Oncolytic Viruses in the Era of Omics, Computational Technologies, and Modeling: Thesis, Antithesis, and Synthesis
Abstract
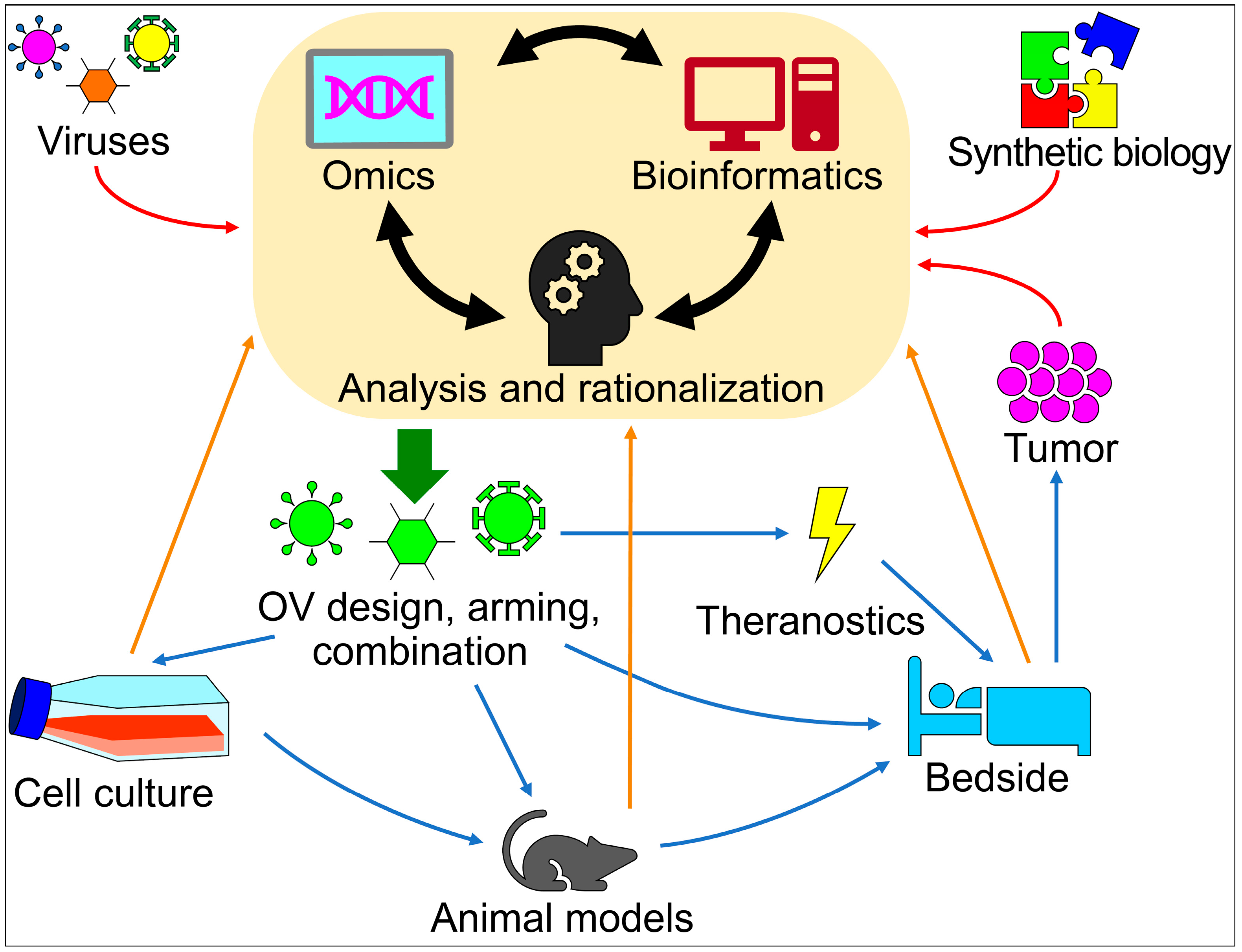
:1. Introduction
2. Mathematical and Computational Models
3. Systems and Synthetic Biology for OV Therapies
4. Next-Generation Combination Therapies to Improve OV Efficacy
5. Genomics, Proteomics, and Computational Tools for Oncolytic Immunovirotherapy
6. Large-Scale Assays for Oncolytic Immunovirotherapy
7. OVs as Theranostic Agents
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fukuhara, H.; Ino, Y.; Todo, T. Oncolytic virus therapy: A new era of cancer treatment at dawn. Cancer Sci. 2016, 107, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.J.; Peng, K.W.; Bell, J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012, 30, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Martuza, R.L.; Malick, A.; Markert, J.M.; Ruffner, K.L.; Coen, D.M. Experimental therapy of human glioma by means of a genetically engineered virus mutant. Science 1991, 252, 854–856. [Google Scholar] [CrossRef] [PubMed]
- Andreansky, S.S.; He, B.; Gillespie, G.Y.; Soroceanu, L.; Markert, J.; Chou, J.; Roizman, B.; Whitley, R.J. The application of genetically engineered herpes simplex viruses to the treatment of experimental brain tumors. Proc. Natl. Acad. Sci. USA 1996, 93, 11313–11318. [Google Scholar] [CrossRef] [PubMed]
- Andtbacka, R.H.; Kaufman, H.L.; Collichio, F.; Amatruda, T.; Senzer, N.; Chesney, J.; Delman, K.A.; Spitler, L.E.; Puzanov, I.; Agarwala, S.S.; et al. Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma. J. Clin. Oncol. 2015, 33, 2780–2788. [Google Scholar] [CrossRef]
- Chen, N.G.; Szalay, A.A.; Buller, R.M.; Lauer, U.M. Oncolytic viruses. Adv. Virol. 2012, 2012, 320206. [Google Scholar] [CrossRef]
- Feola, S.; Russo, S.; Ylosmaki, E.; Cerullo, V. Oncolytic ImmunoViroTherapy: A long history of crosstalk between viruses and immune system for cancer treatment. Pharmacol. Ther. 2022, 236, 108103. [Google Scholar] [CrossRef]
- Ma, R.; Li, Z.; Chiocca, E.A.; Caligiuri, M.A.; Yu, J. The emerging field of oncolytic virus-based cancer immunotherapy. Trends Cancer 2023, 9, 122–139. [Google Scholar] [CrossRef]
- Macedo, N.; Miller, D.M.; Haq, R.; Kaufman, H.L. Clinical landscape of oncolytic virus research in 2020. J. Immunother. Cancer 2020, 8, e001486. [Google Scholar] [CrossRef]
- Lawler, S.E.; Speranza, M.C.; Cho, C.F.; Chiocca, E.A. Oncolytic Viruses in Cancer Treatment: A Review. JAMA Oncol. 2017, 3, 841–849. [Google Scholar] [CrossRef]
- Carpenter, A.B.; Carpenter, A.M.; Aiken, R.; Hanft, S. Oncolytic virus in gliomas: A review of human clinical investigations. Ann. Oncol. 2021, 32, 968–982. [Google Scholar] [CrossRef] [PubMed]
- Ghajar-Rahimi, G.; Kang, K.D.; Totsch, S.K.; Gary, S.; Rocco, A.; Blitz, S.; Kachurak, K.; Chambers, M.R.; Li, R.; Beierle, E.A.; et al. Clinical advances in oncolytic virotherapy for pediatric brain tumors. Pharmacol. Ther. 2022, 239, 108193. [Google Scholar] [CrossRef] [PubMed]
- Engeland, C.E.; Ungerechts, G. Measles Virus as an Oncolytic Immunotherapy. Cancers 2021, 13, 544. [Google Scholar] [CrossRef]
- Filley, A.C.; Dey, M. Immune System, Friend or Foe of Oncolytic Virotherapy? Front. Oncol. 2017, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Menotti, L.; Avitabile, E. Herpes Simplex Virus Oncolytic Immunovirotherapy: The Blossoming Branch of Multimodal Therapy. Int. J. Mol. Sci. 2020, 21, 8310. [Google Scholar] [CrossRef]
- Coffin, R.S. From virotherapy to oncolytic immunotherapy: Where are we now? Curr. Opin. Virol. 2015, 13, 93–100. [Google Scholar] [CrossRef]
- Coffin, R. Interview with Robert Coffin, inventor of T-VEC: The first oncolytic immunotherapy approved for the treatment of cancer. Immunotherapy 2016, 8, 103–106. [Google Scholar] [CrossRef]
- Liu, B.L.; Robinson, M.; Han, Z.Q.; Branston, R.H.; English, C.; Reay, P.; McGrath, Y.; Thomas, S.K.; Thornton, M.; Bullock, P.; et al. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003, 10, 292–303. [Google Scholar] [CrossRef]
- Hu, J.C.; Coffin, R.S.; Davis, C.J.; Graham, N.J.; Groves, N.; Guest, P.J.; Harrington, K.J.; James, N.D.; Love, C.A.; McNeish, I.; et al. A phase I study of OncoVEXGM-CSF, a second-generation oncolytic herpes simplex virus expressing granulocyte macrophage colony-stimulating factor. Clin. Cancer Res. 2006, 12, 6737–6747. [Google Scholar] [CrossRef]
- Senzer, N.N.; Kaufman, H.L.; Amatruda, T.; Nemunaitis, M.; Reid, T.; Daniels, G.; Gonzalez, R.; Glaspy, J.; Whitman, E.; Harrington, K.; et al. Phase II clinical trial of a granulocyte-macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J. Clin. Oncol. 2009, 27, 5763–5771. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Bines, S.D. OPTIM trial: A Phase III trial of an oncolytic herpes virus encoding GM-CSF for unresectable stage III or IV melanoma. Future Oncol. 2010, 6, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, H.L.; Kim, D.W.; DeRaffele, G.; Mitcham, J.; Coffin, R.S.; Kim-Schulze, S. Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GM-CSF in patients with stage IIIc and IV melanoma. Ann. Surg. Oncol. 2010, 17, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed]
- Monie, D.D.; Bhandarkar, A.R.; Parney, I.F.; Correia, C.; Sarkaria, J.N.; Vile, R.G.; Li, H. Synthetic and systems biology principles in the design of programmable oncolytic virus immunotherapies for glioblastoma. Neurosurg. Focus 2021, 50, E10. [Google Scholar] [CrossRef]
- Irmisch, A.; Bonilla, X.; Chevrier, S.; Lehmann, K.V.; Singer, F.; Toussaint, N.C.; Esposito, C.; Mena, J.; Milani, E.S.; Casanova, R.; et al. The Tumor Profiler Study: Integrated, multi-omic, functional tumor profiling for clinical decision support. Cancer Cell 2021, 39, 288–293. [Google Scholar] [CrossRef]
- Crippa, V.; Malighetti, F.; Villa, M.; Graudenzi, A.; Piazza, R.; Mologni, L.; Ramazzotti, D. Characterization of cancer subtypes associated with clinical outcomes by multi-omics integrative clustering. Comput. Biol. Med. 2023, 162, 107064. [Google Scholar] [CrossRef]
- Berg, D.R.; Offord, C.P.; Kemler, I.; Ennis, M.K.; Chang, L.; Paulik, G.; Bajzer, Z.; Neuhauser, C.; Dingli, D. In vitro and in silico multidimensional modeling of oncolytic tumor virotherapy dynamics. PLoS Comput. Biol. 2019, 15, e1006773. [Google Scholar] [CrossRef]
- Bhatt, D.K.; Janzen, T.; Daemen, T.; Weissing, F.J. Modelling the spatial dynamics of oncolytic virotherapy in the presence of virus-resistant tumour cells. PLoS Comput. Biol. 2022, 18, e1010076. [Google Scholar] [CrossRef]
- Almuallem, N.; Trucu, D.; Eftimie, R. Oncolytic viral therapies and the delicate balance between virus-macrophage-tumour interactions: A mathematical approach. Math. Biosci. Eng. 2020, 18, 764–799. [Google Scholar] [CrossRef]
- Eftimie, R.; Eftimie, G. Investigating Macrophages Plasticity Following Tumour-Immune Interactions During Oncolytic Therapies. Acta Biotheor. 2019, 67, 321–359. [Google Scholar] [CrossRef]
- Jenner, A.L.; Frascoli, F.; Coster, A.C.F.; Kim, P.S. Enhancing oncolytic virotherapy: Observations from a Voronoi Cell-Based model. J. Theor. Biol. 2020, 485, 110052. [Google Scholar] [CrossRef] [PubMed]
- Lipatova, A.V.; Soboleva, A.V.; Gorshkov, V.A.; Bubis, J.A.; Solovyeva, E.M.; Krasnov, G.S.; Kochetkov, D.V.; Vorobyev, P.O.; Ilina, I.Y.; Moshkovskii, S.A.; et al. Multi-Omics Analysis of Glioblastoma Cells’ Sensitivity to Oncolytic Viruses. Cancers 2021, 13, 5268. [Google Scholar] [CrossRef] [PubMed]
- Moerdyk-Schauwecker, M.; Shah, N.R.; Murphy, A.M.; Hastie, E.; Mukherjee, P.; Grdzelishvili, V.Z. Resistance of pancreatic cancer cells to oncolytic vesicular stomatitis virus: Role of type I interferon signaling. Virology 2013, 436, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.; Zhao, W.; Jiang, S.; Huang, Y.; Hou, J.; Zhang, X.; Zhai, Z.; Yang, C.; Wang, J.; et al. Integrative multi-omics and drug-response characterization of patient-derived prostate cancer primary cells. Signal Transduct. Target. Ther. 2023, 8, 175. [Google Scholar] [CrossRef] [PubMed]
- Paglino, J.C.; van den Pol, A.N. Vesicular stomatitis virus has extensive oncolytic activity against human sarcomas: Rare resistance is overcome by blocking interferon pathways. J. Virol. 2011, 85, 9346–9358. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, D.K.; Chammas, R.; Daemen, T. Resistance Mechanisms Influencing Oncolytic Virotherapy, a Systematic Analysis. Vaccines 2021, 9, 1166. [Google Scholar] [CrossRef] [PubMed]
- van Asten, S.D.; Raaben, M.; Nota, B.; Spaapen, R.M. Secretome Screening Reveals Fibroblast Growth Factors as Novel Inhibitors of Viral Replication. J. Virol. 2018, 92, e00260-18. [Google Scholar] [CrossRef]
- Song, T.J.; Haddad, D.; Adusumilli, P.; Kim, T.; Stiles, B.; Hezel, M.; Socci, N.D.; Gonen, M.; Fong, Y. Molecular network pathways and functional analysis of tumor signatures associated with development of resistance to viral gene therapy. Cancer Gene Ther. 2012, 19, 38–48. [Google Scholar] [CrossRef]
- Noll, M.; Berchtold, S.; Lampe, J.; Malek, N.P.; Bitzer, M.; Lauer, U.M. Primary resistance phenomena to oncolytic measles vaccine viruses. Int. J. Oncol. 2013, 43, 103–112. [Google Scholar] [CrossRef]
- Mazzacurati, L.; Marzulli, M.; Reinhart, B.; Miyagawa, Y.; Uchida, H.; Goins, W.F.; Li, A.; Kaur, B.; Caligiuri, M.; Cripe, T.; et al. Use of miRNA response sequences to block off-target replication and increase the safety of an unattenuated, glioblastoma-targeted oncolytic HSV. Mol. Ther. 2015, 23, 99–107. [Google Scholar] [CrossRef]
- Ling, A.L.; Solomon, I.H.; Landivar, A.M.; Nakashima, H.; Woods, J.K.; Santos, A.; Masud, N.; Fell, G.; Mo, X.; Yilmaz, A.S.; et al. Clinical trial links oncolytic immunoactivation to survival in glioblastoma. Nature 2023, 623, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Menotti, L.; Avitabile, E.; Gatta, V.; Malatesta, P.; Petrovic, B.; Campadelli-Fiume, G. HSV as A Platform for the Generation of Retargeted, Armed, and Reporter-Expressing Oncolytic Viruses. Viruses 2018, 10, 352. [Google Scholar] [CrossRef]
- Vannini, A.; Parenti, F.; Forghieri, C.; Barboni, C.; Zaghini, A.; Campadelli-Fiume, G.; Gianni, T. Innovative retargeted oncolytic herpesvirus against nectin4-positive cancers. Front. Mol. Biosci. 2023, 10, 1149973. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, Y.; Liu, X.; Li, Z.; Dai, X. The Application of CRISPR/Cas9 Technology for Cancer Immunotherapy: Current Status and Problems. Front. Oncol. 2021, 11, 704999. [Google Scholar] [CrossRef] [PubMed]
- Harsha, H.C.; Pandey, A. Phosphoproteomics in cancer. Mol. Oncol. 2010, 4, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Higgins, L.; Gerdes, H.; Cutillas, P.R. Principles of phosphoproteomics and applications in cancer research. Biochem. J. 2023, 480, 403–420. [Google Scholar] [CrossRef]
- Feola, S.; Chiaro, J.; Martins, B.; Russo, S.; Fusciello, M.; Ylosmaki, E.; Bonini, C.; Ruggiero, E.; Hamdan, F.; Feodoroff, M.; et al. A novel immunopeptidomic-based pipeline for the generation of personalized oncolytic cancer vaccines. eLife 2022, 11, e71156. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Haggstrom, I.; Szczypinski, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Mencattini, A.; Lansche, C.; Veith, I.; Erbs, P.; Balloul, J.M.; Quemeneur, E.; Descroix, S.; Mechta-Grigoriou, F.; Zalcman, G.; Zaupa, C.; et al. Direct imaging and automatic analysis in tumor-on-chip reveal cooperative antitumoral activity of immune cells and oncolytic vaccinia virus. Biosens. Bioelectron. 2022, 215, 114571. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Guz-Montgomery, K.; Saha, D. Oncolytic Virus Encoding a Master Pro-Inflammatory Cytokine Interleukin 12 in Cancer Immunotherapy. Cells 2020, 9, 400. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Wang, H.; Ren, J.; Liu, W.; Chen, L.; Chen, H.; Ye, J.; Dai, E.; Ma, C.; Ju, S.; et al. Oncolytic vaccinia virus delivering tethered IL-12 enhances antitumor effects with improved safety. J. Immunother. Cancer 2020, 8, e000710. [Google Scholar] [CrossRef] [PubMed]
- Markl, F.; Huynh, D.; Endres, S.; Kobold, S. Utilizing chemokines in cancer immunotherapy. Trends Cancer 2022, 8, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Hamid, O.; Ismail, R.; Puzanov, I. Intratumoral Immunotherapy-Update 2019. Oncologist 2020, 25, e423–e438. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, P.F.; Pala, L.; Conforti, F.; Cocorocchio, E. Talimogene Laherparepvec (T-VEC): An Intralesional Cancer Immunotherapy for Advanced Melanoma. Cancers 2021, 13, 1383. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Shalhout, S.Z.; Iodice, G. Talimogene Laherparepvec: Moving From First-In-Class to Best-In-Class. Front. Mol. Biosci. 2022, 9, 834841. [Google Scholar] [CrossRef] [PubMed]
- Tomita, Y.; Kurozumi, K.; Yoo, J.Y.; Fujii, K.; Ichikawa, T.; Matsumoto, Y.; Uneda, A.; Hattori, Y.; Shimizu, T.; Otani, Y.; et al. Oncolytic Herpes Virus Armed with Vasculostatin in Combination with Bevacizumab Abrogates Glioma Invasion via the CCN1 and AKT Signaling Pathways. Mol. Cancer Ther. 2019, 18, 1418–1429. [Google Scholar] [CrossRef]
- Ho, T.Y.; Mealiea, D.; Okamoto, L.; Stojdl, D.F.; McCart, J.A. Deletion of immunomodulatory genes as a novel approach to oncolytic vaccinia virus development. Mol. Ther. Oncolytics 2021, 22, 85–97. [Google Scholar] [CrossRef]
- Fukuhara, H.; Takeshima, Y.; Todo, T. Triple-mutated oncolytic herpes virus for treating both fast- and slow-growing tumors. Cancer Sci. 2021, 112, 3293–3301. [Google Scholar] [CrossRef]
- Russell, S.J.; Barber, G.N. Oncolytic Viruses as Antigen-Agnostic Cancer Vaccines. Cancer Cell 2018, 33, 599–605. [Google Scholar] [CrossRef]
- Ramelyte, E.; Tastanova, A.; Balazs, Z.; Ignatova, D.; Turko, P.; Menzel, U.; Guenova, E.; Beisel, C.; Krauthammer, M.; Levesque, M.P.; et al. Oncolytic virotherapy-mediated anti-tumor response: A single-cell perspective. Cancer Cell 2021, 39, 394–406.e4. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.F.; Mardis, E.R. The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 2018, 15, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Monie, D.D.; Correia, C.; Zhang, C.; Ung, C.Y.; Vile, R.G.; Li, H. Modular network mechanism of CCN1-associated resistance to HSV-1-derived oncolytic immunovirotherapies for glioblastomas. Sci. Rep. 2021, 11, 11198. [Google Scholar] [CrossRef] [PubMed]
- Tripodi, L.; Sasso, E.; Feola, S.; Coluccino, L.; Vitale, M.; Leoni, G.; Szomolay, B.; Pastore, L.; Cerullo, V. Systems Biology Approaches for the Improvement of Oncolytic Virus-Based Immunotherapies. Cancers 2023, 15, 1297. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, Y.; Liao, W.; Cao, Y.; Liu, Q.; Guo, Y.; Lu, Y.; Xie, Z. Oncolytic adenovirus programmed by synthetic gene circuit for cancer immunotherapy. Nat. Commun. 2019, 10, 4801. [Google Scholar] [CrossRef] [PubMed]
- Azad, T.; Rezaei, R.; Singaravelu, R.; Pelin, A.; Boulton, S.; Petryk, J.; Onsu, K.A.; Martin, N.T.; Hoskin, V.; Ghahremani, M.; et al. Synthetic virology approaches to improve the safety and efficacy of oncolytic virus therapies. Nat. Commun. 2023, 14, 3035. [Google Scholar] [CrossRef] [PubMed]
- Haseley, A.; Boone, S.; Wojton, J.; Yu, L.; Yoo, J.Y.; Yu, J.; Kurozumi, K.; Glorioso, J.C.; Caligiuri, M.A.; Kaur, B. Extracellular matrix protein CCN1 limits oncolytic efficacy in glioma. Cancer Res. 2012, 72, 1353–1362. [Google Scholar] [CrossRef]
- Lorenzo-Sanz, L.; Munoz, P. Tumor-Infiltrating Immunosuppressive Cells in Cancer-Cell Plasticity, Tumor Progression and Therapy Response. Cancer Microenviron. 2019, 12, 119–132. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Romero, D. Immunotherapy: Oncolytic viruses prime antitumour immunity. Nat. Rev. Clin. Oncol. 2018, 15, 135. [Google Scholar] [CrossRef]
- Leber, M.F.; Neault, S.; Jirovec, E.; Barkley, R.; Said, A.; Bell, J.C.; Ungerechts, G. Engineering and combining oncolytic measles virus for cancer therapy. Cytokine Growth Factor Rev. 2020, 56, 39–48. [Google Scholar] [CrossRef]
- Tian, L.; Xu, B.; Teng, K.Y.; Song, M.; Zhu, Z.; Chen, Y.; Wang, J.; Zhang, J.; Feng, M.; Kaur, B.; et al. Targeting Fc Receptor-Mediated Effects and the “Don’t Eat Me” Signal with an Oncolytic Virus Expressing an Anti-CD47 Antibody to Treat Metastatic Ovarian Cancer. Clin. Cancer Res. 2022, 28, 201–214. [Google Scholar] [CrossRef]
- Senior, M. Checkpoint inhibitors go viral. Nat. Biotechnol. 2019, 37, 12–17. [Google Scholar] [CrossRef]
- Chesney, J.A.; Ribas, A.; Long, G.V.; Kirkwood, J.M.; Dummer, R.; Puzanov, I.; Hoeller, C.; Gajewski, T.F.; Gutzmer, R.; Rutkowski, P.; et al. Randomized, Double-Blind, Placebo-Controlled, Global Phase III Trial of Talimogene Laherparepvec Combined With Pembrolizumab for Advanced Melanoma. J. Clin. Oncol. 2023, 41, 528–540. [Google Scholar] [CrossRef]
- Patel, M.R.; Dash, A.; Jacobson, B.A.; Ji, Y.; Baumann, D.; Ismail, K.; Kratzke, R.A. JAK/STAT inhibition with ruxolitinib enhances oncolytic virotherapy in non-small cell lung cancer models. Cancer Gene Ther. 2019, 26, 411–418. [Google Scholar] [CrossRef]
- Selman, M.; Ou, P.; Rousso, C.; Bergeron, A.; Krishnan, R.; Pikor, L.; Chen, A.; Keller, B.A.; Ilkow, C.; Bell, J.C.; et al. Dimethyl fumarate potentiates oncolytic virotherapy through NF-kappaB inhibition. Sci. Transl. Med. 2018, 10, eaao1613. [Google Scholar] [CrossRef]
- Wakimoto, H.; Fulci, G.; Tyminski, E.; Chiocca, E.A. Altered expression of antiviral cytokine mRNAs associated with cyclophosphamide’s enhancement of viral oncolysis. Gene Ther. 2004, 11, 214–223. [Google Scholar] [CrossRef]
- Peng, K.W.; Myers, R.; Greenslade, A.; Mader, E.; Greiner, S.; Federspiel, M.J.; Dispenzieri, A.; Russell, S.J. Using clinically approved cyclophosphamide regimens to control the humoral immune response to oncolytic viruses. Gene Ther. 2013, 20, 255–261. [Google Scholar] [CrossRef]
- Koch, J.; Schober, S.J.; Hindupur, S.V.; Schoning, C.; Klein, F.G.; Mantwill, K.; Ehrenfeld, M.; Schillinger, U.; Hohnecker, T.; Qi, P.; et al. Targeting the Retinoblastoma/E2F repressive complex by CDK4/6 inhibitors amplifies oncolytic potency of an oncolytic adenovirus. Nat. Commun. 2022, 13, 4689. [Google Scholar] [CrossRef]
- Schober, S.J.; Schoening, C.; Eck, J.; Middendorf, C.; Lutsch, J.; Knoch, P.; von Ofen, A.J.; Gassmann, H.; Thiede, M.; Hauer, J.; et al. The Oncolytic Adenovirus XVir-N-31 Joins Forces with CDK4/6 Inhibition Augmenting Innate and Adaptive Antitumor Immunity in Ewing Sarcoma. Clin. Cancer Res. 2023, 29, 1996–2011. [Google Scholar] [CrossRef]
- Zheng, Y.; Stamminger, T.; Hearing, P. E2F/Rb Family Proteins Mediate Interferon Induced Repression of Adenovirus Immediate Early Transcription to Promote Persistent Viral Infection. PLoS Pathog. 2016, 12, e1005415. [Google Scholar] [CrossRef]
- Kuryk, L.; Haavisto, E.; Garofalo, M.; Capasso, C.; Hirvinen, M.; Pesonen, S.; Ranki, T.; Vassilev, L.; Cerullo, V. Synergistic anti-tumor efficacy of immunogenic adenovirus ONCOS-102 (Ad5/3-D24-GM-CSF) and standard of care chemotherapy in preclinical mesothelioma model. Int. J. Cancer 2016, 139, 1883–1893. [Google Scholar] [CrossRef]
- McKenna, M.K.; Englisch, A.; Brenner, B.; Smith, T.; Hoyos, V.; Suzuki, M.; Brenner, M.K. Mesenchymal stromal cell delivery of oncolytic immunotherapy improves CAR-T cell antitumor activity. Mol. Ther. 2021, 29, 1808–1820. [Google Scholar] [CrossRef]
- Evgin, L.; Kottke, T.; Tonne, J.; Thompson, J.; Huff, A.L.; van Vloten, J.; Moore, M.; Michael, J.; Driscoll, C.; Pulido, J.; et al. Oncolytic virus-mediated expansion of dual-specific CAR T cells improves efficacy against solid tumors in mice. Sci. Transl. Med. 2022, 14, eabn2231. [Google Scholar] [CrossRef]
- Chen, X.; Han, J.; Chu, J.; Zhang, L.; Zhang, J.; Chen, C.; Chen, L.; Wang, Y.; Wang, H.; Yi, L.; et al. A combinational therapy of EGFR-CAR NK cells and oncolytic herpes simplex virus 1 for breast cancer brain metastases. Oncotarget 2016, 7, 27764–27777. [Google Scholar] [CrossRef]
- Todo, T.; Ino, Y.; Ohtsu, H.; Shibahara, J.; Tanaka, M. A phase I/II study of triple-mutated oncolytic herpes virus G47∆ in patients with progressive glioblastoma. Nat. Commun. 2022, 13, 4119. [Google Scholar] [CrossRef]
- Todo, T.; Ito, H.; Ino, Y.; Ohtsu, H.; Ota, Y.; Shibahara, J.; Tanaka, M. Intratumoral oncolytic herpes virus G47∆ for residual or recurrent glioblastoma: A phase 2 trial. Nat. Med. 2022, 28, 1630–1639. [Google Scholar] [CrossRef]
- Yamada, T.; Tateishi, R.; Iwai, M.; Tanaka, M.; Ijichi, H.; Sano, M.; Koike, K.; Todo, T. Overcoming resistance of stroma-rich pancreatic cancer with focal adhesion kinase inhibitor combined with G47Delta and immune checkpoint inhibitors. Mol. Ther. Oncolytics 2023, 28, 31–43. [Google Scholar] [CrossRef]
- Rivera-Caraballo, K.A.; Nair, M.; Lee, T.J.; Kaur, B.; Yoo, J.Y. The complex relationship between integrins and oncolytic herpes Simplex Virus 1 in high-grade glioma therapeutics. Mol. Ther. Oncolytics 2022, 26, 63–75. [Google Scholar] [CrossRef]
- Wang, P.Y.; Cripe, T.P. Gene Editing Thumbs a Ride with Oncolytic Virotherapy. Mol. Ther. 2020, 28, 2103–2104. [Google Scholar] [CrossRef]
- Phelps, M.P.; Yang, H.; Patel, S.; Rahman, M.M.; McFadden, G.; Chen, E. Oncolytic Virus-Mediated RAS Targeting in Rhabdomyosarcoma. Mol. Ther. Oncolytics 2018, 11, 52–61. [Google Scholar] [CrossRef]
- Yoon, A.R.; Jung, B.K.; Choi, E.; Chung, E.; Hong, J.; Kim, J.S.; Koo, T.; Yun, C.O. CRISPR-Cas12a with an oAd Induces Precise and Cancer-Specific Genomic Reprogramming of EGFR and Efficient Tumor Regression. Mol. Ther. 2020, 28, 2286–2296. [Google Scholar] [CrossRef]
- D’Alise, A.M.; Leoni, G.; Cotugno, G.; Troise, F.; Langone, F.; Fichera, I.; De Lucia, M.; Avalle, L.; Vitale, R.; Leuzzi, A.; et al. Adenoviral vaccine targeting multiple neoantigens as strategy to eradicate large tumors combined with checkpoint blockade. Nat. Commun. 2019, 10, 2688. [Google Scholar] [CrossRef]
- Shapiro, I.E.; Bassani-Sternberg, M. The impact of immunopeptidomics: From basic research to clinical implementation. Semin. Immunol. 2023, 66, 101727. [Google Scholar] [CrossRef]
- Ouspenskaia, T.; Law, T.; Clauser, K.R.; Klaeger, S.; Sarkizova, S.; Aguet, F.; Li, B.; Christian, E.; Knisbacher, B.A.; Le, P.M.; et al. Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat. Biotechnol. 2022, 40, 209–217. [Google Scholar] [CrossRef]
- Chong, C.; Coukos, G.; Bassani-Sternberg, M. Identification of tumor antigens with immunopeptidomics. Nat. Biotechnol. 2022, 40, 175–188. [Google Scholar] [CrossRef]
- Rajaraman, S.; Canjuga, D.; Ghosh, M.; Codrea, M.C.; Sieger, R.; Wedekink, F.; Tatagiba, M.; Koch, M.; Lauer, U.M.; Nahnsen, S.; et al. Measles Virus-Based Treatments Trigger a Pro-inflammatory Cascade and a Distinctive Immunopeptidome in Glioblastoma. Mol. Ther. Oncolytics 2019, 12, 147–161. [Google Scholar] [CrossRef]
- Lathwal, A.; Kumar, R.; Raghava, G.P.S. Computer-aided designing of oncolytic viruses for overcoming translational challenges of cancer immunotherapy. Drug Discov. Today 2020, 25, 1198–1205. [Google Scholar] [CrossRef]
- Nhan, N.T.T.; Yamada, T.; Yamada, K.H. Peptide-Based Agents for Cancer Treatment: Current Applications and Future Directions. Int. J. Mol. Sci. 2023, 24, 12931. [Google Scholar] [CrossRef]
- Hilchie, A.L.; Hoskin, D.W.; Power Coombs, M.R. Anticancer Activities of Natural and Synthetic Peptides. Adv. Exp. Med. Biol. 2019, 1117, 131–147. [Google Scholar] [CrossRef]
- Feng, P.; Wang, Z. Recent Advances in Computational Methods for Identifying Anticancer Peptides. Curr. Drug Targets 2019, 20, 481–487. [Google Scholar] [CrossRef]
- Gautam, A.; Kapoor, P.; Chaudhary, K.; Kumar, R.; Open Source Drug Discovery Consortium; Raghava, G.P.S. Tumor homing peptides as molecular probes for cancer therapeutics, diagnostics and theranostics. Curr. Med. Chem. 2014, 21, 2367–2391. [Google Scholar] [CrossRef]
- Kumar, S.; Li, H. In Silico Design of Anticancer Peptides. Methods Mol. Biol. 2017, 1647, 245–254. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhagat, D.; Mahalwal, M.; Sharma, N.; Raghava, G.P.S. AntiCP 2.0: An updated model for predicting anticancer peptides. Brief. Bioinform. 2021, 22, bbaa153. [Google Scholar] [CrossRef]
- Schaduangrat, N.; Nantasenamat, C.; Prachayasittikul, V.; Shoombuatong, W. ACPred: A Computational Tool for the Prediction and Analysis of Anticancer Peptides. Molecules 2019, 24, 1973. [Google Scholar] [CrossRef]
- Rao, B.; Zhou, C.; Zhang, G.; Su, R.; Wei, L. ACPred-Fuse: Fusing multi-view information improves the prediction of anticancer peptides. Brief. Bioinform. 2020, 21, 1846–1855. [Google Scholar] [CrossRef]
- Wei, L.; Zhou, C.; Chen, H.; Song, J.; Su, R. ACPred-FL: A sequence-based predictor using effective feature representation to improve the prediction of anti-cancer peptides. Bioinformatics 2018, 34, 4007–4016. [Google Scholar] [CrossRef]
- Manavalan, B.; Basith, S.; Shin, T.H.; Choi, S.; Kim, M.O.; Lee, G. MLACP: Machine-learning-based prediction of anticancer peptides. Oncotarget 2017, 8, 77121–77136. [Google Scholar] [CrossRef]
- Thi Phan, L.; Woo Park, H.; Pitti, T.; Madhavan, T.; Jeon, Y.J.; Manavalan, B. MLACP 2.0: An updated machine learning tool for anticancer peptide prediction. Comput. Struct. Biotechnol. J. 2022, 20, 4473–4480. [Google Scholar] [CrossRef]
- Sharma, A.; Kapoor, P.; Gautam, A.; Chaudhary, K.; Kumar, R.; Chauhan, J.S.; Tyagi, A.; Raghava, G.P.S. Computational approach for designing tumor homing peptides. Sci. Rep. 2013, 3, 1607. [Google Scholar] [CrossRef]
- Romero, M.; Marrero-Ponce, Y.; Rodriguez, H.; Aguero-Chapin, G.; Antunes, A.; Aguilera-Mendoza, L.; Martinez-Rios, F. A Novel Network Science and Similarity-Searching-Based Approach for Discovering Potential Tumor-Homing Peptides from Antimicrobials. Antibiotics 2022, 11, 401. [Google Scholar] [CrossRef]
- Peng, M.; Mo, Y.; Wang, Y.; Wu, P.; Zhang, Y.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; Li, X.; et al. Neoantigen vaccine: An emerging tumor immunotherapy. Mol. Cancer 2019, 18, 128. [Google Scholar] [CrossRef]
- Nielsen, M.; Andreatta, M.; Peters, B.; Buus, S. Immunoinformatics: Predicting Peptide-MHC Binding. Annu. Rev. Biomed. Data Sci. 2020, 3, 191–215. [Google Scholar] [CrossRef]
- Roudko, V.; Greenbaum, B.; Bhardwaj, N. Computational Prediction and Validation of Tumor-Associated Neoantigens. Front. Immunol. 2020, 11, 27. [Google Scholar] [CrossRef]
- Lang, F.; Schrors, B.; Lower, M.; Tureci, O.; Sahin, U. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat. Rev. Drug Discov. 2022, 21, 261–282. [Google Scholar] [CrossRef]
- Xie, N.; Shen, G.; Gao, W.; Huang, Z.; Huang, C.; Fu, L. Neoantigens: Promising targets for cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 9. [Google Scholar] [CrossRef]
- Swift, S.L.; Stojdl, D.F. Big Data Offers Novel Insights for Oncolytic Virus Immunotherapy. Viruses 2016, 8, 45. [Google Scholar] [CrossRef]
- Wu, Q.; Hu, X.; Zhang, X.; Kong, D.; Yang, Z.; Li, G.; Gu, Z.; Zhang, Q.; Wan, D.; Cheng, S.; et al. Single-cell transcriptomics of peripheral blood reveals anti-tumor systemic immunity induced by oncolytic virotherapy. Theranostics 2022, 12, 7371–7389. [Google Scholar] [CrossRef]
- Herz, J.; Johnson, K.R.; McGavern, D.B. Therapeutic antiviral T cells noncytopathically clear persistently infected microglia after conversion into antigen-presenting cells. J. Exp. Med. 2015, 212, 1153–1169. [Google Scholar] [CrossRef]
- Best, J.A.; Blair, D.A.; Knell, J.; Yang, E.; Mayya, V.; Doedens, A.; Dustin, M.L.; Goldrath, A.W.; Immunological Genome Project Consortium. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat. Immunol. 2013, 14, 404–412. [Google Scholar] [CrossRef]
- Jha, A.K.; Huang, S.C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef]
- Hedberg, J.; Studebaker, A.; Smith, L.; Chen, C.Y.; Westfall, J.J.; Cam, M.; Gross, A.; Hernandez-Aguirre, I.; Martin, A.; Kim, D.; et al. Oncolytic virus-driven immune remodeling revealed in mouse medulloblastomas at single cell resolution. Mol. Ther. Oncolytics 2023, 30, 39–55. [Google Scholar] [CrossRef]
- Valiente, G. The Landscape of Virus-Host Protein-Protein Interaction Databases. Front. Microbiol. 2022, 13, 827742. [Google Scholar] [CrossRef]
- Saha, D.; Iannuccelli, M.; Brun, C.; Zanzoni, A.; Licata, L. The Intricacy of the Viral-Human Protein Interaction Networks: Resources, Data, and Analyses. Front. Microbiol. 2022, 13, 849781. [Google Scholar] [CrossRef]
- Mahoney, D.J.; Lefebvre, C.; Allan, K.; Brun, J.; Sanaei, C.A.; Baird, S.; Pearce, N.; Gronberg, S.; Wilson, B.; Prakesh, M.; et al. Virus-tumor interactome screen reveals ER stress response can reprogram resistant cancers for oncolytic virus-triggered caspase-2 cell death. Cancer Cell 2011, 20, 443–456. [Google Scholar] [CrossRef]
- Mahoney, D.J.; Stojdl, D.F. Functional genomic screening to enhance oncolytic virotherapy. Br. J. Cancer 2013, 108, 245–249. [Google Scholar] [CrossRef]
- Allan, K.J.; Mahoney, D.J.; Baird, S.D.; Lefebvre, C.A.; Stojdl, D.F. Genome-wide RNAi Screening to Identify Host Factors That Modulate Oncolytic Virus Therapy. J. Vis. Exp. 2018, 134, e56913. [Google Scholar] [CrossRef]
- Varble, A.; Benitez, A.A.; Schmid, S.; Sachs, D.; Shim, J.V.; Rodriguez-Barrueco, R.; Panis, M.; Crumiller, M.; Silva, J.M.; Sachidanandam, R.; et al. An in vivo RNAi screening approach to identify host determinants of virus replication. Cell Host Microbe 2013, 14, 346–356. [Google Scholar] [CrossRef]
- Al-Yahya, S.; Mahmoud, L.; Al-Zoghaibi, F.; Al-Tuhami, A.; Amer, H.; Almajhdi, F.N.; Polyak, S.J.; Khabar, K.S. Human Cytokinome Analysis for Interferon Response. J. Virol. 2015, 89, 7108–7119. [Google Scholar] [CrossRef]
- Muscolini, M.; Hiscott, J.; Tassone, E. A Genome-Wide CRISPR-Cas9 Loss-of-Function Screening to Identify Host Restriction Factors Modulating Oncolytic Virotherapy. Methods Mol. Biol. 2023, 2589, 379–399. [Google Scholar] [CrossRef]
- Liikanen, I.; Monsurro, V.; Ahtiainen, L.; Raki, M.; Hakkarainen, T.; Diaconu, I.; Escutenaire, S.; Hemminki, O.; Dias, J.D.; Cerullo, V.; et al. Induction of interferon pathways mediates in vivo resistance to oncolytic adenovirus. Mol. Ther. 2011, 19, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Diallo, J.S.; Le Boeuf, F.; Lai, F.; Cox, J.; Vaha-Koskela, M.; Abdelbary, H.; MacTavish, H.; Waite, K.; Falls, T.; Wang, J.; et al. A high-throughput pharmacoviral approach identifies novel oncolytic virus sensitizers. Mol. Ther. 2010, 18, 1123–1129. [Google Scholar] [CrossRef]
- Zloza, A.; Kim, D.W.; Kim-Schulze, S.; Jagoda, M.C.; Monsurro, V.; Marincola, F.M.; Kaufman, H.L. Immunoglobulin-like transcript 2 (ILT2) is a biomarker of therapeutic response to oncolytic immunotherapy with vaccinia viruses. J. Immunother. Cancer 2014, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- McGray, A.J.; Hallett, R.; Bernard, D.; Swift, S.L.; Zhu, Z.; Teoderascu, F.; Vanseggelen, H.; Hassell, J.A.; Hurwitz, A.A.; Wan, Y.; et al. Immunotherapy-induced CD8+ T cells instigate immune suppression in the tumor. Mol. Ther. 2014, 22, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Zamarin, D.; Holmgaard, R.B.; Ricca, J.; Plitt, T.; Palese, P.; Sharma, P.; Merghoub, T.; Wolchok, J.D.; Allison, J.P. Intratumoral modulation of the inducible co-stimulator ICOS by recombinant oncolytic virus promotes systemic anti-tumour immunity. Nat. Commun. 2017, 8, 14340. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kang, X.; Chen, K.S.; Jehng, T.; Jones, L.; Chen, J.; Huang, X.F.; Chen, S.Y. An engineered oncolytic virus expressing PD-L1 inhibitors activates tumor neoantigen-specific T cell responses. Nat. Commun. 2020, 11, 1395. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Wu, M.; Liang, M.; Xiong, S.; Dong, C. A novel oncolytic virus engineered with PD-L1 scFv effectively inhibits tumor growth in a mouse model. Cell. Mol. Immunol. 2019, 16, 780–782. [Google Scholar] [CrossRef] [PubMed]
- Ravirala, D.; Pei, G.; Zhao, Z.; Zhang, X. Single-cell RNA sequencing reveals a strong connection between Gadd45g upregulation and oncolytic HSV infection in tumor tissue. Mol. Ther. Oncolytics 2021, 23, 330–341. [Google Scholar] [CrossRef]
- Samouha, A.; Fogel, E.J.; Goel, S.; Maitra, R. Oncolytic Virus Affects the RAS Pathway in Cancer: RNA Sequence Analysis. J. Oncol. Res. Ther. 2021, 6, 10118. [Google Scholar] [CrossRef]
- Jiang, T.; Yang, T.; Chen, Y.; Miao, Y.; Xu, Y.; Jiang, H.; Yang, M.; Mao, C. Emulating interactions between microorganisms and tumor microenvironment to develop cancer theranostics. Theranostics 2022, 12, 2833–2859. [Google Scholar] [CrossRef]
- Veeranarayanan, S.; Azam, A.H.; Kiga, K.; Watanabe, S.; Cui, L. Bacteriophages as Solid Tumor Theragnostic Agents. Int. J. Mol. Sci. 2021, 23, 402. [Google Scholar] [CrossRef] [PubMed]
- Bortot, B.; Apollonio, M.; Baj, G.; Andolfi, L.; Zupin, L.; Crovella, S.; di Giosia, M.; Cantelli, A.; Saporetti, R.; Ulfo, L.; et al. Advanced photodynamic therapy with an engineered M13 phage targeting EGFR: Mitochondrial localization and autophagy induction in ovarian cancer cell lines. Free Radic. Biol. Med. 2022, 179, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Ulfo, L.; Cantelli, A.; Petrosino, A.; Costantini, P.E.; Nigro, M.; Starinieri, F.; Turrini, E.; Zadran, S.K.; Zuccheri, G.; Saporetti, R.; et al. Orthogonal nanoarchitectonics of M13 phage for receptor targeted anticancer photodynamic therapy. Nanoscale 2022, 14, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Ulfo, L.; Costantini, P.E.; Di Giosia, M.; Danielli, A.; Calvaresi, M. EGFR-Targeted Photodynamic Therapy. Pharmaceutics 2022, 14, 241. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, M.; Sabbaghi, A.; Miri, S.M.; Rezaeyan, A.; Arjeini, Y.; Ghaemi, A. Virotheranostics, a double-barreled viral gun pointed toward cancer; ready to shoot? Cancer Cell Int. 2020, 20, 131. [Google Scholar] [CrossRef] [PubMed]
- Semenova, A.V.; Sivolobova, G.F.; Grazhdantseva, A.A.; Agafonov, A.P.; Kochneva, G.V. Reporter Transgenes for Monitoring the Antitumor Efficacy of Recombinant Oncolytic Viruses. Acta Naturae 2022, 14, 46–56. [Google Scholar] [CrossRef]
- Godlewski, J.; Farhath, M.; Ricklefs, F.L.; Passaro, C.; Kiel, K.; Nakashima, H.; Chiocca, E.A.; Bronisz, A. Oncolytic Virus Therapy Alters the Secretome of Targeted Glioblastoma Cells. Cancers 2021, 13, 1287. [Google Scholar] [CrossRef]
- Rodrigues, M.; Fan, J.; Lyon, C.; Wan, M.; Hu, Y. Role of Extracellular Vesicles in Viral and Bacterial Infections: Pathogenesis, Diagnostics, and Therapeutics. Theranostics 2018, 8, 2709–2721. [Google Scholar] [CrossRef]
- Chaurasiya, S.; Kim, S.I.; O’Leary, M.; Park, A.K.; Lu, J.; Kang, S.; Zhang, Z.; Yang, A.; Woo, Y.; Fong, Y.; et al. Toward comprehensive imaging of oncolytic viroimmunotherapy. Mol. Ther. Oncolytics 2021, 23, 303–310. [Google Scholar] [CrossRef]
- Dingli, D.; Peng, K.W.; Harvey, M.E.; Greipp, P.R.; O’Connor, M.K.; Cattaneo, R.; Morris, J.C.; Russell, S.J. Image-guided radiovirotherapy for multiple myeloma using a recombinant measles virus expressing the thyroidal sodium iodide symporter. Blood 2004, 103, 1641–1646. [Google Scholar] [CrossRef]
- Penheiter, A.R.; Griesmann, G.E.; Federspiel, M.J.; Dingli, D.; Russell, S.J.; Carlson, S.K. Pinhole micro-SPECT/CT for noninvasive monitoring and quantitation of oncolytic virus dispersion and percent infection in solid tumors. Gene Ther. 2012, 19, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Aref, S.; Bailey, K.; Fielding, A. Measles to the Rescue: A Review of Oncolytic Measles Virus. Viruses 2016, 8, 294. [Google Scholar] [CrossRef] [PubMed]
- Rajecki, M.; Kangasmaki, A.; Laasonen, L.; Escutenaire, S.; Hakkarainen, T.; Haukka, J.; Ristimaki, A.; Kairemo, K.; Kangasniemi, L.; Kiljunen, T.; et al. Sodium iodide symporter SPECT imaging of a patient treated with oncolytic adenovirus Ad5/3-Delta24-hNIS. Mol. Ther. 2011, 19, 629–631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, A.; Chaurasiya, S.; Park, A.K.; Kim, S.I.; Lu, J.; Olafsen, T.; Warner, S.G.; Fong, Y.; Woo, Y. PET imaging and treatment of pancreatic cancer peritoneal carcinomatosis after subcutaneous intratumoral administration of a novel oncolytic virus, CF33-hNIS-antiPDL1. Mol. Ther. Oncolytics 2022, 24, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Haddad, D.; Chen, N.G.; Zhang, Q.; Chen, C.H.; Yu, Y.A.; Gonzalez, L.; Carpenter, S.G.; Carson, J.; Au, J.; Mittra, A.; et al. Insertion of the human sodium iodide symporter to facilitate deep tissue imaging does not alter oncolytic or replication capability of a novel vaccinia virus. J. Transl. Med. 2011, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Rojas, J.J.; Thorne, S.H. Theranostic potential of oncolytic vaccinia virus. Theranostics 2012, 2, 363–373. [Google Scholar] [CrossRef]
- Zhang, L.; Steele, M.B.; Jenks, N.; Grell, J.; Suksanpaisan, L.; Naik, S.; Federspiel, M.J.; Lacy, M.Q.; Russell, S.J.; Peng, K.W. Safety Studies in Tumor and Non-Tumor-Bearing Mice in Support of Clinical Trials Using Oncolytic VSV-IFNbeta-NIS. Hum. Gene Ther. Clin. Dev. 2016, 27, 111–122. [Google Scholar] [CrossRef]
- Barton, K.N.; Stricker, H.; Brown, S.L.; Elshaikh, M.; Aref, I.; Lu, M.; Pegg, J.; Zhang, Y.; Karvelis, K.C.; Siddiqui, F.; et al. Phase I study of noninvasive imaging of adenovirus-mediated gene expression in the human prostate. Mol. Ther. 2008, 16, 1761–1769. [Google Scholar] [CrossRef]
- Dispenzieri, A.; Tong, C.; LaPlant, B.; Lacy, M.Q.; Laumann, K.; Dingli, D.; Zhou, Y.; Federspiel, M.J.; Gertz, M.A.; Hayman, S.; et al. Phase I trial of systemic administration of Edmonston strain of measles virus genetically engineered to express the sodium iodide symporter in patients with recurrent or refractory multiple myeloma. Leukemia 2017, 31, 2791–2798. [Google Scholar] [CrossRef]
- Galanis, E.; Atherton, P.J.; Maurer, M.J.; Knutson, K.L.; Dowdy, S.C.; Cliby, W.A.; Haluska, P., Jr.; Long, H.J.; Oberg, A.; Aderca, I.; et al. Oncolytic measles virus expressing the sodium iodide symporter to treat drug-resistant ovarian cancer. Cancer Res. 2015, 75, 22–30. [Google Scholar] [CrossRef]
- Peikert, T.; Mandrekar, S.; Mansfield, A.; Tan, A.; Tippmann-Peikert, M.; Galanis, E. Intrapleural Modified Vaccine Strain Measles Virus Therapy for Patients with Malignant Pleural Mesotheliomajourna of thoraci. J. Thorac. Oncol. 2017, 12, S296. [Google Scholar] [CrossRef]
- Kuruppu, D.; Bhere, D.; Farrar, C.T.; Shah, K.; Brownell, A.L.; Mahmood, U.; Tanabe, K.K. Oncolytic HSV1 targets different growth phases of breast cancer leptomeningeal metastases. Cancer Gene Ther. 2023, 30, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Penuelas, I.; Mazzolini, G.; Boan, J.F.; Sangro, B.; Marti-Climent, J.; Ruiz, M.; Ruiz, J.; Satyamurthy, N.; Qian, C.; Barrio, J.R.; et al. Positron emission tomography imaging of adenoviral-mediated transgene expression in liver cancer patients. Gastroenterology 2005, 128, 1787–1795. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.; Tazawa, H.; Kishimoto, H.; Kagawa, S.; Fujiwara, T.; Hoffman, R.M. Real-Time Fluorescence Image-Guided Oncolytic Virotherapy for Precise Cancer Treatment. Int. J. Mol. Sci. 2021, 22, 879. [Google Scholar] [CrossRef]
- Stritzker, J.; Kirscher, L.; Scadeng, M.; Deliolanis, N.C.; Morscher, S.; Symvoulidis, P.; Schaefer, K.; Zhang, Q.; Buckel, L.; Hess, M.; et al. Vaccinia virus-mediated melanin production allows MR and optoacoustic deep tissue imaging and laser-induced thermotherapy of cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 3316–3320. [Google Scholar] [CrossRef]
| Virus Backbone | OV Name | Number of Clinical Trials 2 |
|---|---|---|
| Adenovirus (Ad, AdV) | DNX-2401; ONCOS-102; Enadenotucirev; OBP-301; Icovir-5; NSC-crAd; Ad-E6A7; Ad-MA3; Ad-MAGEA3; VCN-01; CG0070; CG7870; ONYX015; H101; TILT-123; LOAd703; ADV/HSV-tk; Ad5-yCD/mutTKSR39rep-ADP; DNX-2440; CAdVEC; NG-350A; ORCA-010; AdAPT-001; MEM-288; L-IFN; Ad-TD-nsIL12; ICVB-1042; NG-641 | 84 |
| Coxsackievirus | V937 | 15 |
| Herpes simplex virus 1 (HSV-1) | T-VEC; OrienX010; HSV1716; HF10; M032; G207; NV1020; rQNestin; C134; Rp1; Rp2; Rp3; ONCR-177; STI-1386; VG2025; T3011; R130; VG161; G47; Delytact | 105 |
| Herpes simplex virus 1 (HSV-2) | OH2; BS-006 | 16 |
| Influenza | CodaLytic | 1 |
| Lymphocytic choriomeningitis virus (LCMV) | HB-302; HB-301 | 1 |
| Maraba virus | MG1-E6A7; MG1-MA3; MG1-MAGEA3 | 4 |
| Measles (MV) | MV-CEA; MV-NIS; MV-CD; MV-NAP | 17 |
| Newcastle disease virus (NDV) | NDV-HUJ; PV701; MTH-68H; MEDI5395; MEDI9253 | 6 |
| Parvovirus | Parvoryx | 2 |
| Poliovirus-rhinovirus | PVSripo | 11 |
| Reovirus | Reolysin | 36 |
| Sendai virus | GEN0101 | 2 |
| Seneca Valley virus | NTX-010; SVV-001 | 3 |
| Vaccinia virus (VV) | Pexavec; GL-ONC1; JX-594; vvDD; TG6002; T601; TBio-6517; BT-001; OVV-01; RGV004; PF-07263689; VV-GMCSF-Lact; KM1; TG6050; hV01; CF33-CD19; JX-594; ACAM2000; MQ710; TG4023; CF33-hNIS; ASP9801 | 48 |
| Vesicular stomatitis virus (VSV) | VSV-IFNβ; VSV-IFNβ-NIS; Revottack | 9 |
| Other (alphavirus; picornavirus; unspecified) | M1-c6v1; IVX037; RT-01 | 9 |
| Cancer Type | Number of Clinical Trials 2 |
|---|---|
| Brain | 43 |
| Breast | 33 |
| Colorectal (CRC) | 32 |
| Gastrointestinal | 16 |
| Gynecological | 32 |
| Head and neck | 29 |
| Kidney | 4 |
| Liver | 26 |
| Lung | 28 |
| Mesothelioma | 7 |
| Neuroendocrine and peripheral nervous system | 5 |
| Pancreas | 25 |
| Prostate | 8 |
| Sarcoma | 22 |
| Skin | 96 |
| Urinary tract | 21 |
| Other solid tumors | 43 |
| Hematological tumors | 18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menotti, L.; Vannini, A. Oncolytic Viruses in the Era of Omics, Computational Technologies, and Modeling: Thesis, Antithesis, and Synthesis. Int. J. Mol. Sci. 2023, 24, 17378. https://doi.org/10.3390/ijms242417378
Menotti L, Vannini A. Oncolytic Viruses in the Era of Omics, Computational Technologies, and Modeling: Thesis, Antithesis, and Synthesis. International Journal of Molecular Sciences. 2023; 24(24):17378. https://doi.org/10.3390/ijms242417378
Chicago/Turabian StyleMenotti, Laura, and Andrea Vannini. 2023. "Oncolytic Viruses in the Era of Omics, Computational Technologies, and Modeling: Thesis, Antithesis, and Synthesis" International Journal of Molecular Sciences 24, no. 24: 17378. https://doi.org/10.3390/ijms242417378
APA StyleMenotti, L., & Vannini, A. (2023). Oncolytic Viruses in the Era of Omics, Computational Technologies, and Modeling: Thesis, Antithesis, and Synthesis. International Journal of Molecular Sciences, 24(24), 17378. https://doi.org/10.3390/ijms242417378






