How Do Phages Disrupt the Structure of Enterococcus faecalis Biofilm?
Abstract
:1. Introduction
2. Results
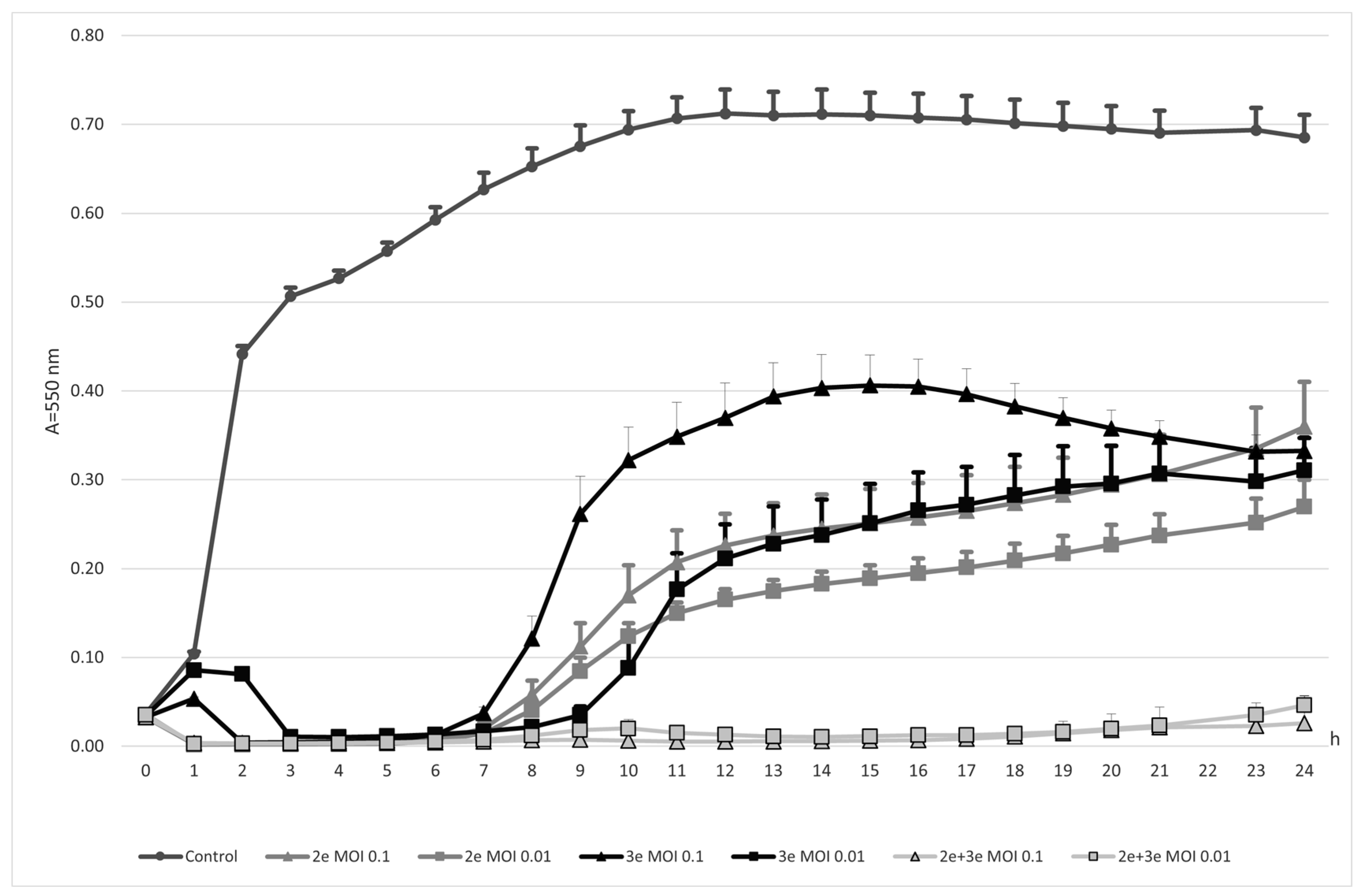
2.1. Bacteriophages Action against E. faecalis Cells
2.1.1. Planktonic Forms
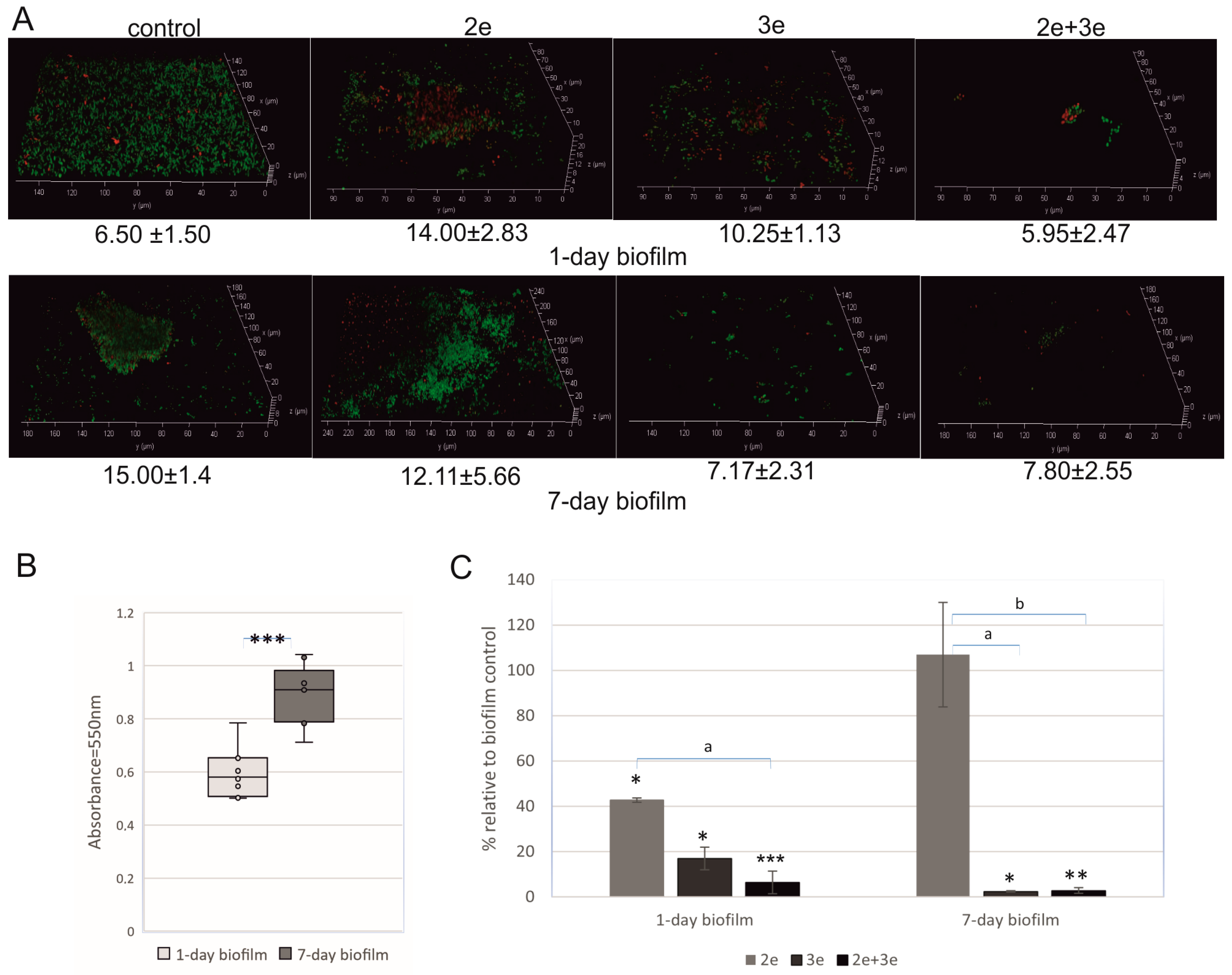
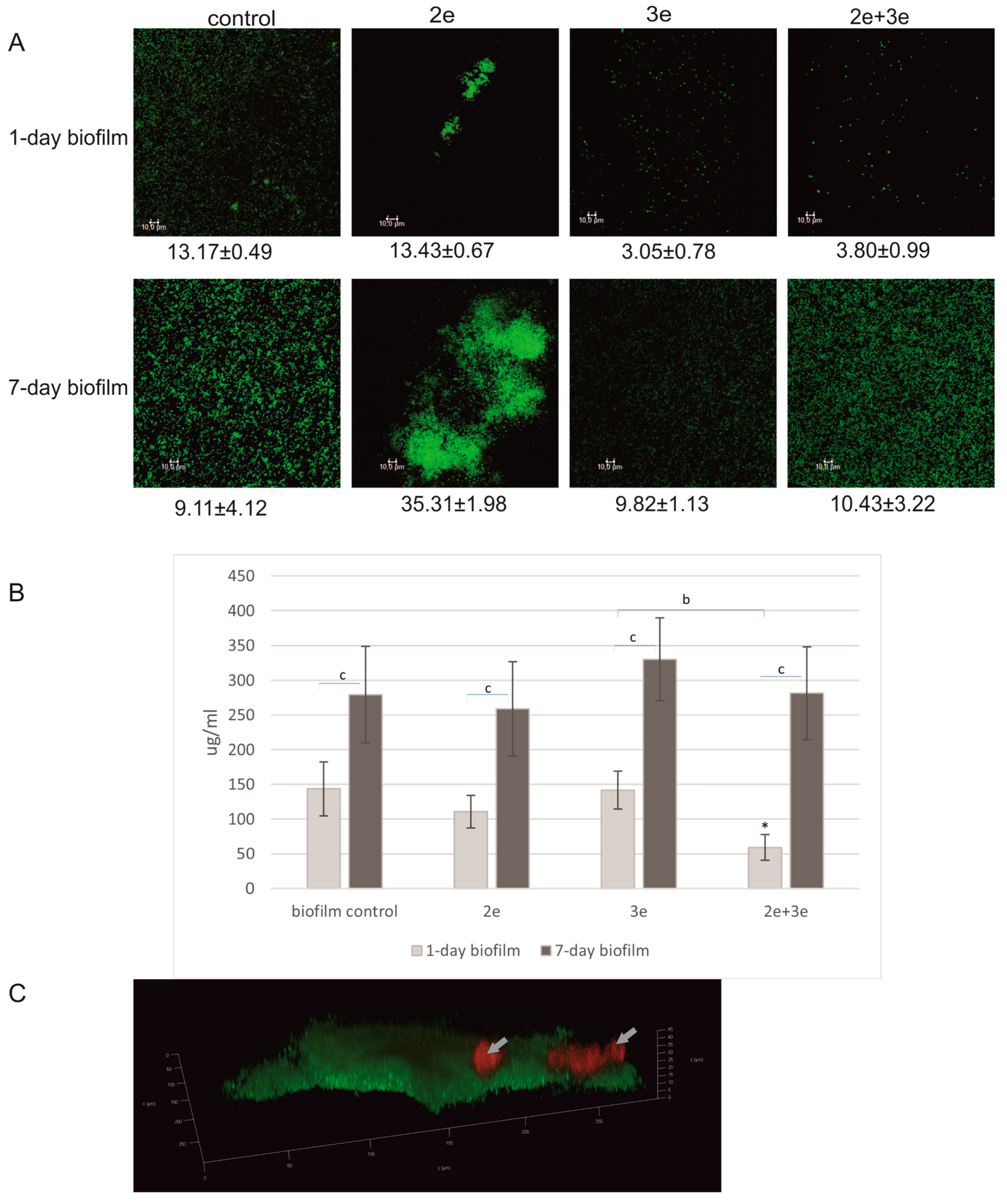
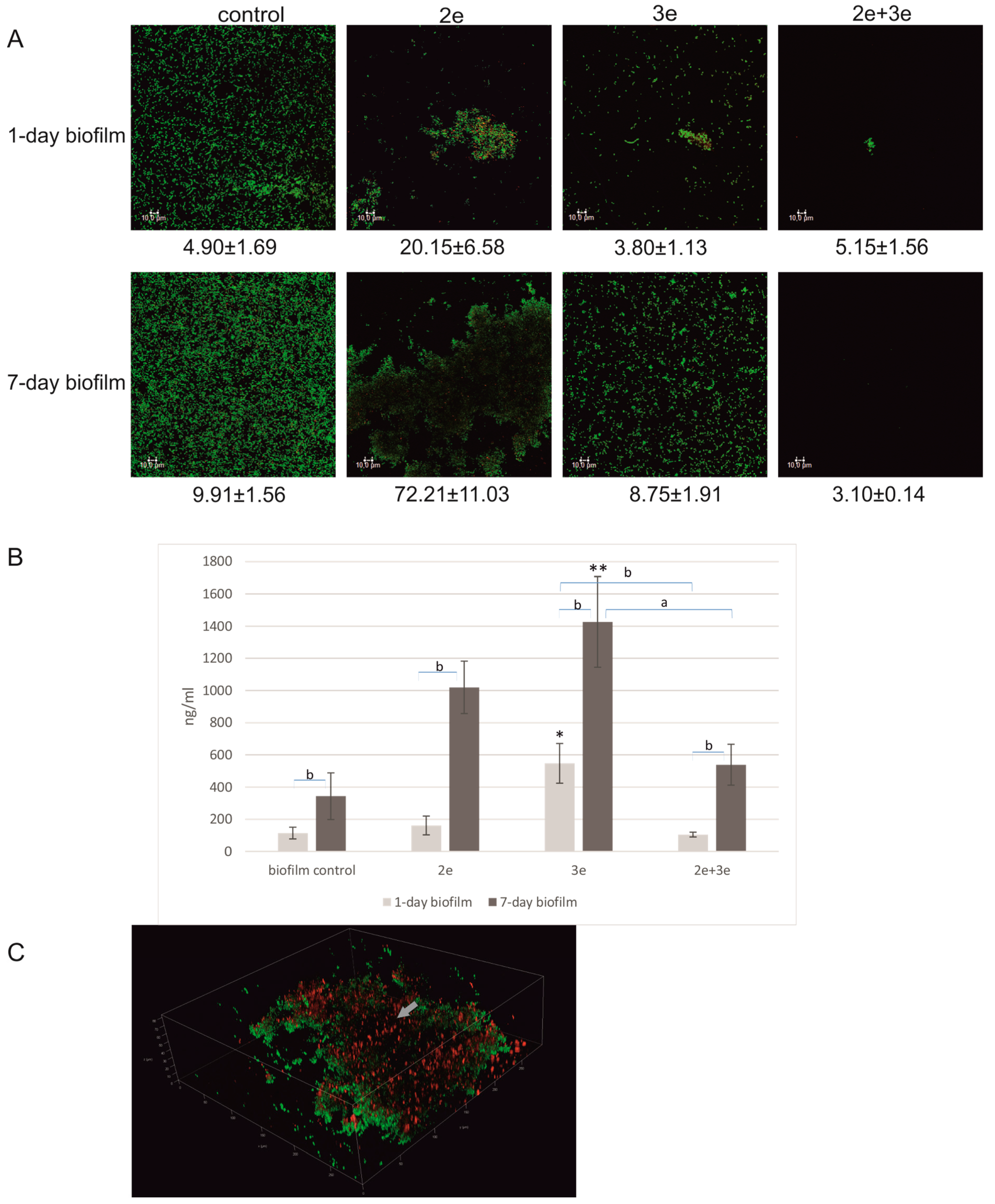
2.1.2. Biofilm Decomposition
2.2. Bacteriophages Influence on EPSs in E. faecalis Biofilm
2.2.1. Polysaccharides
2.2.2. Proteins
2.2.3. eDNA
3. Discussion
4. Materials and Methods
4.1. Bacteriophages and Bacteria
4.2. Time Kills Assay—Bacteriophages Action against Planktonic Cells
4.3. Bacteriophages Action against Sessile Cells in Biofilm
4.3.1. Biofilm Cultivation
4.3.2. Bacteriophage Application on Preformed Biofilm
4.3.3. Biofilm Visualization
4.4. Estimation of Extracellular Polymers: Polysaccharides, Proteins, and eDNA in Biofilm
4.5. CLSM Visualization
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prazmo, E.; Godlewska, R.; Kwasny, M.; Mielczarek, A. Virulence Factors of Enterococcus faecalis in Relation to Pulp Diseases and Periapical Infections. Adv. Mocrobiol. 2016, 55, 247–254. [Google Scholar]
- Lee, D.; Im, J.; Na, H.; Ryu, S.; Yun, C.H.; Han, S.H. The Novel Enterococcus Phage VB_EfaS_HEf13 Has Broad Lytic Activity Against Clinical Isolates of Enterococcus faecalis. Front. Microbiol. 2019, 10, 2877. [Google Scholar] [CrossRef] [PubMed]
- Bolocan, A.S.; Upadrasta, A.; De Almeida Bettio, P.H.; Clooney, A.G.; Draper, L.A.; Ross, R.P.; Hill, C. Evaluation of Phage Therapy in the Context of Enterococcus faecalis and Its Associated Diseases. Viruses 2019, 11, 366. [Google Scholar] [CrossRef] [PubMed]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef]
- Al-Zubidi, M.; Widziolek, M.; Court, E.K.; Gains, A.F.; Smith, R.E.; Ansbro, K.; Alrafaie, A.; Evans, C.; Murdoch, C.; Mesnage, S.; et al. Identification of Novel Bacteriophages with Therapeutic Potential That Target Enterococcus faecalis. Infect. Immun. 2019, 87, e00512-19. [Google Scholar] [CrossRef]
- Coenye, T. Biofilms. Ref. Modul. Life Sci. 2022. [Google Scholar] [CrossRef]
- Moryl, M.; Kaleta, A.; Strzelecki, K.; Różalska, S.; Różalski, A. Effect of Nutrient and Stress Factors on Polysaccharides Synthesis in Proteus Mirabilis Biofilm. Acta Biochim. Pol. 2014, 61, 133–139. [Google Scholar] [CrossRef]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.J.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The Clinical Impact of Bacterial Biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Mah, T.F.; O’Toole, G.A. Mechanisms of Biofilm Resistance to Antimicrobial Agents. Trends Microbiol. 2015, 9, 34–39. [Google Scholar] [CrossRef]
- Khalifa, L.; Brosh, Y.; Gelman, D.; Coppenhagen-Glazer, S.; Beyth, S.; Poradosu-Cohen, R.; Que, Y.A.; Beyth, N.; Hazan, R. Targeting Enterococcus faecalis Biofilms with Phage Therapy. Appl. Environ. Microbiol. 2015, 81, 2696–2705. [Google Scholar] [CrossRef]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol. J. 2017, 11, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Dharmaraj, T.; Cai, P.C.; Burgener, E.B.; Haddock, N.L.; Spakowitz, A.J.; Bollyky, P.L. Bacteriophage and Bacterial Susceptibility, Resistance, and Tolerance to Antibiotics. Pharmaceutics 2022, 14, 1425. [Google Scholar] [CrossRef] [PubMed]
- Sadekuzzaman, M.; Mizan, M.F.R.; Yang, S.; Kim, H.S.; Ha, S.D. Application of Bacteriophages for the Inactivation of Salmonella Spp. in Biofilms. Food Sci. Technol. Int. 2018, 24, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Maszewska, A. Phage Associated Polysaccharide Depolymerases-Characteristics and Application. Adv. Clin. Exp. Med. 2015, 69, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lu, H.; Zhang, S.; Shi, Y.; Chen, Q. Phages against Pathogenic Bacterial Biofilms and Biofilm-Based Infections: A Review. Pharmaceutics 2022, 14, 427. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Du, F.; Long, M.; Li, P. Limitations of Phage Therapy and Corresponding Optimization Strategies: A Review. Molecules 2022, 27, 1857. [Google Scholar] [CrossRef] [PubMed]
- Borin, J.M.; Avrani, S.; Barrick, J.E.; Petrie, K.L.; Meyer, J.R. Coevolutionary Phage Training Leads to Greater Bacterial Suppression and Delays the Evolution of Phage Resistance. Proc. Natl. Acad. Sci. USA 2021, 118, 32104592118. [Google Scholar] [CrossRef]
- Elois, M.A.; da Silva, R.; Pilati, G.V.T.; Rodríguez-Lázaro, D.; Fongaro, G. Bacteriophages as Biotechnological Tools. Viruses 2023, 15, 349. [Google Scholar] [CrossRef]
- Sharma, U.; Vipra, A.; Channabasappa, S. Phage-Derived Lysins as Potential Agents for Eradicating Biofilms and Persisters. Drug Discov. Today 2018, 23, 848–856. [Google Scholar] [CrossRef]
- Meneses, L.; Brandão, A.C.; Coenye, T.; Braga, A.C.; Pires, D.P.; Azeredo, J. A Systematic Review of the Use of Bacteriophages for in Vitro Biofilm Control. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 919–928. [Google Scholar] [CrossRef]
- Moryl, M.; Palatyńska-Ulatowska, A.; Maszewska, A.; Grzejdziak, I.; Dias de Oliveira, S.; Pradebon, M.C.; Steier, L.; Różalski, A.; Poli de Figueiredo, J.A. Benefits and Challenges of the Use of Two Novel VB_Efa29212_2e and VB_Efa29212_3e Bacteriophages in Biocontrol of the Root Canal Enterococcus faecalis Infections. J. Clin. Med. 2022, 11, 6494. [Google Scholar] [CrossRef] [PubMed]
- Vilas Boas, D.; Almeida, C.; Sillankorva, S.; Nicolau, A.; Azeredo, J.; Azevedo, N.F. Discrimination of Bacteriophage Infected Cells Using Locked Nucleic Acid Fluorescent in Situ Hybridization (LNA-FISH). Biofouling 2016, 32, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Kifelew, L.G.; Warner, M.S.; Morales, S.; Thomas, N.; Gordon, D.L.; Mitchell, J.G.; Speck, P.G. Efficacy of Lytic Phage Cocktails on Staphylococcus aureus and Pseudomonas aeruginosa in Mixed-Species Planktonic Cultures and Biofilms. Viruses 2020, 12, 559. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Vandenheuvel, D.; Martínez, B.; Rodríguez, A.; Lavigne, R.; García, P. Two Phages, PhiIPLA-RODI and PhiIPLA-C1C, Lyse Mono-and Dual-Species Staphylococcal Biofilms. Appl. Environ. Microbiol. 2015, 81, 3336–3348. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.K.; Erwin, T.C.; McLean, R.J.C.; Aron, G.M. Bacteriophage Ecology in Escherichia coli and Pseudomonas aeruginosa Mixed-Biofilm Communities. Appl. Environ. Microbiol. 2011, 77, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Alves, D.R.; Gaudion, A.; Bean, J.E.; Perez Esteban, P.; Arnot, T.C.; Harper, D.R.; Kot, W.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T.A. Combined Use of Bacteriophage K and a Novel Bacteriophage to Reduce Staphylococcus aureus Biofilm Formation. Appl. Environ. Microbiol. 2014, 80, 6694–6703. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; González, S.; Campelo, A.B.; Martínez, B.; Rodríguez, A.; García, P. Low-Level Predation by Lytic Phage PhiIPLA-RODI Promotes Biofilm Formation and Triggers the Stringent Response in Staphylococcus aureus. Sci. Rep. 2017, 7, 40965. [Google Scholar] [CrossRef]
- Goodarzi, F.; Hallajzadeh, M.; Sholeh, M.; Talebi, M.; Mahabadi, V.P.; Amirmozafari, N. Anti-Biofilm Activity of a Lytic Phage Against Vancomycin-Resistant Enterococcus faecalis. Iran. J. Pathol. 2022, 17, 285–293. [Google Scholar] [CrossRef]
- Hanlon, G.W.; Denyer, S.P.; Olliff, C.J.; Ibrahim, L.J. Reduction in Exopolysaccharide Viscosity as an Aid to Bacteriophage Penetration through Pseudomonas aeruginosa Biofilms. Appl. Environ. Microbiol. 2001, 67, 2746–2753. [Google Scholar] [CrossRef]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J. Phage Cocktail Development for Bacteriophage Therapy: Toward Improving Spectrum of Activity Breadth and Depth. Pharmaceuticals 2021, 14, 1019. [Google Scholar] [CrossRef]
- Chadha, P.; Katare, O.P.; Chhibber, S. In Vivo Efficacy of Single Phage versus Phage Cocktail in Resolving Burn Wound Infection in BALB/c Mice. Microb. Pathog. 2016, 99, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.; Pereira, C.; Gomes, A.T.P.C.; Almeida, A. Efficiency of Single Phage Suspensions and Phage Cocktail in the Inactivation of Escherichia coli and Salmonella typhimurium: An In Vitro Preliminary Study. Microorganisms 2019, 7, 94. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Werier, J.M.; Abdelbary, H. Bacteriophage Cocktail Is More Efficacious in Treating MRSA Biofilm on Plasma Spray Titanium Surface Compared to Vancomycin or a Single Bacteriophage. Orthop. Proc. 2021, 103 (Suppl. S3), 1. [Google Scholar] [CrossRef]
- Konopacki, M.; Grygorcewicz, B.; Gliźniewicz, M.; Miłek, D.; Kordas, M.; Rakoczy, R. Dynamic Modelling of Bacteriophage Production Process. Chem. Process Eng. 2022, 43, 471–482. [Google Scholar] [CrossRef]
- Zhou, Y.; Bao, H.; Zhang, H.; Wang, R. Isolation and Characterization of Lytic Phage VB_EcoM_JS09 against Clinically Isolated Antibiotic-Resistant Avian Pathogenic Escherichia coli and Enterotoxigenic Escherichia coli. Intervirology 2015, 58, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Zhu, L.; Chen, L.; Qiu, Y.; Wang, J. Detection Techniques for Extracellular Polymeric Substances in Biofilms: A Review. BioRes. 2016, 11, 8092–8115. [Google Scholar] [CrossRef]
- Flemming, H.C.; Neu, T.R.; Wozniak, D.J. The EPS Matrix: The “House of Biofilm Cells”. J. Bacteriol. 2007, 189, 7945. [Google Scholar] [CrossRef]
- Campoccia, D.; Montanaro, L.; Arciola, C.R. Tracing the Origins of Extracellular DNA in Bacterial Biofilms: Story of Death and Predation to Community Benefit. Biofouling 2021, 37, 1022–1039. [Google Scholar] [CrossRef]
- Di Martino, P. Extracellular Polymeric Substances, a Key Element in Understanding Biofilm Phenotype. AIMS Microbiol. 2018, 4, 274–288. [Google Scholar] [CrossRef]
- Mahto, K.U.; Vandana; Priyadarshanee, M.; Samantaray, D.P.; Das, S. Bacterial Biofilm and Extracellular Polymeric Substances in the Treatment of Environmental Pollutants: Beyond the Protective Role in Survivability. J. Clean. Prod. 2022, 379, 134759. [Google Scholar] [CrossRef]
- Chug, R.; Khosla, B.; Singh, M. Modulation of the Extracellular Polymeric Substances (EPS) Production by Quorum Sensing (QS) in Bacteria. Int. J. Curr. Microbiol. App. Sci. 2015, 4, 884–896. [Google Scholar]
- Ferriol-González, C.; Domingo-Calap, P. Phages for Biofilm Removal. Antibiotics 2020, 9, 268. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, R.; Xu, M.; Liu, Y.; Zhu, X.; Qiu, J.; Liu, Q.; He, P.; Li, Q. A Novel Polysaccharide Depolymerase Encoded by the Phage SH-KP152226 Confers Specific Activity Against Multidrug-Resistant Klebsiella pneumoniae via Biofilm Degradation. Front. Microbiol. 2019, 10, 2768. [Google Scholar] [CrossRef] [PubMed]
- Catalog, F. FilmTracerTM SYPRO® Ruby Biofilm Matrix Stain. Image. 2009, pp. 3–5. Available online: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2Fmp10318.pdf (accessed on 30 October 2023).
- Yi, S.; Sahni, N.; Daniels, K.J.; Lu, K.L.; Huang, G.; Garnaas, A.M.; Pujol, C.; Srikantha, T.; Soll, D.R.; Yi, C.; et al. Utilization of the Mating Scaffold Protein in the Evolution of a New Signal Transduction Pathway for Biofilm Development. MBio 2011, 2, e00237-10. [Google Scholar] [CrossRef] [PubMed]
- Mlynek, K.D.; Bulock, L.L.; Stone, C.J.; Curran, L.J.; Sadykov, M.R.; Bayles, K.W.; Brinsmade, S.R. Genetic and Biochemical Analysis of CodY-Mediated Cell Aggregation in Staphylococcus aureus Reveals an Interaction between Extracellular DNA and Polysaccharide in the Extracellular Matrix. J. Bacteriol. 2020, 202, 593–612. [Google Scholar] [CrossRef]
- Leroy, S.; Lebert, I.; Andant, C.; Micheau, P.; Talon, R. Investigating Extracellular DNA Release in Staphylococcus xylosus Biofilm In Vitro. Microorganisms 2021, 9, 2192. [Google Scholar] [CrossRef] [PubMed]
- Moryl, M.; Spetana, M.; Dziubek, K.; Paraszkiewicz, K.; Różalska, S.; Płaza, G.A.; Różalski, A. Antimicrobial, Antiadhesive and Antibiofilm Potential of Lipopeptides Synthesised by Bacillus subtilis, on Uropathogenic Bacteria. Acta Biochim. Pol. 2015, 62, 725–732. [Google Scholar] [CrossRef]
- Plota, M.; Sazakli, E.; Giormezis, N.; Gkartziou, F.; Kolonitsiou, F.; Leotsinidis, M.; Antimisiaris, S.G.; Spiliopoulou, I. In Vitro Anti-Biofilm Activity of Bacteriophage k (Atcc 19685-B1) and Daptomycin against Staphylococci. Microorganisms 2021, 9, 1853. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.I.; Lee, Y.C. Carbohydrate Analysis by a Phenol–Sulfuric Acid Method in Microplate Format. Anal. Biochem. 2005, 339, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Schlafer, S.; Meyer, R.L. Confocal Microscopy Imaging of the Biofilm Matrix. J. Microbiol. Methods 2017, 138, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Mann, E.E.; Rice, K.C.; Boles, B.R.; Endres, J.L.; Ranjit, D.; Chandramohan, L.; Tsang, L.H.; Smeltzer, M.S.; Horswill, A.R.; Bayles, K.W. Modulation of EDNA Release and Degradation Affects Staphylococcus aureus Biofilm Maturation. PLoS ONE 2009, 4, e5822. [Google Scholar] [CrossRef] [PubMed]
- Catalog, L. FilmTracerTM LIVE/DEAD® Biofilm Viability Kit. Image. 2009, pp. 1–4. Available online: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2Fmp10316.pdf (accessed on 30 October 2023).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moryl, M.; Różalski, A.; de Figueiredo, J.A.P.; Palatyńska-Ulatowska, A. How Do Phages Disrupt the Structure of Enterococcus faecalis Biofilm? Int. J. Mol. Sci. 2023, 24, 17260. https://doi.org/10.3390/ijms242417260
Moryl M, Różalski A, de Figueiredo JAP, Palatyńska-Ulatowska A. How Do Phages Disrupt the Structure of Enterococcus faecalis Biofilm? International Journal of Molecular Sciences. 2023; 24(24):17260. https://doi.org/10.3390/ijms242417260
Chicago/Turabian StyleMoryl, Magdalena, Antoni Różalski, Jose Antonio Poli de Figueiredo, and Aleksandra Palatyńska-Ulatowska. 2023. "How Do Phages Disrupt the Structure of Enterococcus faecalis Biofilm?" International Journal of Molecular Sciences 24, no. 24: 17260. https://doi.org/10.3390/ijms242417260
APA StyleMoryl, M., Różalski, A., de Figueiredo, J. A. P., & Palatyńska-Ulatowska, A. (2023). How Do Phages Disrupt the Structure of Enterococcus faecalis Biofilm? International Journal of Molecular Sciences, 24(24), 17260. https://doi.org/10.3390/ijms242417260





