Two-Component System Genes in Brassica napus: Identification, Analysis, and Expression Patterns in Response to Abiotic and Biotic Stresses
Abstract
:1. Introduction
2. Results
2.1. Identification of Two-Component System Genes in Brassica napus and Its Two Diploid Progenitors
2.1.1. Histidine Kinase Proteins in Rapeseed
2.1.2. Histidine Phosphotransfer Proteins in Rapeseed
2.1.3. Response Regulators in Rapeseed
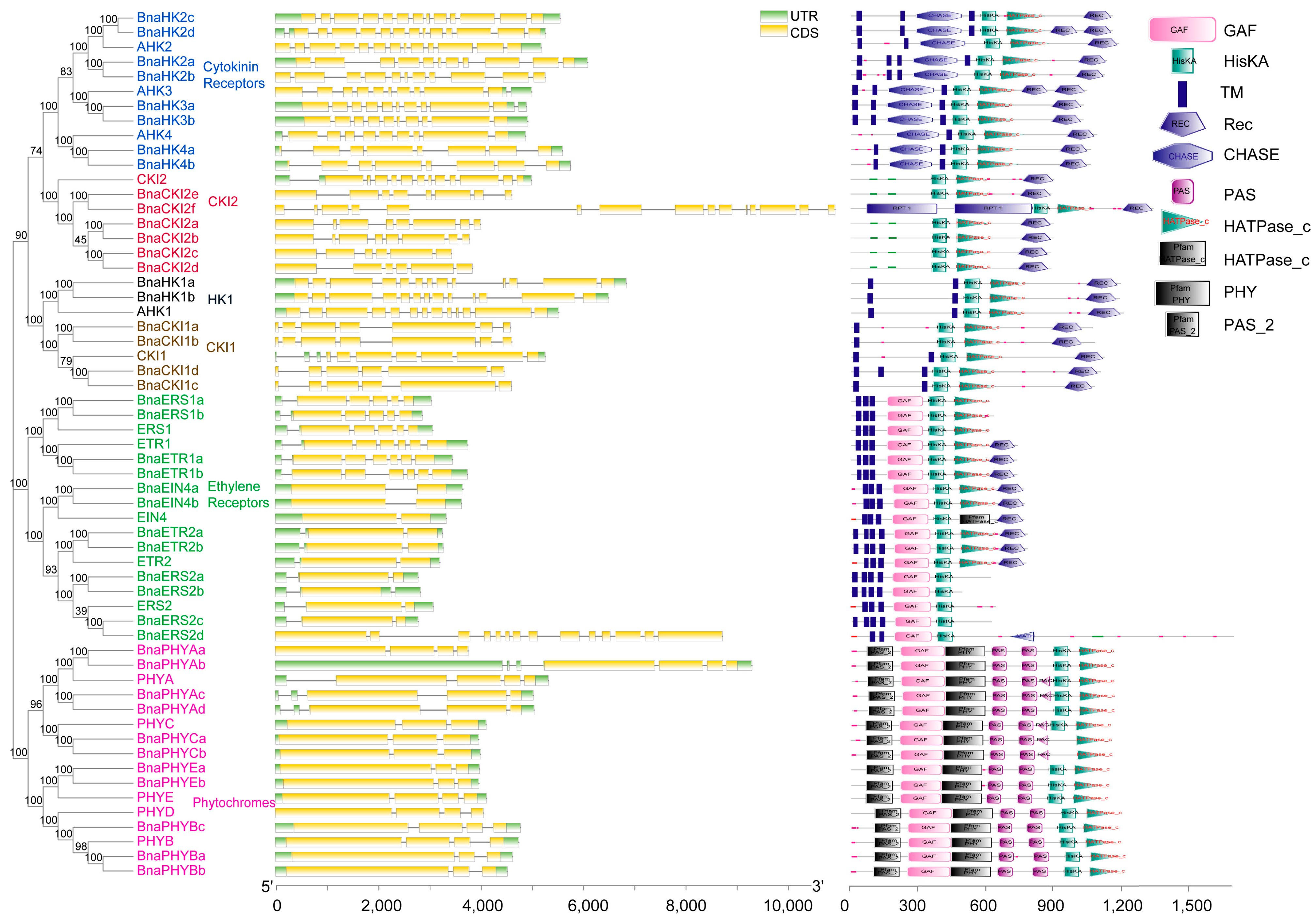
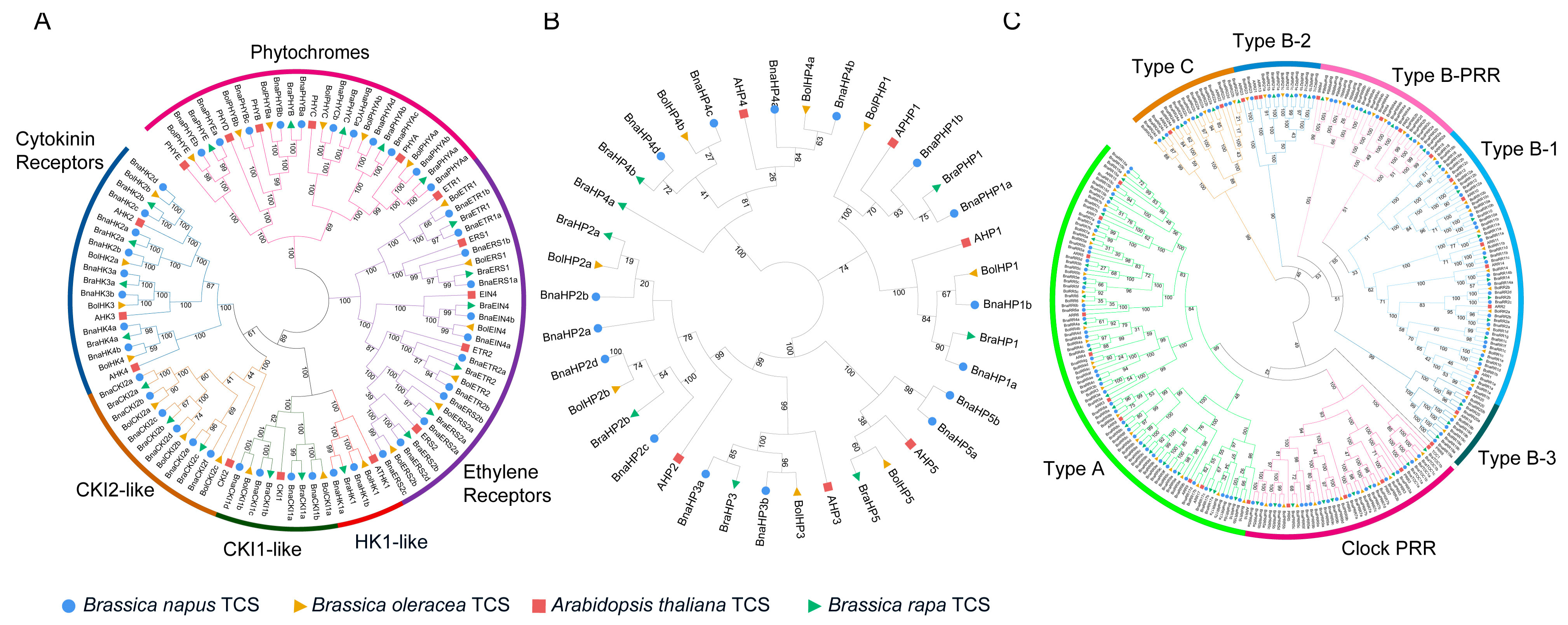
2.2. Phylogenetic Analysis
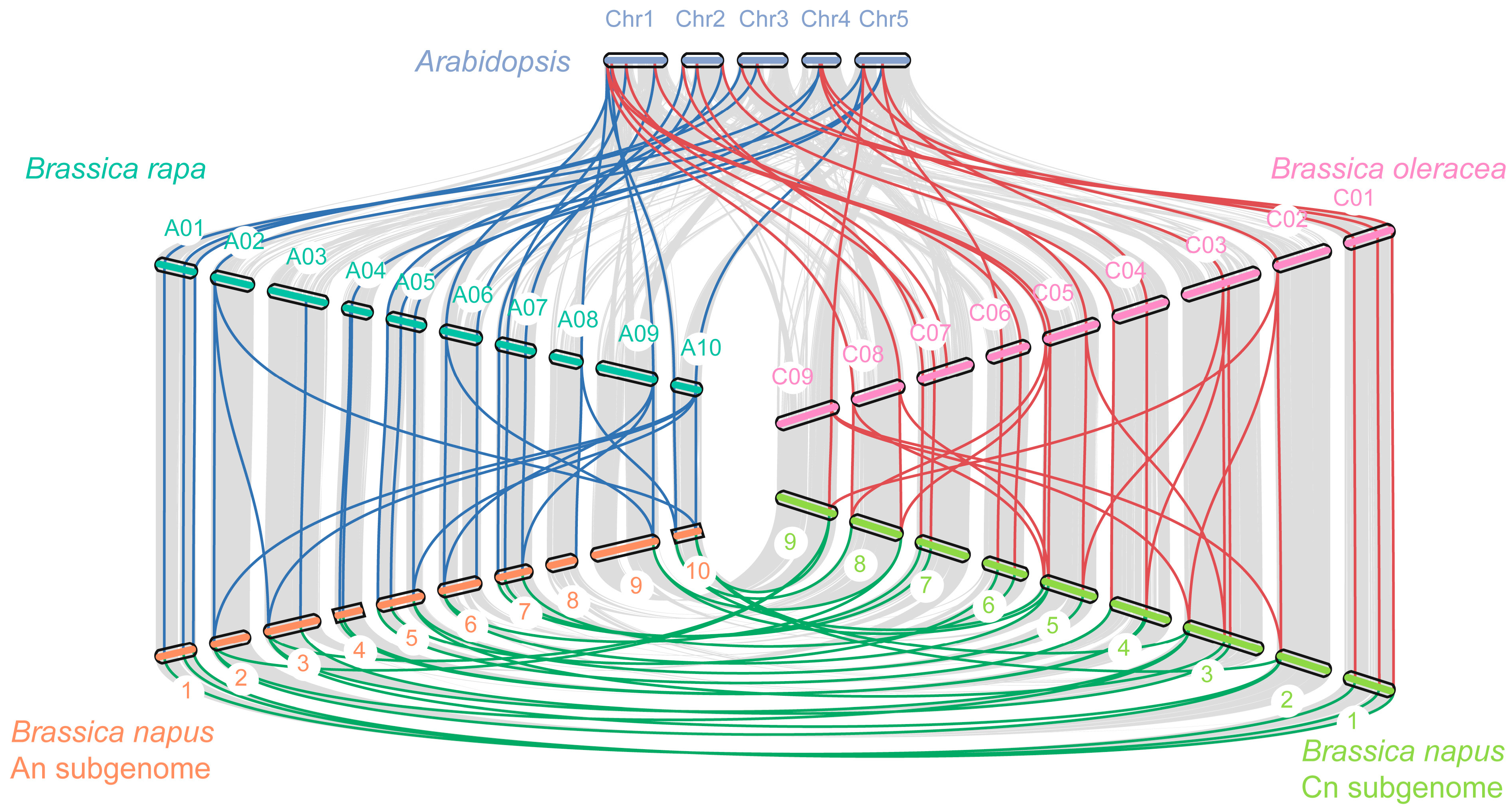
2.3. Genomic Distribution, Gene Duplication, and Synteny Analysis
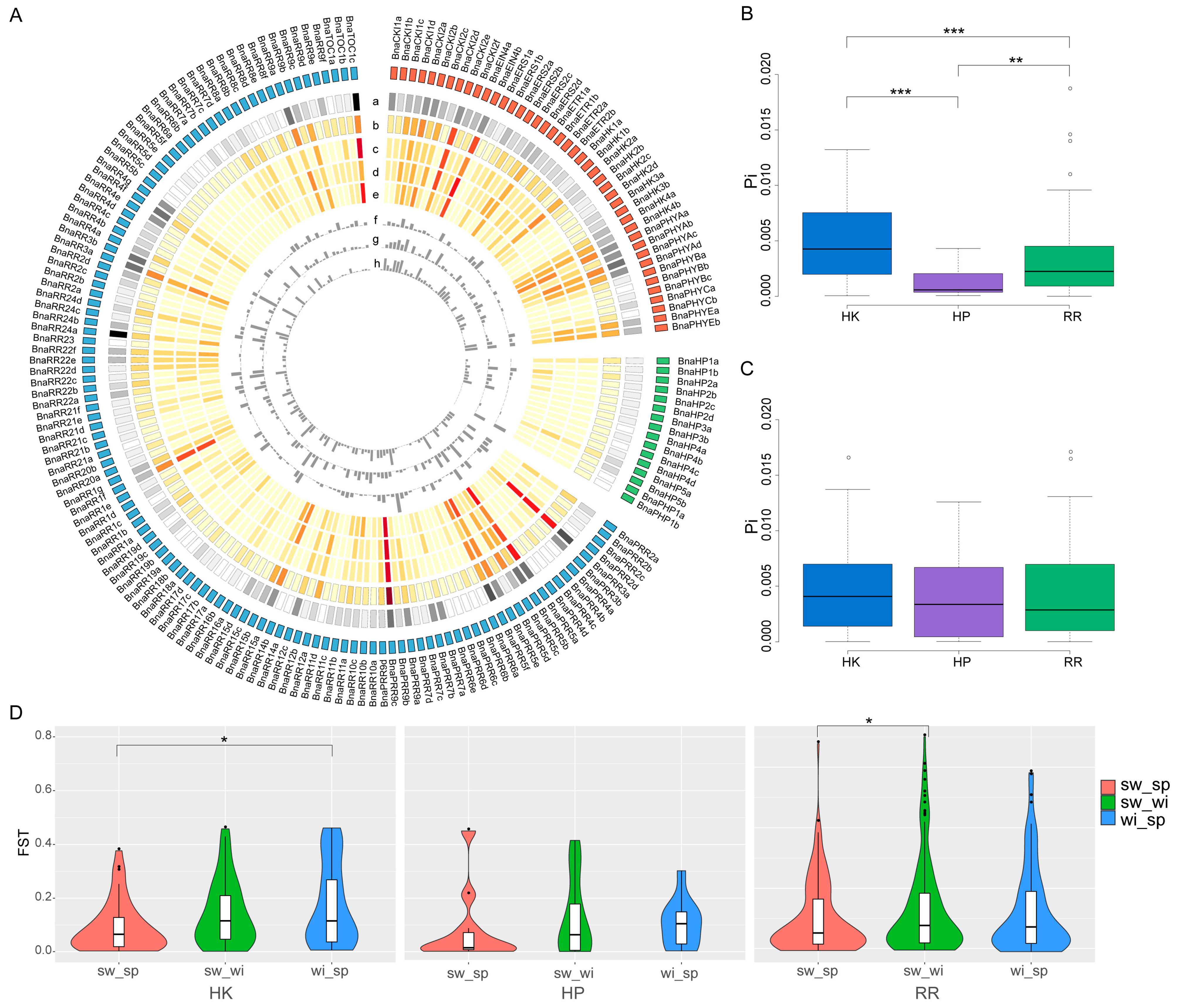
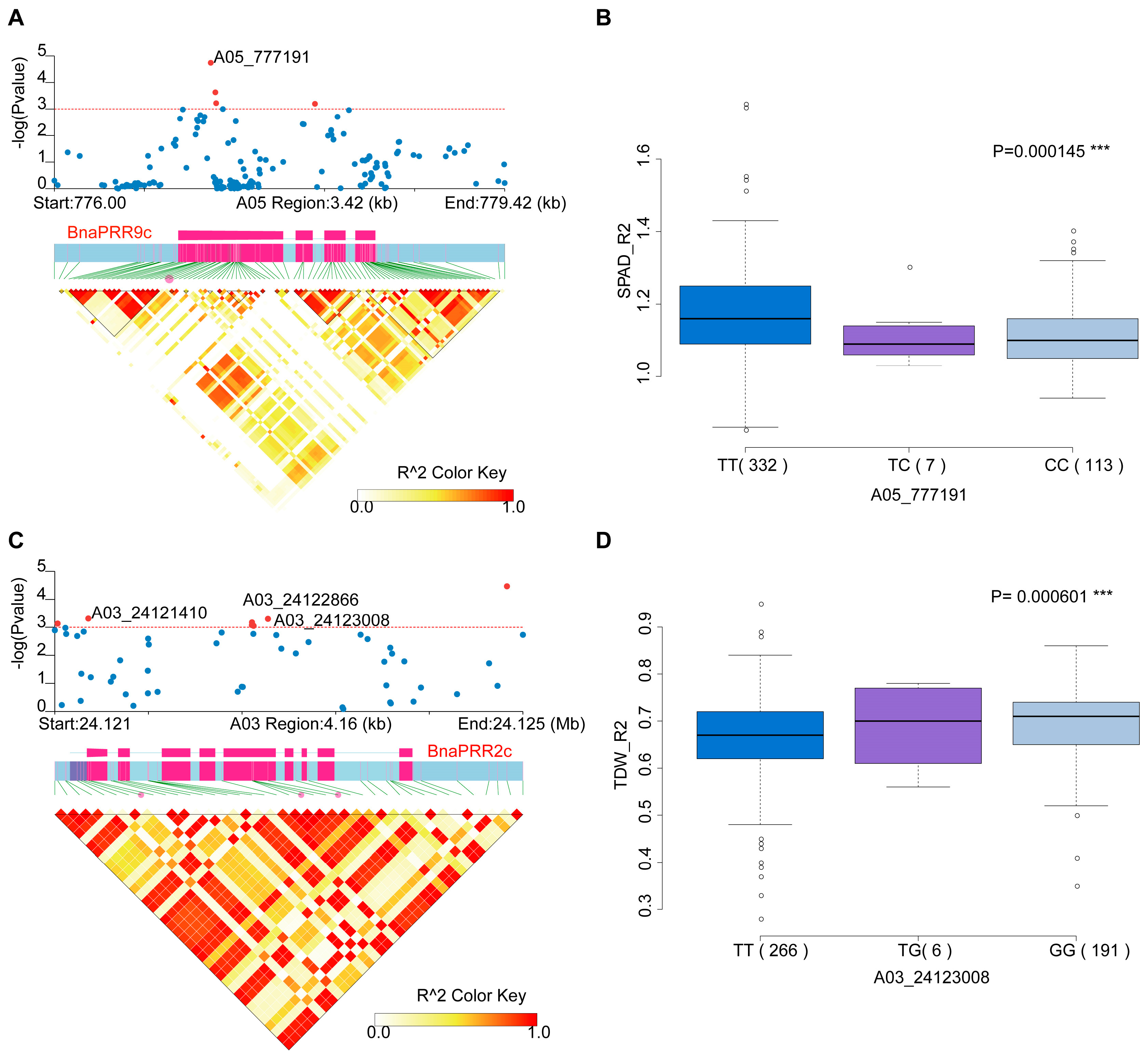
2.4. Natural Variations of the BnaTCS Genes
2.5. Cis-Elements Analysis of the BnaTCS Genes
2.6. Expression Analysis of BnaTCS Genes
2.6.1. Expression Profiles in Various B. napus Organs
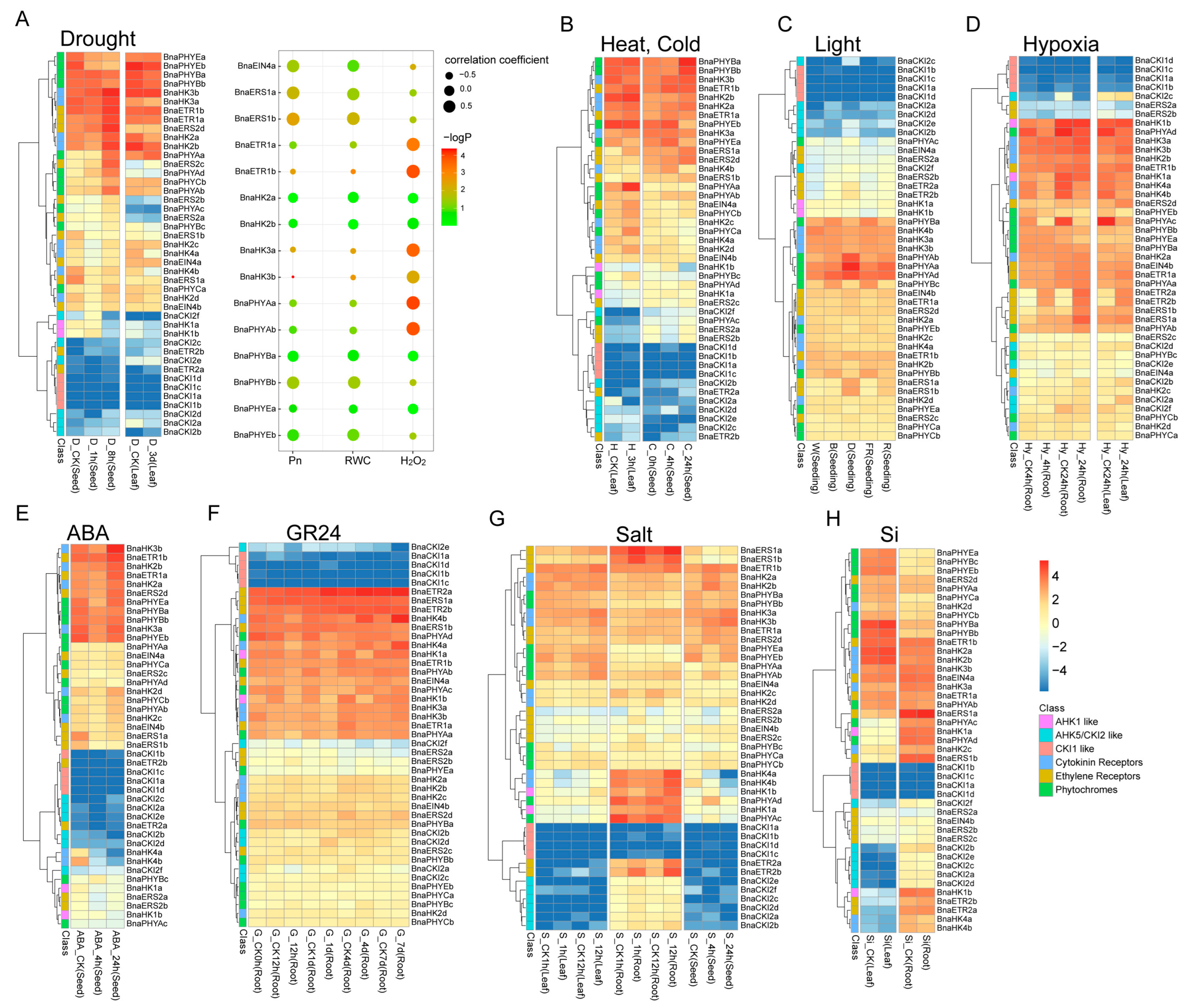
2.6.2. Expression of BnaTCS Genes in Response to Abiotic Stresses
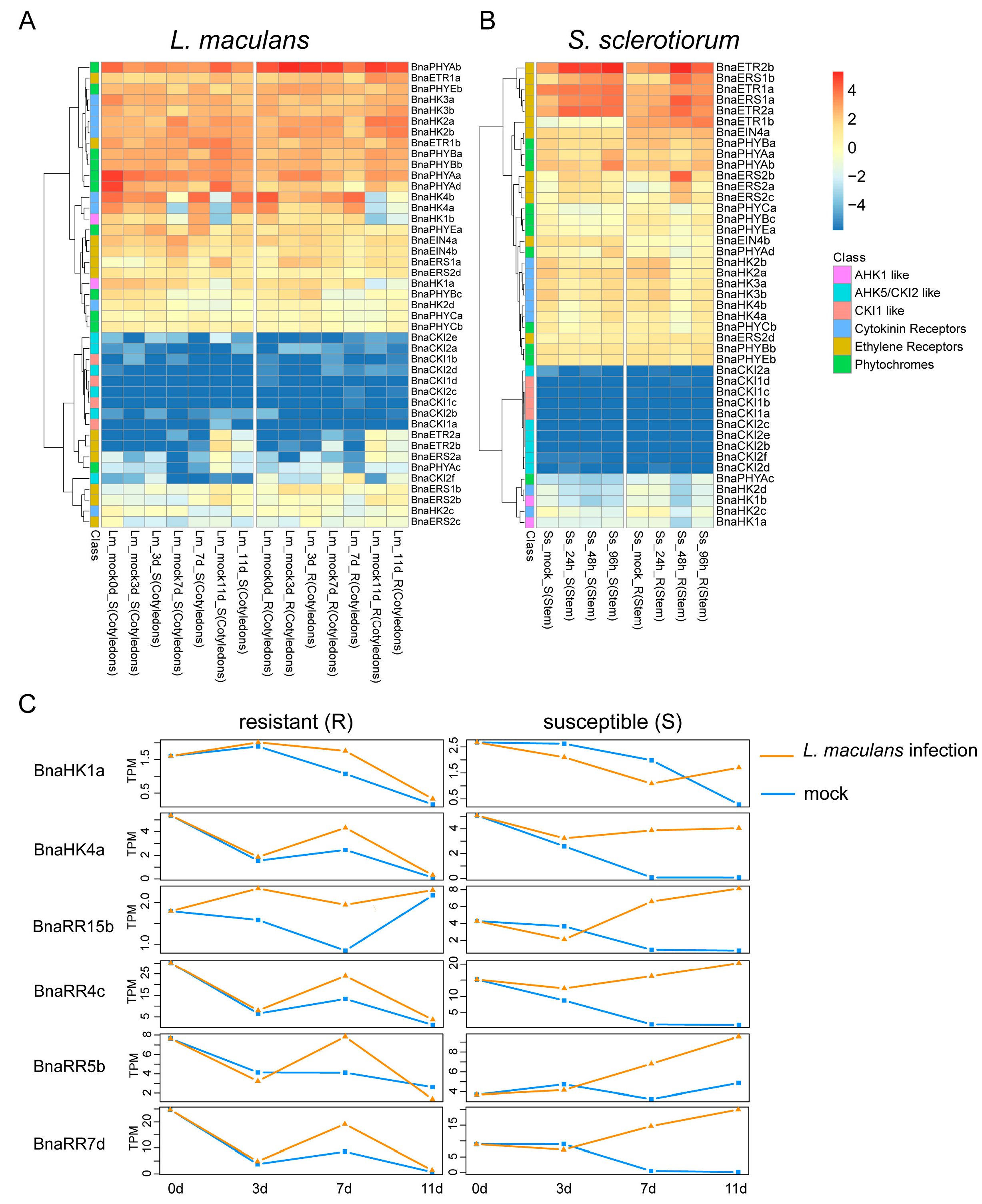
2.6.3. Expression of BnaTCS Genes in Response to Biotic Stresses
3. Discussion
4. Materials and Methods
4.1. Identification and Property Analysis of BnaTCS Genes
4.2. Phylogenetic Analysis, Genetic Structure, and Conserved Motifs Analysis
4.3. Chromosome Location, Gene Synteny, and Duplicate Analysis
4.4. Cis-Acting Regulatory Elements Analysis
4.5. Expression Analyses Based on RNA-seq Data
4.6. Nucleotide Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mizuno, T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997, 4, 161–168. [Google Scholar] [CrossRef]
- Schaller, G.E.; Kieber, J.J.; Shiu, S.-H. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arab. Book 2008, 6, e0112. [Google Scholar] [CrossRef] [PubMed]
- Kamberov, E.S.; Atkinson, M.R.; Chandran, P.; Ninfa, A.J. Effect of mutations in Escherichia coli glnL (ntrB), encoding nitrogen regulator II (NRII or NtrB), on the phosphatase activity involved in bacterial nitrogen regulation. J. Biol. Chem. 1994, 269, 28294–28299. [Google Scholar] [CrossRef] [PubMed]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef]
- Hwang, I. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002, 129, 500–515. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.E.; Kieber, J.J. Cytokinin signaling in Arabidopsis. Plant Cell 2002, 14, S47–S59. [Google Scholar] [CrossRef] [PubMed]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef]
- Hoang, Q.T.N.; Han, Y.J.; Kim, J.I. Plant phytochromes and their phosphorylation. Int. J. Mol. Sci. 2019, 20, 3450. [Google Scholar] [CrossRef]
- Imamura, A.; Hanaki, N.; Umeda, H.; Nakamura, A.; Suzuki, T.; Ueguchi, C.; Mizuno, T. Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 2691–2696. [Google Scholar] [CrossRef]
- Sakai, H.; Aoyama, T.; Oka, A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 2000, 24, 703–711. [Google Scholar] [CrossRef]
- Huo, R.; Liu, Z.; Yu, X.; Li, Z. The interaction network and signaling specificity of two-component system in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 4898. [Google Scholar] [CrossRef]
- Pareek, A.; Singh, A.; Kumar, M.; Kushwaha, H.R.; Lynn, A.M.; Singla-Pareek, S.L. Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 2006, 142, 380–397. [Google Scholar] [CrossRef] [PubMed]
- Mochida, K.; Yoshida, T.; Sakurai, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 2010, 17, 303–324. [Google Scholar] [CrossRef]
- Chu, Z.X.; Ma, Q.; Lin, Y.X.; Tang, X.L.; Zhou, Y.Q.; Zhu, S.W.; Fan, J.; Cheng, B.J. Genome-wide identification, classification, and analysis of two-component signal system genes in maize. GMR Genet. Mol. Res. 2011, 10, 3316–3330. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, M.; Kong, L.; Lv, Y.; Zou, M.; Lu, G.; Cao, J.; Yu, X. Genome-wide identification, phylogeny, duplication, and expression analyses of two-component system genes in Chinese cabbage (Brassica rapa ssp. pekinensis). DNA Res. 2014, 21, 379–396. [Google Scholar] [CrossRef] [PubMed]
- Gahlaut, V.; Mathur, S.; Dhariwal, R.; Khurana, J.P.; Tyagi, A.K.; Balyan, H.S.; Gupta, P.K. A multi-step phosphorelay two-component system impacts on tolerance against dehydration stress in common wheat. Funct. Integr. Genom. 2014, 14, 707–716. [Google Scholar] [CrossRef]
- Zameer, R.; Sadaqat, M.; Fatima, K.; Fiaz, S.; Rasul, S.; Zafar, H.; Qayyum, A.; Nashat, N.; Raza, A.; Shah, A.N.; et al. Two-Component System Genes in Sorghum bicolor: Genome-wide identification and expression profiling in response to environmental stresses. Front. Genet. 2021, 12, 794305. [Google Scholar] [CrossRef]
- Huo, R.; Zhao, Y.; Liu, T.; Xu, M.; Wang, X.; Xu, P.; Dai, S.; Cui, X.; Han, Y.; Liu, Z.; et al. Genome-wide identification and expression analysis of two-component system genes in sweet potato (Ipomoea batatas L.). Front. Plant Sci. 2023, 13, 1091620. [Google Scholar] [CrossRef]
- He, Y.; Liu, X.; Zou, T.; Pan, C.; Qin, L.; Chen, L.; Lu, G. Genome-wide identification of two-component system genes in Cucurbitaceae Crops and expression profiling analyses in Cucumber. Front. Plant Sci. 2016, 7, 899. [Google Scholar] [CrossRef]
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef]
- Kinoshita-Tsujimura, K.; Kakimoto, T. Cytokinin receptors in sporophytes are essential for male and female functions in Arabidopsis thaliana. Plant Signal. Behav. 2011, 6, 66–71. [Google Scholar] [CrossRef]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis Cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Bartrina, I.; Jensen, H.; Novák, O.; Strnad, M.; Werner, T.; Schmülling, T. Gain-of-function mutants of the cytokinin receptors AHK2 and AHK3 regulate plant organ size, flowering time and plant longevity. Plant Physiol. 2017, 173, 1783–1797. [Google Scholar] [CrossRef] [PubMed]
- Burr, C.A.; Sun, J.; Yamburenko, M.V.; Willoughby, A.; Hodgens, C.; Boeshore, S.L.; Elmore, A.; Atkinson, J.; Nimchuk, Z.L.; Bishopp, A.; et al. The HK5 and HK6 cytokinin receptors mediate diverse developmental pathways in rice. Development 2020, 147, dev191734. [Google Scholar] [CrossRef] [PubMed]
- Argyros, R.D.; Mathews, D.E.; Chiang, Y.H.; Palmer, C.M.; Thibault, D.M.; Etheridge, N.; Argyros, D.A.; Mason, M.G.; Kieber, J.J.; Schaller, G.E. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 2008, 20, 2102–2116. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Role of cytokinin responsive two-component system in ABA and osmotic stress signalings. Plant Signaling Behav. 2010, 5, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Kim, N.Y.; Kim, S.; Kang, N.Y.; Novák, O.; Ku, S.J.; Cho, C.; Lee, D.J.; Lee, E.J.; Strnad, M.; et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 2010, 285, 23371–23386. [Google Scholar] [CrossRef]
- Pham, J.; Liu, J.; Bennett, M.H.; Mansfield, J.W.; Desikan, R. Arabidopsis histidine kinase 5 regulates salt sensitivity and resistance against bacterial and fungal infection. New Phytol. 2012, 194, 168–180. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J. Arabidopsis response Regulator1 and Arabidopsis histidine phosphotransfer Protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol. 2013, 161, 408–424. [Google Scholar] [CrossRef]
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Watanabe, Y.; Leyva-Gonzalez, M.A.; Ha, C.V.; Fujita, Y.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Herrera-Estrella, L.; et al. Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. USA 2013, 110, 4840–4845. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.V.; Mostofa, M.G.; Nguyen, K.H.; Tran, C.D.; Watanabe, Y.; Li, W.; Osakabe, Y.; Sato, M.; Toyooka, K.; Tanaka, M.; et al. The histidine phosphotransfer AHP4 plays a negative role in Arabidopsis plant response to drought. Plant J. 2022, 111, 1732–1752. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.H.; Ha, C.V.; Nishiyama, R.; Watanabe, Y.; Leyva-González, M.A.; Fujita, Y.; Tran, U.T.; Li, W.; Tanaka, M.; Seki, M.; et al. Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. USA 2016, 113, 3090–3095. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kusano, M.; Fukushima, A.; Kita, M.; Ito, S.; Yamashino, T.; Saito, K.; Sakakibara, H.; Mizuno, T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009, 50, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, Q.; Wu, J.; Zhang, L.; Jiao, X.; Zhang, S.; Zhang, Z.; Sun, D.; Lu, T.; Sun, Y. Two rice authentic histidine phosphotransfer proteins, OsAHP1 and OsAHP2, mediate cytokinin signaling and stress responses in rice. Plant Physiol. 2014, 165, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-C.; Lin, T.-C.; Kieber, J.; Tsai, Y.-C. Response regulators 9 and 10 negatively regulate salinity tolerance in rice. Plant Cell Physiol. 2019, 60, 2549–2563. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, A.; Paul, L.K.; Sharma, E.; Jha, S.; Jain, M.; Khurana, J.P. OsRR6, a Type-A response regulator in rice, mediates cytokinin, light and stress responses when over-expressed in Arabidopsis. Plant Physiol. Biochem. 2021, 161, 98–112. [Google Scholar] [CrossRef]
- Zhao, L.; Guo, L.; Lu, X.; Malik, W.A.; Zhang, Y.; Wang, J.; Chen, X.; Wang, S.; Wang, J.; Wang, D.; et al. Structure and character analysis of cotton response regulator genes family reveals that GhRR7 responses to draught stress. Biol. Res. 2022, 55, 27. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hall, A.E.; O’Malley, R.; Bleecker, A.B. Canonical histidine kinase activity of the transmitter fomain of the ETR1 Ethylene Receptor from Arabidopsis is not required for signal transmission. Proc. Natl. Acad. Sci. USA 2003, 100, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Binder, B.M.; O’Malley, R.C.; Wang, W.; Moore, J.M.; Parks, B.M.; Spalding, E.P.; Bleecker, A.B. Arabidopsis seedling growth response and recovery to ethylene. A kinetic analysis. Plant Physiol. 2004, 136, 2913–2920. [Google Scholar] [CrossRef] [PubMed]
- Nagatani, A. Phytochrome: Structural basis for its functions. Curr. Opin. Plant Biol. 2010, 13, 565–570. [Google Scholar] [CrossRef]
- Xiong, A.; Jayaswal, R.K. Molecular characterization of a chromosomal determinant conferring resistance to zinc and cobalt ions in Staphylococcus aureus. J. Bacteriol. 1998, 180, 4024–4029. [Google Scholar] [CrossRef]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Zhang, G.; Zhou, J.; Peng, Y.; Tan, Z.; Li, L.; Yu, L.; Jin, C.; Fang, S.; Lu, S.; Guo, L.; et al. Genome-Wide association studies of salt tolerance at seed germination and seedling stages in Brassica napus. Front. Plant Sci. 2022, 12, 772708. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, S.; Wei, L.; Huang, Y.; Liu, D.; Jia, Y.; Luo, C.; Lin, Y.; Liang, C.; Hu, Y.; et al. BnIR: A multi-omics database with various tools for Brassica napus research and breeding. Mol. Plant 2023, 16, 775–789. [Google Scholar] [CrossRef]
- Deng, Y.; Dong, H.; Mu, J.; Ren, B.; Zheng, B.; Ji, Z.; Yang, W.-C.; Liang, Y.; Zuo, J. Arabidopsis Histidine Kinase CKI1 Acts Upstream of HISTIDINE PHOSPHOTRANSFER PROTEINS to Regulate Female Gametophyte Development and Vegetative Growth. Plant Cell 2010, 22, 1232–1248. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.-J.; Wang, D.; Zhu, L.-H.; Fu, B.-Y.; Li, Z.-K. Differential expressions of two-component element genes in rice under drought stress. Acta Agron. Sin. 2009, 35, 1628–1636. [Google Scholar] [CrossRef]
- Kang, N.Y.; Cho, C.; Kim, N.Y.; Kim, J. Cytokinin receptor-dependent and receptor-independent pathways in the dehydration response of Arabidopsis Thaliana. J. Plant Physiol. 2012, 169, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, J.; Deng, L.; Liu, H.; Liu, H.; Zhao, W.; Zhao, Y.; Sun, X.; Fan, S.; Wang, H.; et al. An intrinsically disordered region-containing protein mitigates the drought-growth trade-off to boost yields. Plant Physiol. 2023, 192, 274–292. [Google Scholar] [CrossRef]
- Zhang, X.; Fang, T.; Huang, Y.; Sun, W.; Cai, S. Transcriptional regulation of photomorphogenesis in seedlings of Brassica Napus under different light qualities. Planta 2022, 256, 77. [Google Scholar] [CrossRef]
- Ambros, S.; Kotewitsch, M.; Wittig, P.R.; Bammer, B.; Mustroph, A. Transcriptional response of two Brassica napus cultivars to short-term hypoxia in the root zone. Front. Plant Sci. 2022, 13, 897673. [Google Scholar] [CrossRef]
- Guntzer, F.; Keller, C.; Meunier, J.-D. Benefits of plant silicon for crops: A review. Agron. Sustain. Dev. 2012, 32, 201–213. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.-J.; Bressan, R.A.; Song, C.-P.; Zhu, J.-K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Trasoletti, M.; Visentin, I.; Campo, E.; Schubert, A.; Cardinale, F. Strigolactones as a hormonal hub for the acclimation and priming to environmental stress in plants. Plant Cell Environ. 2022, 45, 3611–3630. [Google Scholar] [CrossRef]
- Becker, M.G.; Zhang, X.; Walker, P.L.; Wan, J.C.; Millar, J.L.; Khan, D.; Granger, M.J.; Cavers, J.D.; Chan, A.C.; Fernando, D.W.G.; et al. Transcriptome analysis of the Brassica napus–Leptosphaeria maculans pathosystem identifies receptor, signaling and structural genes underlying plant resistance. Plant J. 2017, 90, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Girard, I.J.; Tong, C.; Becker, M.G.; Mao, X.; Huang, J.; de Kievit, T.; Fernando, W.G.D.; Liu, S.; Belmonte, M.F. RNA sequencing of Brassica napus reveals cellular redox control of Sclerotinia Infection. J. Exp. Bot. 2017, 68, 5079–5091. [Google Scholar] [CrossRef] [PubMed]
- Tadege, M.; Sheldon, C.C.; Helliwell, C.A.; Stoutjesdijk, P.; Dennis, E.S.; Peacock, W.J. Control of flowering time by FLC orthologues in Brassica napus. Plant J. 2001, 28, 545–553. [Google Scholar] [CrossRef]
- Tan, Z.; Xie, Z.; Dai, L.; Zhang, Y.; Zhao, H.; Tang, S.; Wan, L.; Yao, X.; Guo, L.; Hong, D. Genome- and transcriptome-wide association studies reveal the genetic basis and the breeding history of seed glucosinolate content in Brassica napus. Plant Biotechnol. J. 2022, 20, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Song, J.M.; Guan, Z.; Hu, J.; Guo, C.; Yang, Z.; Wang, S.; Liu, D.; Wang, B.; Lu, S.; Zhou, R.; et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chang, L.; Zhang, T.; Chen, H.; Zhang, L.; Lin, R.; Liang, J.; Wu, J.; Freeling, M.; Wang, X. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021, 22, 166. [Google Scholar] [CrossRef]
- Guo, N.; Liu, F. Genome assemblies and annotations of cauliflower (Brassica Oleracea Var. Botrytis Cv. Korso) and Cabbage (Brassica Oleracea Var. Capitata Cv. OX-Heart). 2021. Available online: https://figshare.com/collections/Korso_and_OX_heart_genome_assemblies_and_annotations/5392466/2 (accessed on 2 January 2022). [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the expasy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Haddad, C.; Trouverie, J.; Arkoun, M.; Yvin, J.-C.; Caïus, J.; Brunaud, V.; Laîné, P.; Etienne, P. Silicon supply affects the root transcriptome of Brassica napus L. Planta 2019, 249, 1645–1651. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.; Li, T.; Luo, C.; Huang, H.; Ruan, Y.; Li, X.; Niu, Y.; Fan, Y.; Sun, W.; Zhang, K.; et al. BrassicaEDB: A gene expression database for Brassica Crops. Int. J. Mol. Sci. 2020, 21, 5831. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Wan, L.; Zhao, W.; Liu, H.; Li, J.; Zhang, C. Exogenous strigolactones promote lateral root growth by reducing the endogenous auxin level in rapeseed. J. Integr. Agric. 2020, 19, 465–482. [Google Scholar] [CrossRef]
- Zhang, Y.; Ali, U.; Zhang, G.; Yu, L.; Fang, S.; Iqbal, S.; Li, H.; Lu, S.; Guo, L. Transcriptome analysis reveals genes commonly responding to multiple abiotic stresses in rapeseed. Mol. Breed. 2019, 39, 158. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, J.; Sun, X.; Kuang, C.; Liu, H.; Zhang, L.; Zheng, Q.; Liu, J.; Li, J.; Wang, H.; et al. Stress-induced higher vein density in the C3–C4 intermediate Moricandia suffruticosa under Drought and Heat Stress. J. Exp. Bot. 2022, 73, 6334–6351. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Niwa, Y.; Yamashino, T.; Mizuno, T. A Genome-Wide compilation of the two-component systems in Lotus Japonicus. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2009, 16, 237–247. [Google Scholar] [CrossRef]
- Ishida, K.; Yamashino, T.; Nakanishi, H.; Mizuno, T. Classification of the genes involved in the two-component system of the moss Physcomitrella Patens. Biosci. Biotechnol. Biochem. 2010, 74, 2542–2545. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, X.; Ye, L.; Pan, C.; Chen, L.; Zou, T.; Lu, G. Genome-wide identification and expression analysis of two-component system genes in tomato. Int. J. Mol. Sci. 2016, 17, 1204. [Google Scholar] [CrossRef]
- Ahmad, B.; Azeem, F.; Ali, M.A.; Nawaz, M.A.; Nadeem, H.; Abbas, A.; Batool, R.; Atif, R.M.; Ijaz, U.; Nieves-Cordones, M.; et al. Genome-wide identification and expression analysis of two component system genes in Cicer arietinum. Genomics 2020, 112, 1371–1383. [Google Scholar] [CrossRef]
- Liu, P.; Wang, S.; Wang, X.; Yang, X.; Li, Q.; Wang, C.; Chen, C.; Shi, Q.; Ren, Z.; Wang, L. Genome-wide characterization of two-component system (TCS) genes in melon (Cucumis melo L.). Plant Physiol. Biochem. 2020, 151, 197–213. [Google Scholar] [CrossRef]
- He, L.; Zhang, F.; Wu, X.; Hu, Y.; Dong, L.; Dewitte, W.; Wen, B. Genome-wide characterization and expression of two-component system genes in cytokinin-regulated gall formation in Zizania latifolia. Plants 2020, 9, 1409. [Google Scholar] [CrossRef]

| Gene Family 1 | Gene Pairs | Number of Genes Involved in Gene Pairs | Percentages (%) |
|---|---|---|---|
| AHK(L)/BraHK(L) | 17 | 14/17 | 87.5/81.0 |
| AHK(L)/BolHK(L) | 19 | 14/19 | 87.5/86.4 |
| BraHK(L)/BnaAHK(L) | 29 | 20/21 | 95.2/100 |
| BolHK(L)/BnaCHK(L) | 34 | 21/21 | 95.5/95.5 |
| AHP/BraHP | 6 | 4/6 | 66.7/75.0 |
| AHP/BolHP | 6 | 4/6 | 66.7/75.0 |
| BraHP/BnaAHP | 15 | 8/8 | 100/100 |
| BolHP/BnaCHP | 15 | 8/8 | 100/100 |
| ARR/BraRR | 67 | 28/54 | 84.8/91.5 |
| ARR/BolRR | 66 | 28/55 | 84.8/84.6 |
| BraRR/BnaARR | 158 | 53/54 | 98.8/93.1 |
| BolRR/BnaCRR | 168 | 62/62 | 95.4/95.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Liu, N.; Peng, C.; Huang, J.; Hua, W.; Fu, Z.; Liu, J. Two-Component System Genes in Brassica napus: Identification, Analysis, and Expression Patterns in Response to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2023, 24, 17308. https://doi.org/10.3390/ijms242417308
Liu H, Liu N, Peng C, Huang J, Hua W, Fu Z, Liu J. Two-Component System Genes in Brassica napus: Identification, Analysis, and Expression Patterns in Response to Abiotic and Biotic Stresses. International Journal of Molecular Sciences. 2023; 24(24):17308. https://doi.org/10.3390/ijms242417308
Chicago/Turabian StyleLiu, Hongfang, Nian Liu, Chen Peng, Jiaquan Huang, Wei Hua, Zhengwei Fu, and Jing Liu. 2023. "Two-Component System Genes in Brassica napus: Identification, Analysis, and Expression Patterns in Response to Abiotic and Biotic Stresses" International Journal of Molecular Sciences 24, no. 24: 17308. https://doi.org/10.3390/ijms242417308
APA StyleLiu, H., Liu, N., Peng, C., Huang, J., Hua, W., Fu, Z., & Liu, J. (2023). Two-Component System Genes in Brassica napus: Identification, Analysis, and Expression Patterns in Response to Abiotic and Biotic Stresses. International Journal of Molecular Sciences, 24(24), 17308. https://doi.org/10.3390/ijms242417308







