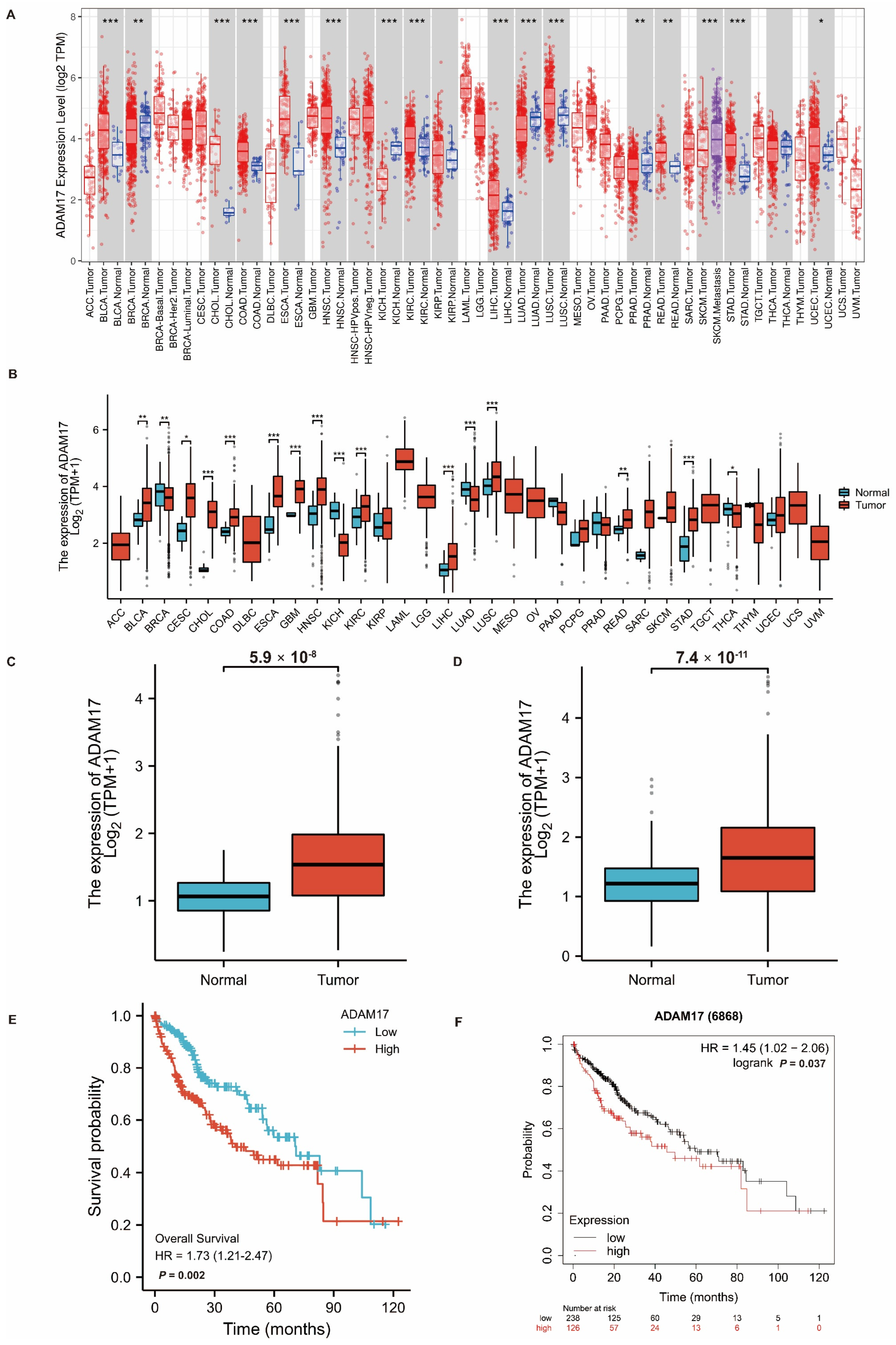
2.1. Expression of ADAM17 in Pan-Cancer and HCC
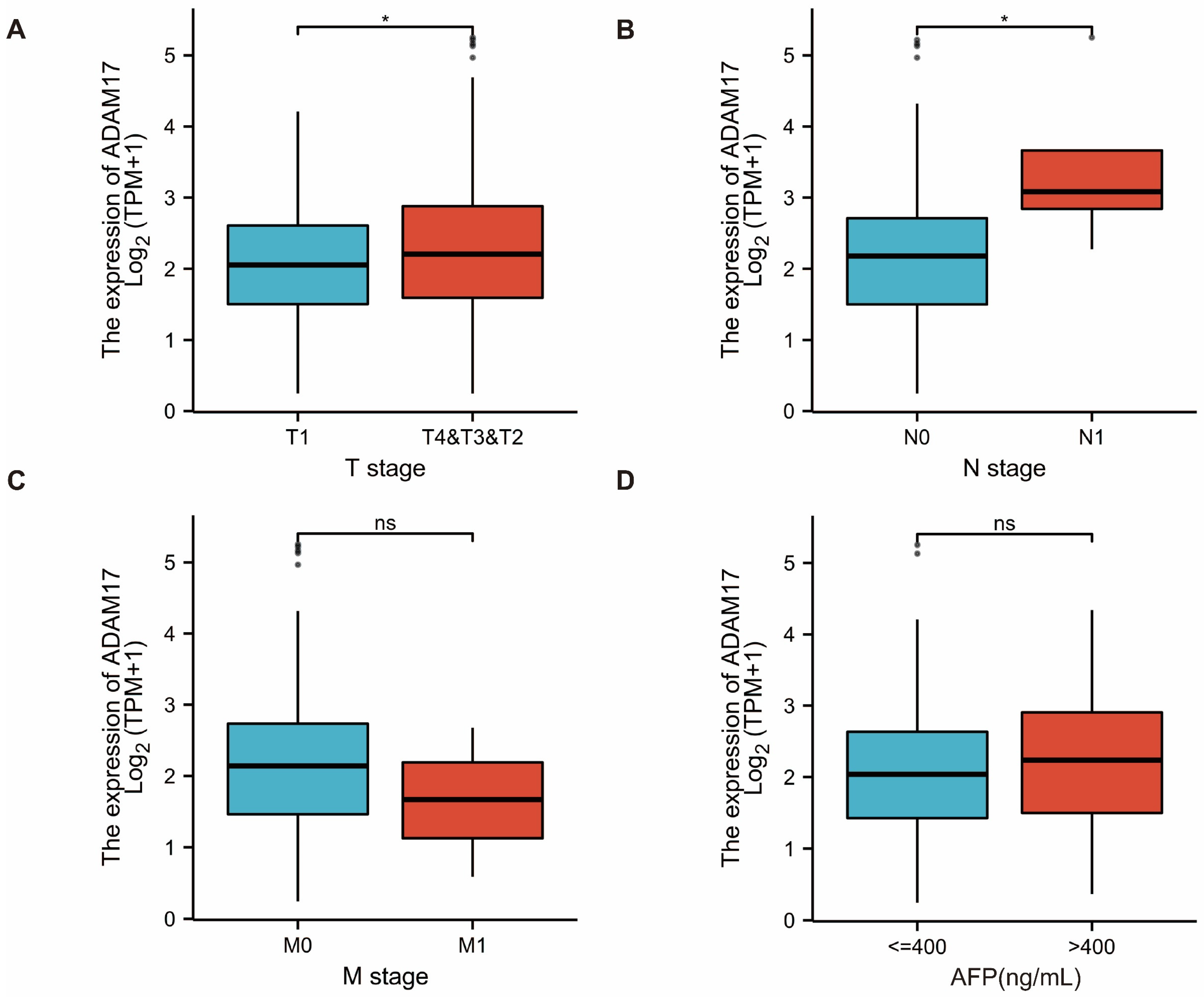
2.2. Prognostic Role of ADAM17 and Its Correlation with HCC Clinicopathological Features
2.3. Investigating the Function of ADAM17 and Related Pathways
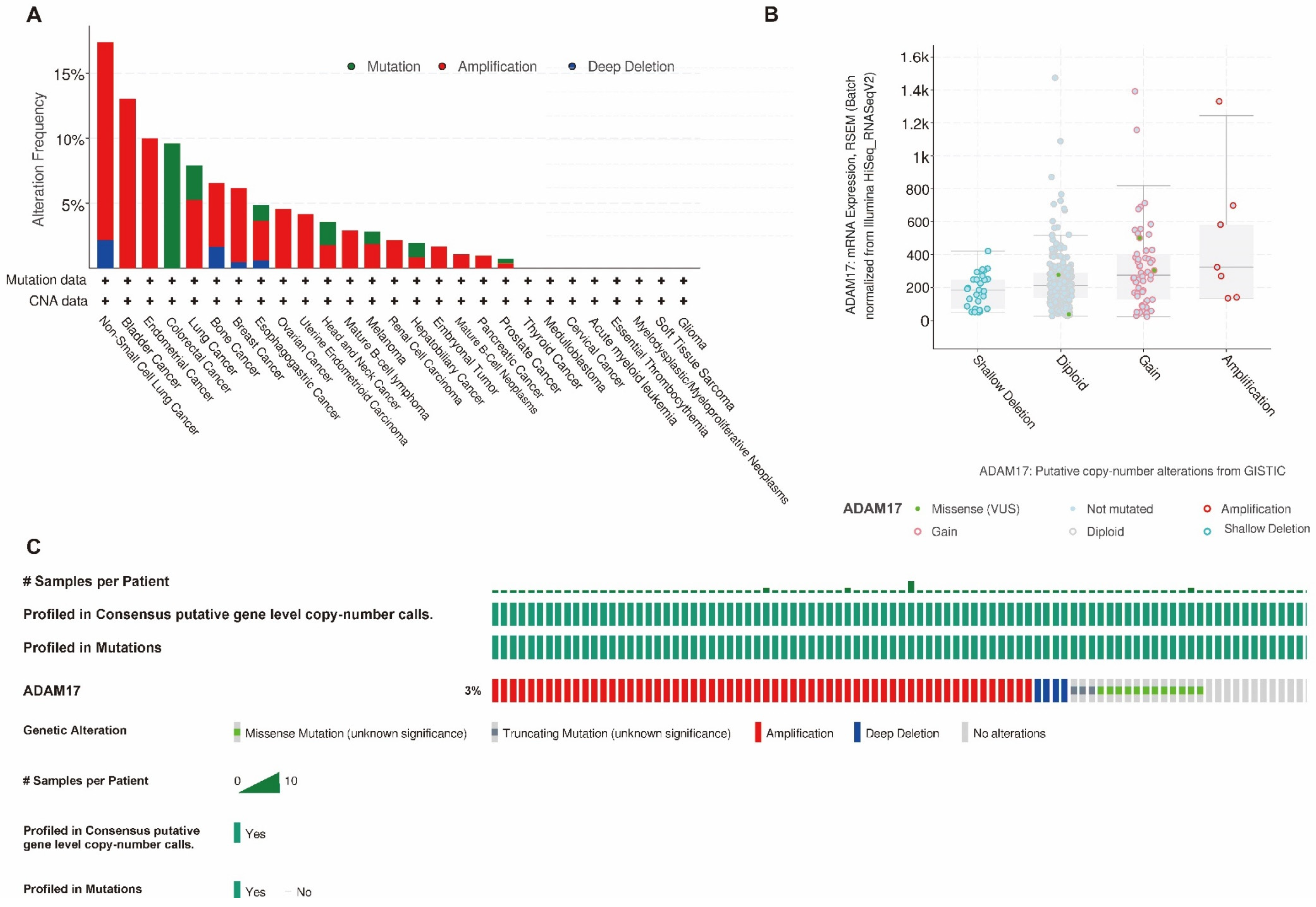
2.4. Association between ADAM17 Expression, Immune Infiltration, and Copy Number
2.5. Gene Profile Analysis of ADAM17
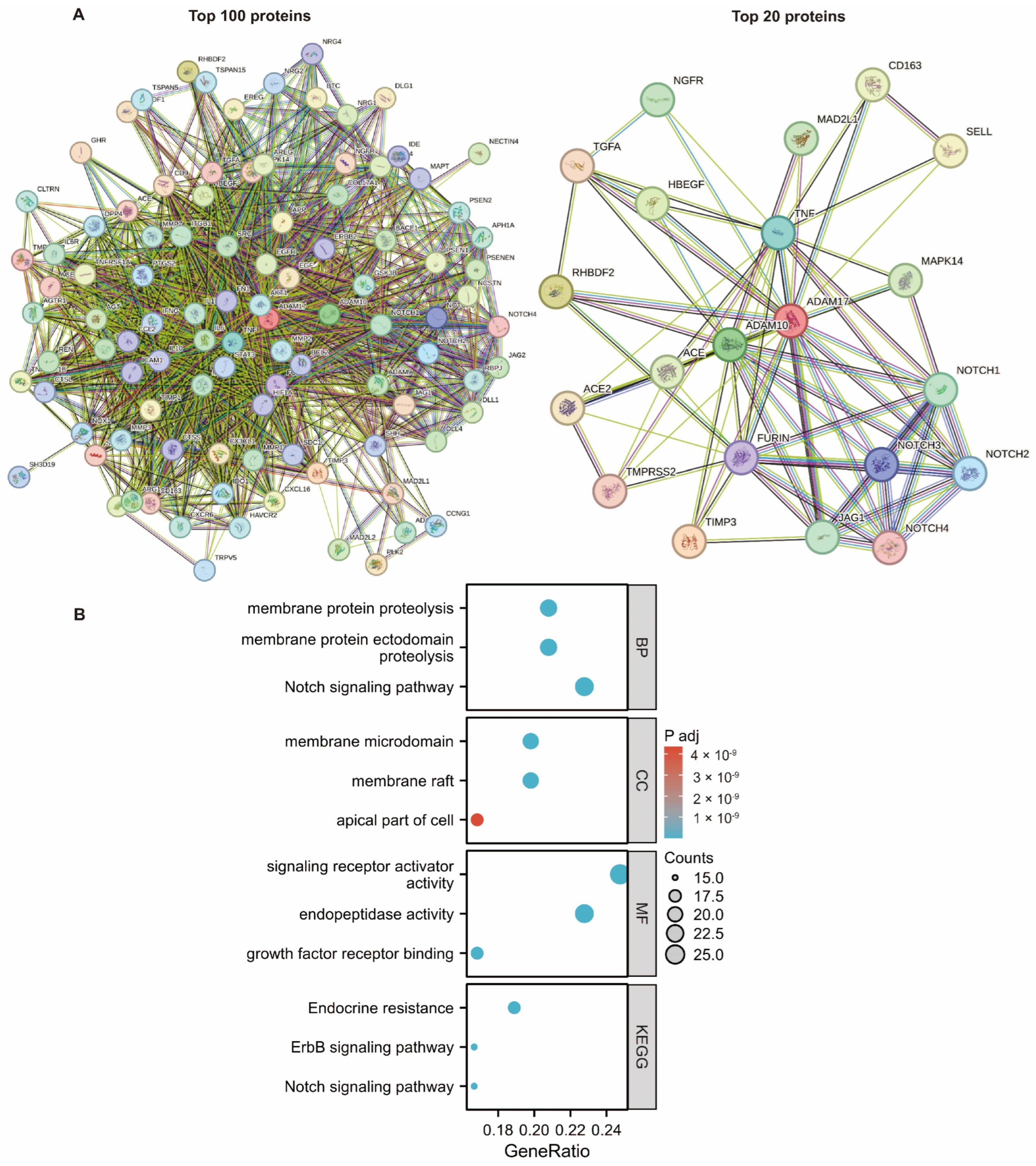
2.6. PPI Network Analysis
2.7. Verification of ADAM17 Expression and Its Prognostic Roles in HCC
2.8. In Vitro Roles of ADAM17 in HCC Cell Growth
2.9. In Vitro Roles of ADAM17 in HCC Metastasis
2.10. In Vivo Investigation of the Role of ADAM17 in HCC Growth
2.11. Verification of the Correlation between ADAM17 and Immune Cell Infiltration
3. Discussion
4. Materials and Methods
4.1. Datasets and Data Preprocessing
4.2. ADAM17 Association with Patient Survival and Clinicopathological Features
4.3. Enrichment Analysis
4.4. Immune Cell Infiltration Analysis
4.5. Mutation Analysis of ADAM17
4.6. PPI Network Analysis
4.7. Patients and Tumour Samples
4.8. Immunohistochemistry
4.9. Cell Culture and Stable Cell Construction
4.10. Western Blotting
4.11. Cell Proliferation and Colony Formation Assays
4.12. Wound Healing Assays
4.13. Transwell Matrigel Invasion and Migration Assay
4.14. In Vivo Growth Assays
4.15. Statistical Analyses
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.F.; Liu, T.Q.; Zhi, X.T.; Zou, J.; Zhong, J.T.; Li, T.; Mo, X.L.; Zhou, W.; Guo, W.W.; Liu, X.; et al. COX-2/PGE2 Axis Regulates HIF2alpha Activity to Promote Hepatocellular Carcinoma Hypoxic Response and Reduce the Sensitivity of Sorafenib Treatment. Clin. Cancer Res. 2018, 24, 3204–3216. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.R.; Sun, D.; Yang, Y.F.; Zhou, W.; Wu, R.; Wang, X.W.; Shi, K.; Yan, Y.C.; Yan, L.J.; Yao, C.Y.; et al. TMPRSS4 Drives Angiogenesis in Hepatocellular Carcinoma by Promoting HB-EGF Expression and Proteolytic Cleavage. Hepatology 2020, 72, 923–939. [Google Scholar] [CrossRef] [PubMed]
- Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; Jessup, J.M.; Brierley, J.D.; Gaspar, L.E.; et al. AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017. [Google Scholar]
- Flynn, M.J.; Sayed, A.A.; Sharma, R.; Siddique, A.; Pinato, D.J. Challenges and Opportunities in the Clinical Development of Immune Checkpoint Inhibitors for Hepatocellular Carcinoma. Hepatology 2019, 69, 2258–2270. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.I.; Rose-John, S.; Jenkins, B.J. ADAM17: An Emerging Therapeutic Target for Lung Cancer. Cancers 2019, 11, 1218. [Google Scholar] [CrossRef] [PubMed]
- Dusterhoft, S.; Lokau, J.; Garbers, C. The metalloprotease ADAM17 in inflammation and cancer. Pathol. Res. Pract. 2019, 215, 152410. [Google Scholar] [CrossRef]
- Black, R.A.; Rauch, C.T.; Kozlosky, C.J.; Peschon, J.J.; Slack, J.L.; Wolfson, M.F.; Castner, B.J.; Stocking, K.L.; Reddy, P.; Srinivasan, S.; et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 1997, 385, 729–733. [Google Scholar] [CrossRef]
- Saad, M.I.; Jenkins, B.J. The protease ADAM17 at the crossroads of disease: Revisiting its significance in inflammation, cancer, and beyond. FEBS J. 2023. [Google Scholar] [CrossRef]
- Zunke, F.; Rose-John, S. The shedding protease ADAM17: Physiology and pathophysiology. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- McGowan, P.M.; Ryan, B.M.; Hill, A.D.; McDermott, E.; O’Higgins, N.; Duffy, M.J. ADAM-17 expression in breast cancer correlates with variables of tumor progression. Clin. Cancer Res. 2007, 13, 2335–2343. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.I.; Alhayyani, S.; McLeod, L.; Yu, L.; Alanazi, M.; Deswaerte, V.; Tang, K.; Jarde, T.; Smith, J.A.; Prodanovic, Z.; et al. ADAM17 selectively activates the IL-6 trans-signaling/ERK MAPK axis in KRAS-addicted lung cancer. EMBO Mol. Med. 2019, 11, e9976. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.I.; McLeod, L.; Yu, L.; Ebi, H.; Ruwanpura, S.; Sagi, I.; Rose-John, S.; Jenkins, B.J. The ADAM17 protease promotes tobacco smoke carcinogen-induced lung tumorigenesis. Carcinogenesis 2020, 41, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Qian, J.; Wu, Q.; Chen, Y.; Yu, G. ADAM-17 expression is enhanced by FoxM1 and is a poor prognostic sign in gastric carcinoma. J. Surg. Res. 2017, 220, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Merchant, N.B.; Voskresensky, I.; Rogers, C.M.; Lafleur, B.; Dempsey, P.J.; Graves-Deal, R.; Revetta, F.; Foutch, A.C.; Rothenberg, M.L.; Washington, M.K.; et al. TACE/ADAM-17: A component of the epidermal growth factor receptor axis and a promising therapeutic target in colorectal cancer. Clin. Cancer Res. 2008, 14, 1182–1191. [Google Scholar] [CrossRef]
- Ringel, J.; Jesnowski, R.; Moniaux, N.; Luttges, J.; Ringel, J.; Choudhury, A.; Batra, S.K.; Kloppel, G.; Lohr, M. Aberrant expression of a disintegrin and metalloproteinase 17/tumor necrosis factor-alpha converting enzyme increases the malignant potential in human pancreatic ductal adenocarcinoma. Cancer Res. 2006, 66, 9045–9053. [Google Scholar] [CrossRef]
- Kornfeld, J.W.; Meder, S.; Wohlberg, M.; Friedrich, R.E.; Rau, T.; Riethdorf, L.; Loning, T.; Pantel, K.; Riethdorf, S. Overexpression of TACE and TIMP3 mRNA in head and neck cancer: Association with tumour development and progression. Br. J. Cancer 2011, 104, 138–145. [Google Scholar] [CrossRef]
- Arneth, B. Tumor Microenvironment. Medicina 2019, 56, 15. [Google Scholar] [CrossRef]
- Khalaf, K.; Hana, D.; Chou, J.T.; Singh, C.; Mackiewicz, A.; Kaczmarek, M. Aspects of the Tumor Microenvironment Involved in Immune Resistance and Drug Resistance. Front. Immunol. 2021, 12, 656364. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, H.; Sun, Y. Tumor microenvironment and cellular senescence: Understanding therapeutic resistance and harnessing strategies. Semin. Cancer Biol. 2022, 86 Pt 3, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Xuan, Z.; Liu, X.; Zheng, M.; Yang, C.; Wang, H. Immunomodulatory role of metalloproteinase ADAM17 in tumor development. Front. Immunol. 2022, 13, 1059376. [Google Scholar] [CrossRef] [PubMed]
- Gnosa, S.P.; Puig Blasco, L.; Piotrowski, K.B.; Freiberg, M.L.; Savickas, S.; Madsen, D.H.; Auf dem Keller, U.; Kronqvist, P.; Kveiborg, M. ADAM17-mediated EGFR ligand shedding directs macrophage-promoted cancer cell invasion. JCI Insight 2022, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Brown, Z.J.; Tsilimigras, D.I.; Ruff, S.M.; Mohseni, A.; Kamel, I.R.; Cloyd, J.M.; Pawlik, T.M. Management of Hepatocellular Carcinoma: A Review. JAMA Surg. 2023, 158, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Luo, Q.; Liang, N.; Liu, H. A prognostic prediction system for hepatocellular carcinoma based on gene co-expression network. Exp. Ther. Med. 2019, 17, 4506–4516. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mishra, H.K.; Walcheck, B. Role of ADAM17 as a regulatory checkpoint of CD16A in NK cells and as a potential target for cancer immunotherapy. J. Leukoc. Biol. 2019, 105, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Mishra, H.K.; Dixon, K.J.; Pore, N.; Felices, M.; Miller, J.S.; Walcheck, B. Activation of ADAM17 by IL-15 Limits Human NK Cell Proliferation. Front. Immunol. 2021, 12, 711621. [Google Scholar] [CrossRef]
- Hedemann, N.; Herz, A.; Schiepanski, J.H.; Dittrich, J.; Sebens, S.; Dempfle, A.; Feuerborn, J.; Rogmans, C.; Tribian, N.; Florkemeier, I.; et al. ADAM17 Inhibition Increases the Impact of Cisplatin Treatment in Ovarian Cancer Spheroids. Cancers 2021, 13, 2039. [Google Scholar] [CrossRef]
- Ye, J.; Yuen, S.M.; Murphy, G.; Xie, R.; Kwok, H.F. Anti-tumor effects of a ‘human & mouse cross-reactive’ anti-ADAM17 antibody in a pancreatic cancer model in vivo. Eur. J. Pharm. Sci. 2017, 110, 62–69. [Google Scholar]
- Bolik, J.; Krause, F.; Stevanovic, M.; Gandrass, M.; Thomsen, I.; Schacht, S.S.; Rieser, E.; Muller, M.; Schumacher, N.; Fritsch, J.; et al. Inhibition of ADAM17 impairs endothelial cell necroptosis and blocks metastasis. J. Exp. Med. 2022, 219, e20201039. [Google Scholar] [CrossRef]
- Romee, R.; Foley, B.; Lenvik, T.; Wang, Y.; Zhang, B.; Ankarlo, D.; Luo, X.; Cooley, S.; Verneris, M.; Walcheck, B.; et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17). Blood 2013, 121, 3599–3608. [Google Scholar] [CrossRef] [PubMed]
- Romero, Y.; Wise, R.; Zolkiewska, A. Proteolytic processing of PD-L1 by ADAM proteases in breast cancer cells. Cancer Immunol. Immunother. 2020, 69, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]


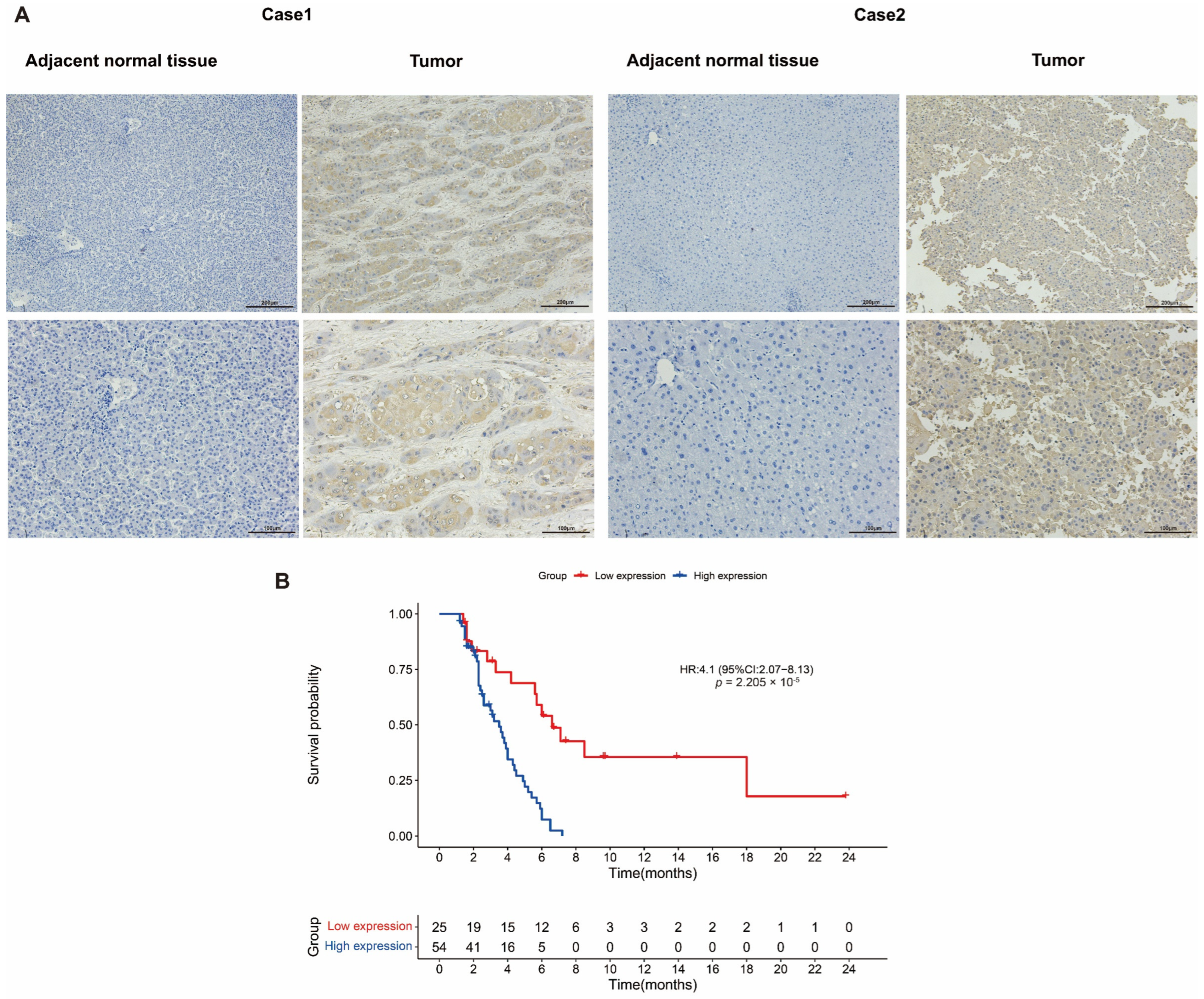
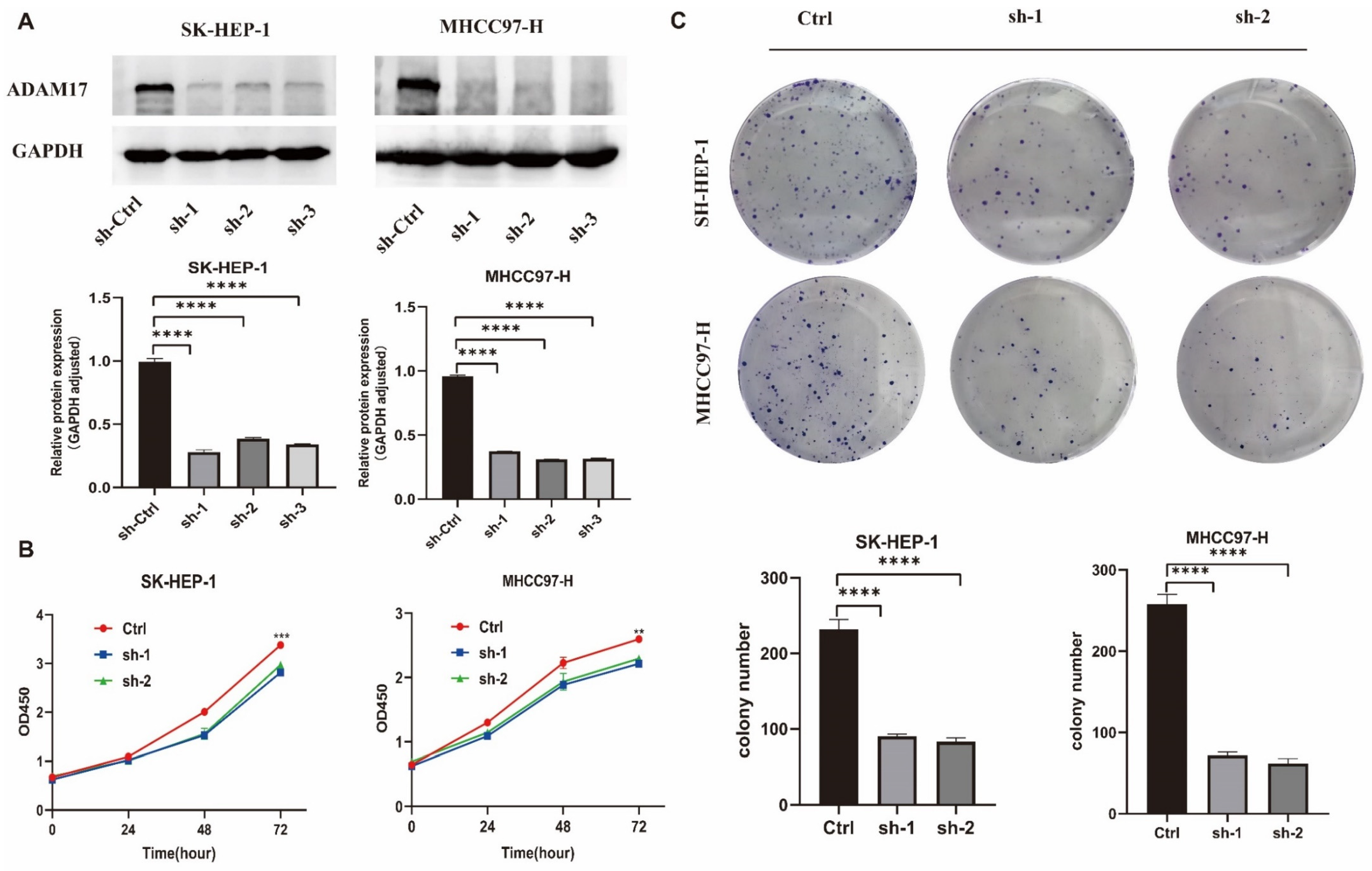
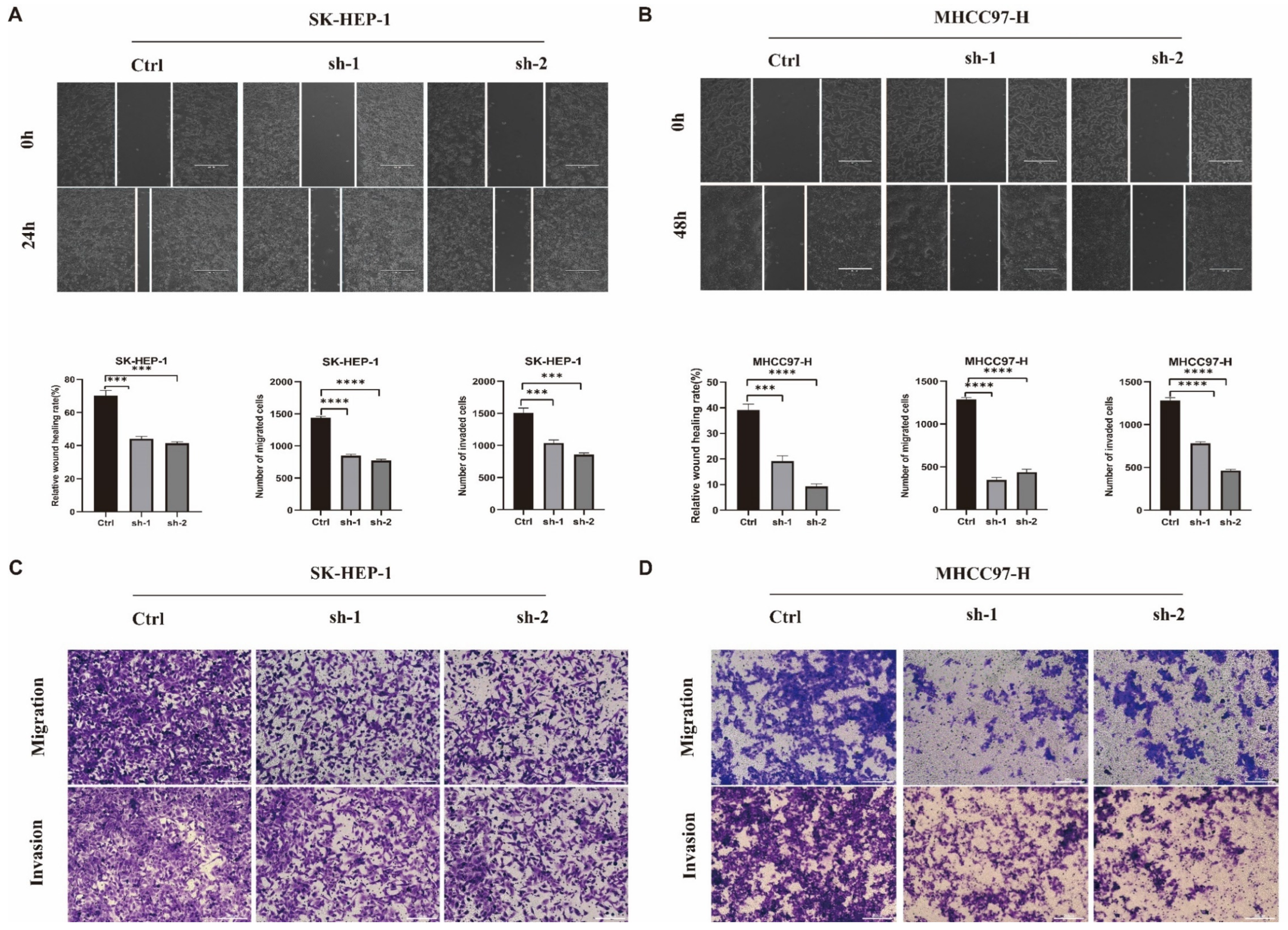
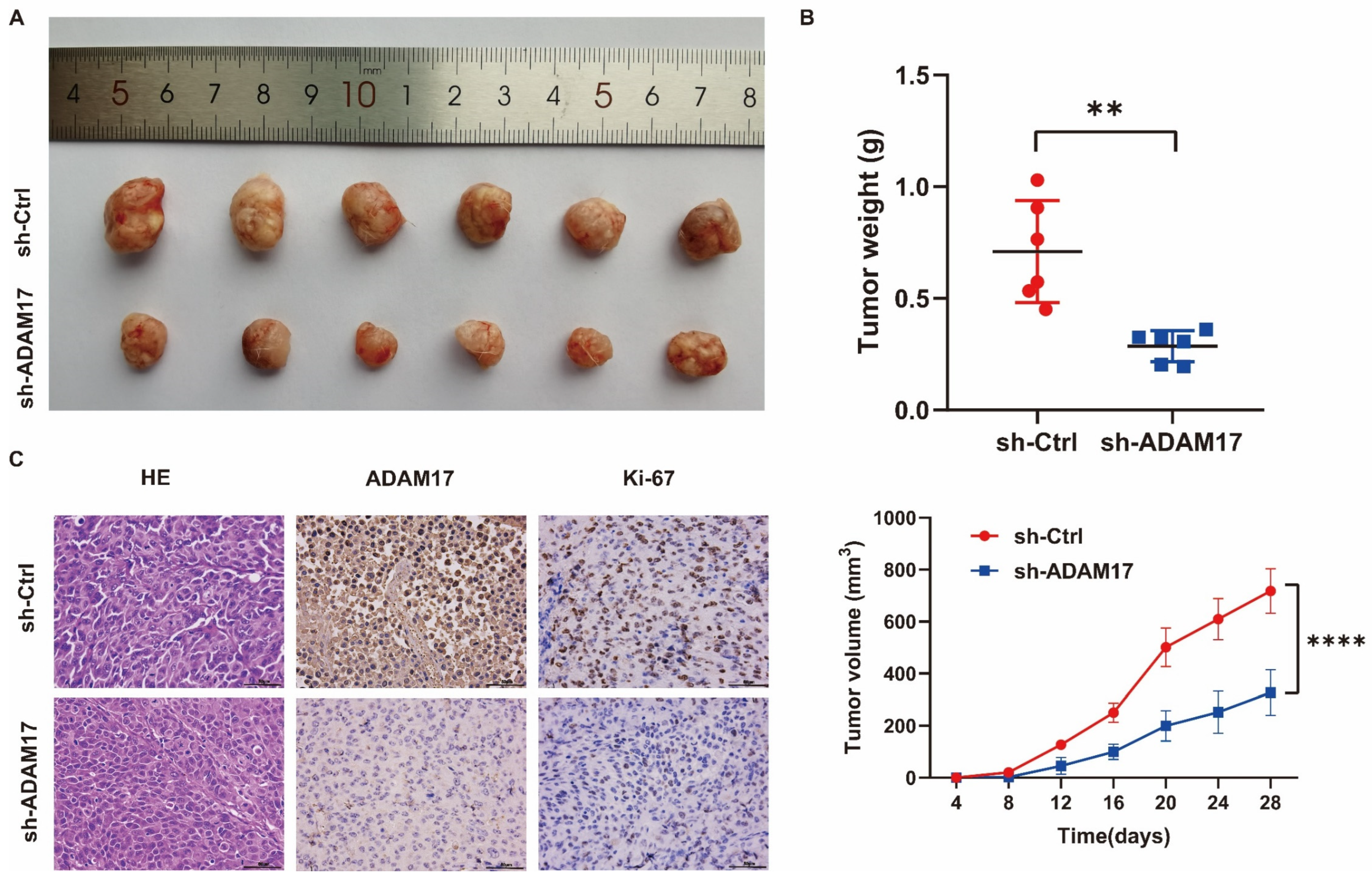

| Characteristics | Overall $ | Characteristics | Overall |
|---|---|---|---|
| Pathologic T stage, n (%) | Gender, n (%) | ||
| T1 | 183 (49.3%) | Female | 121 (32.4%) |
| T2 | 95 (25.6%) | Male | 253 (67.6%) |
| T3 | 80 (21.6%) | Age, n (%) | |
| T4 | 13 (3.5%) | ≤60 | 177 (47.5%) |
| Pathologic N stage, n (%) | >60 | 196 (52.5%) | |
| N0 | 254 (98.4%) | BMI, n (%) | |
| N1 | 4 (1.6%) | ≤25 | 177 (52.5%) |
| Pathologic M stage, n (%) | >25 | 160 (47.5%) | |
| M0 | 268 (98.5%) | Histologic grade, n (%) | |
| M1 | 4 (1.5%) | G1 | 55 (14.9%) |
| Pathologic stage, n (%) | G2 | 178 (48.2%) | |
| Stage I | 173 (49.4%) | G3 | 124 (33.6%) |
| Stage II | 87 (24.9%) | G4 | 12 (3.3%) |
| Stage III | 85 (24.3%) | AFP (ng/mL), n (%) | |
| Stage IV | 5 (1.4%) | ≤400 | 215 (76.8%) |
| >400 | 65 (23.2%) |
| Clinicopathological Parameters | Level | ADAM17 Expression | p-Value | |
|---|---|---|---|---|
| Low Expression | High Expression | |||
| N | 25 | 54 | ||
| Age (median [IQR]) | 51.00 [46.00, 59.00] | 56.00 [49.00, 59.75] | 0.509 | |
| Sex (%) | female | 10 (40.0) | 12 (22.2) | 0.114 |
| male | 15 (60.0) | 42 (77.8) | ||
| Cirrhosis (%) | no | 2 (8.0) | 2 (3.7) | 0.587 |
| yes | 23 (92.0) | 52 (96.3) | ||
| Capsule (%) | incomplete | 2 (8.0) | 10 (18.5) | 0.458 |
| intact | 6 (24.0) | 9 (16.7) | ||
| unclear | 17 (68.0) | 35 (64.8) | ||
| MVI risk (%) | M0 | 14 (56.0) | 24 (44.4) | 0.678 |
| M1 | 9 (36.0) | 25 (46.3) | ||
| M2 | 2 (8.0) | 5 (9.3) | ||
| Differentiation (%) | low | 0 (0.0) | 8 (14.8) | 0.001 |
| moderate | 19 (76.0) | 41 (75.9) | ||
| high | 6 (24.0) | 1 (1.9) | ||
| unclear | 0 (0.0) | 4 (7.4) | ||
| Histologic Grade (%) | G1 | 6 (24.0) | 1 (1.9) | 0.001 |
| G2 | 19 (76.0) | 41 (75.9) | ||
| G3 | 0 (0.0) | 8 (14.8) | ||
| Gx | 0 (0.0) | 4 (7.4) | ||
| Stage (%) | I | 12 (48.0) | 10 (18.5) | 0.021 |
| II | 10 (40.0) | 23 (42.6) | ||
| III | 3 (12.0) | 18 (33.3) | ||
| IV | 0 (0.0) | 3 (5.6) | ||
| History of hepatitis (%) | no | 3 (12.0) | 3 (5.6) | 0.374 |
| yes | 22 (88.0) | 51 (94.4) | ||
| TBIL μmol/L (median [IQR]) | 13.90 [11.80, 21.50] | 16.00 [11.60, 24.60] | 0.602 | |
| ALB g/L (median [IQR]) | 37.70 [34.70, 38.50] | 37.80 [36.20, 41.38] | 0.158 | |
| PT s (median [IQR]) | 12.10 [11.20, 12.60] | 12.70 [12.00, 13.35] | 0.038 | |
| Child Pugh (%) | A | 24 (96.0) | 50 (92.6) | 1 |
| B | 1 (4.0) | 3 (5.6) | ||
| C | 0 (0.0) | 1 (1.9) | ||
| Tumor size (%) | MHCC | 4 (16.0) | 14 (25.9) | 0.702 |
| SHCC | 10 (40.0) | 22 (40.7) | ||
| LHCC | 9 (36.0) | 14 (25.9) | ||
| HHCC | 2 (8.0) | 4 (7.4) | ||
| Serum AFP ng/mL (%) | ≤400 | 19 (76.0) | 44 (81.5) | 0.563 |
| >400 | 6 (24.0) | 10 (18.5) | ||
| Clinicopathological Parameters | Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | ||
| Age | 1.01 | 0.98–1.04 | 0.449 | NA | NA | NA | |
| ALB g/L | 1 | 0.92–1.07 | 0.918 | NA | NA | NA | |
| Capsule | Incomplete | Reference | |||||
| intact | 0.83 | 0.34–2.04 | 0.679 | NA | NA | NA | |
| unclear | 0.86 | 0.42–1.75 | 0.678 | NA | NA | NA | |
| Child Pugh | 3.41 | 1.45–8.02 | 0.005 | 3.48 | 1.09–11.11 | 0.0356 | |
| Cirrhosis | No | Reference | |||||
| yes | 5.56 | 0.77–40.24 | 0.09 | NA | NA | NA | |
| Differentiation | Low | Reference | |||||
| high | 0.24 | 0.05–1.03 | 0.054 | NA | NA | NA | |
| moderate | 0.74 | 0.29–1.92 | 0.535 | NA | NA | NA | |
| unclear | 1.73 | 0.46–6.47 | 0.416 | NA | NA | NA | |
| group | Low expression | Reference | |||||
| High expression | 4.07 | 2.06–8.08 | 0 | 3 | 1.4–6.42 | 0.0047 | |
| Histologic Grade | G1 | Reference | |||||
| G2 | 3.13 | 0.97–10.13 | 0.057 | 1.83 | 0.53–6.37 | 0.341 | |
| G3 | 4.22 | 0.98–18.3 | 0.054 | 1.35 | 0.27–6.88 | 0.7159 | |
| Gx | 7.3 | 1.59–33.56 | 0.011 | 3.44 | 0.67–17.77 | 0.1405 | |
| History of hepatitis | No | Reference | |||||
| yes | 1.39 | 0.55–3.49 | 0.487 | NA | NA | NA | |
| MVI risk | 1.11 | 0.72–1.7 | 0.633 | NA | NA | NA | |
| PT s | 1.31 | 1.1–1.56 | 0.002 | 1.07 | 0.84–1.35 | 0.601 | |
| Serum AFP ng/mL | ≤400 | Reference | |||||
| >400 | 1.37 | 0.72–2.6 | 0.332 | NA | NA | NA | |
| Sex | Female | Reference | |||||
| male | 1.29 | 0.71–2.33 | 0.403 | NA | NA | NA | |
| Stage | 1.45 | 1.04–2.03 | 0.029 | 1.42 | 0.98–2.05 | 0.063 | |
| TBIL μmol/L | 1.02 | 0.99–1.05 | 0.162 | NA | NA | NA | |
| Tumor size | 1.09 | 0.81–1.46 | 0.57 | NA | NA | NA | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, T.; Yu, Y.; Gao, L.; Xiang, L.; Xu, B.; Gu, B.; Chen, H. Predictive Roles of ADAM17 in Patient Survival and Immune Cell Infiltration in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 17069. https://doi.org/10.3390/ijms242317069
Ding T, Yu Y, Gao L, Xiang L, Xu B, Gu B, Chen H. Predictive Roles of ADAM17 in Patient Survival and Immune Cell Infiltration in Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2023; 24(23):17069. https://doi.org/10.3390/ijms242317069
Chicago/Turabian StyleDing, Tianlong, Yang Yu, Lei Gao, Lin Xiang, Bo Xu, Baohong Gu, and Hao Chen. 2023. "Predictive Roles of ADAM17 in Patient Survival and Immune Cell Infiltration in Hepatocellular Carcinoma" International Journal of Molecular Sciences 24, no. 23: 17069. https://doi.org/10.3390/ijms242317069
APA StyleDing, T., Yu, Y., Gao, L., Xiang, L., Xu, B., Gu, B., & Chen, H. (2023). Predictive Roles of ADAM17 in Patient Survival and Immune Cell Infiltration in Hepatocellular Carcinoma. International Journal of Molecular Sciences, 24(23), 17069. https://doi.org/10.3390/ijms242317069






