Integrated Stress Response (ISR) Pathway: Unraveling Its Role in Cellular Senescence
Abstract
1. Introduction
2. An Overview of Cellular Senescence
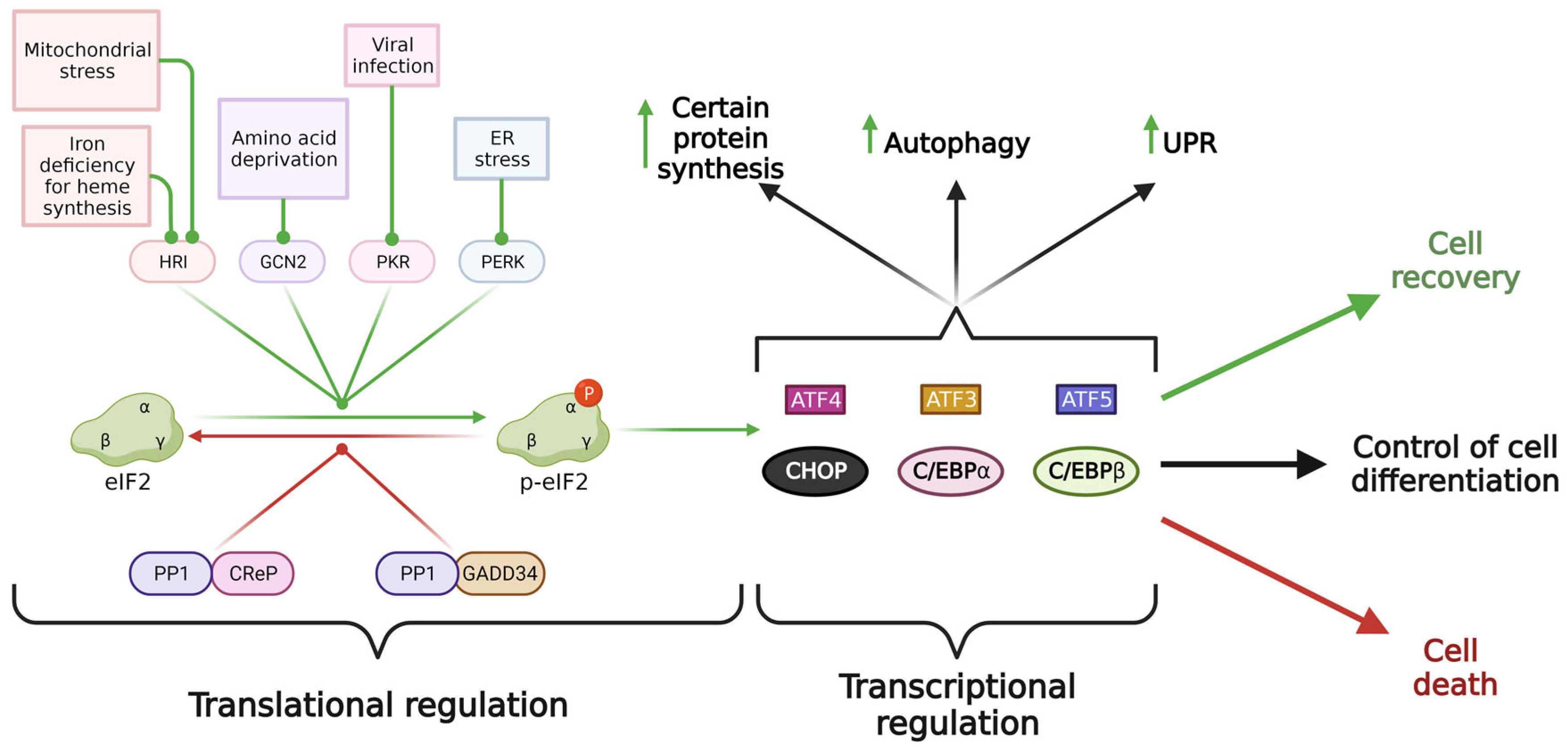
3. Molecular Mechanism of Integrated Stress Response
4. Distinct Outcomes of ISR
5. ISR in Cellular Metabolism
5.1. ISR and Autophagy
5.2. ISR and Mitochondrial Homeostasis
5.3. ISR and Mitochondrial Unfolded Protein Response
6. Interrelationship between ISR and Cellular Senescence
7. Role of ATF4-Interacting Partners in Cellular Senescence
7.1. ATF4-C/EBP Complex
7.2. ATF4-CHOP(DDIT3) Complex
7.3. ATF4-NRF2 (NFE2L2) Complex
7.4. ATF4 and Other bZIP Transcription Factors
7.5. ATF4 and TRIB3
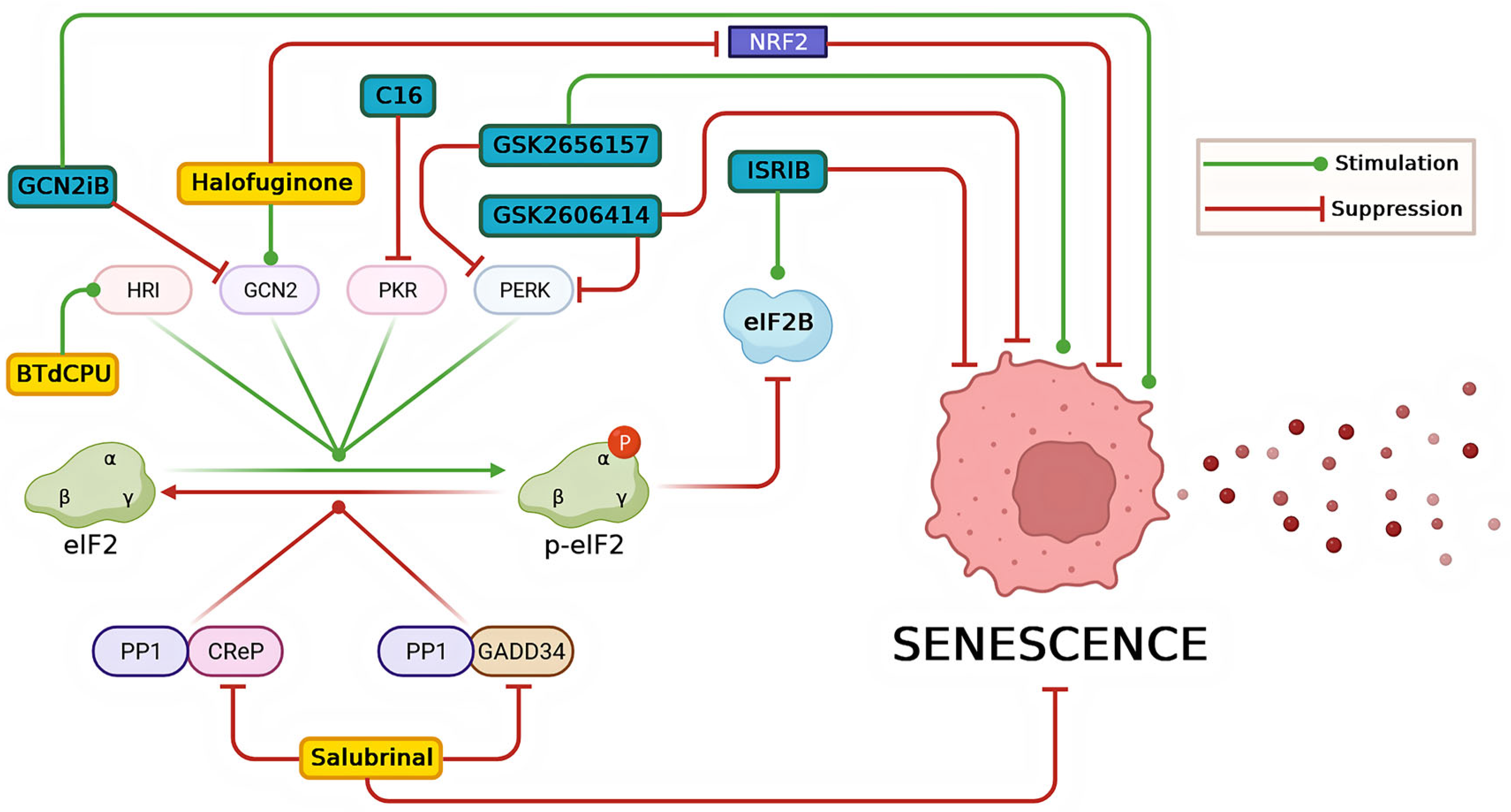
8. Modulators of ISR and Their Impact on Senescence-Related Conditions
8.1. Inhibitors of ISR
8.2. Activators of ISR
9. Take Home Message
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Abbreviations | |||
| ASCs | adipose tissue stem cells | ATF | activating transcription factor |
| ATG | autophagy-related | DS | developmental senescence |
| hAMSCs | human amniotic mesenchymal stem cells | HCCs | hepatocellular carcinoma cells |
| HUVECs | human umbilical vein endothelial cells | IL | interleukin |
| ISR | integrated stress response | MAPK | mitogen associated protein kinase |
| mTORC1 | mechanistic target of rapamycin | NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| OIS | oncogene-induced senescence | OSIS | oxidative stress-induced senescence |
| PIC | preinitiation complex | ROS | reactive oxygen species |
| SAHF | senescence associated heterochromatin foci | SASP | senescence-associated secretory phenotype |
| T2DM | type 2 diabetes mellitus | TIS | therapy-induced senescence |
| uORF | upstream open reading frame | UPRmt | mitochondrial unfolded protein response |
| General Abbreviations | Description | Gene ID (Gene Cards) | Gene Description |
| AMPK | 5′AMP-activated protein kinase | PRKAA1 | Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 |
| ANGPTL4 | angiopoietin like 4 | ANGPTL4 | Hepatic Angiopoietin-Related Protein |
| ASNS | asparagine synthetase | ASNS | Asparagine Synthetase (Glutamine-Hydrolyzing) |
| BECN1 | beclin 1 | BECN1 | Beclin 1, autophagy-related protein |
| BIM | Bcl-2 interacting mediator | BCL2L11 | BCL2 Like 11 |
| CDKN2A | cyclin dependent kinase inhibitor 2A | CDKN2A | Cyclin Dependent Kinase Inhibitor 2A |
| CEBPA | CEBPB, CCAAT/enhancer-binding protein alpha/beta | CEBPA | CCAAT/Enhancer-Binding Protein Alpha |
| cGAS | cyclic GMP-AMP synthase | CGAS | CGAMP Synthase |
| CHOP | C/EBP homologous protein | DDIT3 | DNA Damage Inducible Transcript 3 |
| CReP | constitutive repressor of eIF2α phosphorylation | PPP1R15B | Protein Phosphatase 1 Regulatory Subunit 15B |
| DELE1 | DAP3 binding cell death enhancer 1 | DELE1 | DAP3 Binding Cell Death Enhancer 1 |
| DR4 | death receptor 4 | TNFRSF10A | TNF Receptor Superfamily Member 10a |
| eIF2α | eukaryotic translation initiation factor subunit alpha | EIF2S1 | Eukaryotic Translation Initiation Factor 2 Subunit Alpha |
| EPRS | glutamyl-prolyl-tRNA synthetase | EPRS1 | Glutamyl-Prolyl-TRNA Synthetase 1 |
| FRA1 | Fos-related antigen 1 | FOSL1 | FOS Like 1, AP-1 Transcription Factor Subunit |
| GABARAP | Gamma-aminobutyric acid receptor-associated protein | GABARAP | Gamma-Aminobutyric Acid Receptor-Associated Protein |
| GADD34 | growth arrest and DNA damage-inducible protein 34 | PPP1R15A | Protein Phosphatase 1 Regulatory Subunit 15A |
| GCN2 | general control nonderepressible 2 | EIF2AK4 | Eukaryotic Translation Initiation Factor 2 Alpha Kinase 4 |
| GDF15 | growth differentiation factor 15 | GDF15 | Growth Differentiation Factor 15 |
| HRI | heme-regulated eIFα kinase | EIF2AK1 | Eukaryotic Translation Initiation Factor 2 Alpha Kinase 1 |
| IKK | IkB kinase | IKBKB | Inhibitor Of Nuclear Factor Kappa B Kinase Subunit Beta |
| IRF3 | Interferon regulatory factor 3 | IRF3 | Interferon Regulatory Factor 3 |
| JAK1 | Janus kinase 1 | JAK1 | Janus kinase 1 |
| JDP2 | Jun dimerization protein 2 | JDP2 | Jun Dimerization Protein 2 |
| JNK | c-Jun N-terminal kinase | MAPK8 | Mitogen-Activated Protein Kinase 8 |
| KDM6B | lysin demethylase 6B | KDM6B | Lysine Demethylase 6B |
| KEAP1 | Kelch-like ECH-associated protein 1 | KEAP1 | Kelch Like ECH Associated Protein 1 |
| MAP1LC3B | microtubule-associated protein 1 light chain 3B | MAP1LC3B | Microtubule Associated Protein 1 Light Chain 3 Beta |
| MCL1 | myeloid cell leukemia 1 | MCL1 | MCL1 Apoptosis Regulator, BCL2 Family Member |
| MDM2 | mouse double minute 2 homolog | MDM2 | MDM2, E3-ubiquitin protein ligase |
| mTORC1 | mechanistic target of rapamycin | MTOR | Mechanistic Target Of Rapamycin Kinase |
| NBR1 | neihbor of BRCA1 gene 1 | NBR1 | NBR1 Autophagy Cargo Receptor |
| NFE2 | nuclear factor erythroid 2-related factor 2 | NFE2 | Nuclear Factor, Erythroid 2 |
| Nrf2 | nuclear factor erythroid 2-related factor 2 | NFE2L2 | NFE2 Like BZIP Transcription Factor 2 |
| OMA1 | metalloendopeptidase | OMA1 | OMA1 Zinc Metallopeptidase |
| PAI-1 | plasminogen activator inhibitor type 1 | SERPINE1 | Serpin Family E Member 1 |
| PERK | protein kinase RNA-like ER kinase eIF2α kinase 3 | EIF2AK3 | Eukaryotic Translation Initiation Factor 2 Alpha Kinase 3 |
| PKR | protein kinase | EIF2AK2 | Eukaryotic Translation Initiation Factor 2 Alpha Kinase 2 |
| PP1 | Protein Phosphatase 1 | PPP1CA | Protein Phosphatase 1 Catalytic Subunit Alpha |
| PSPH | phosphoserine phosphatase | PSPH | Phosphoserine Phosphatase |
| PUMA | p53 upregulated modulator of apoptosis | BBC3 | BCL2 Binding Component 3 |
| RAS | small GTPase | KRAS | KRAS Proto-Oncogene, GTPase |
| RIPK1 | Receptor-interacting serine/threonine-protein kinase 1 | RIPK1 | Receptor Interacting Serine/Threonine Kinase 1 |
| SA-βGal | senescence-associated beta-galactosidase | GLB1 | Galactosidase Beta 1 |
| SQSTM1 | sequestosome 1 | SQSTM1 | Sequestosome 1 |
| STING | stimulator of interferon genes | STING1 | Stimulator Of Interferon Response CGAMP Interactor 1 |
| TRAIL | tumor necrosis factor-related apoptosis-inducing ligand | TNFSF10 | TNF Superfamily Member 10 |
| TRIB3 | tribbles preudokinase 3 | TRIB3 | Tribbles Pseudokinase 3 |
| TSC2 | tuberous sclerosis complex 2 protein | TSC2 | TSC Complex Subunit 2 |
| ULK1/2 | Unc-51-like autophagy-activating kinase | ULK1 | Unc-51 Like Autophagy Activating Kinase 1 |
| VEGFA | vascular endothelial factor A | VEGFA | Vascular Endothelial Growth Factor A |
References
- Hayflick, L.; Moorhead, P.S. The Serial Cultivation of Human Diploid Cell Strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Hayflick, L. The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp. Cell Res. 1965, 37, 614–636. [Google Scholar] [CrossRef]
- Storer, M.; Mas, A.; Robert-Moreno, A.; Pecoraro, M.; Ortells, M.C.; Di Giacomo, V.; Yosef, R.; Pilpel, N.; Krizhanovsky, V.; Sharpe, J.; et al. Senescence Is a Developmental Mechanism That Contributes to Embryonic Growth and Patterning. Cell 2013, 155, 1119–1130. [Google Scholar] [CrossRef]
- Sati, S.; Bonev, B.; Szabo, Q.; Jost, D.; Bensadoun, P.; Serra, F.; Loubiere, V.; Papadopoulos, G.L.; Rivera-Mulia, J.-C.; Fritsch, L.; et al. 4D Genome Rewiring during Oncogene-Induced and Replicative Senescence. Mol. Cell 2020, 78, 522–538.e9. [Google Scholar] [CrossRef]
- Basisty, N.; Kale, A.; Jeon, O.H.; Kuehnemann, C.; Payne, T.; Rao, C.; Holtz, A.; Shah, S.; Sharma, V.; Ferrucci, L.; et al. A Proteomic Atlas of Senescence-Associated Secretomes for Aging Biomarker Development. PLoS Biol. 2020, 18, e3000599. [Google Scholar] [CrossRef]
- Pole, A.; Dimri, M.; Dimri, P.G. Oxidative Stress, Cellular Senescence and Ageing. AIMS Mol. Sci. 2016, 3, 300–324. [Google Scholar] [CrossRef]
- Saez-Atienzar, S.; Masliah, E. Cellular Senescence and Alzheimer Disease: The Egg and the Chicken Scenario. Nat. Rev. Neurosci. 2020, 21, 433–444. [Google Scholar] [CrossRef]
- Coryell, P.R.; Diekman, B.O.; Loeser, R.F. Mechanisms and Therapeutic Implications of Cellular Senescence in Osteoarthritis. Nat. Rev. Rheumatol. 2021, 17, 47–57. [Google Scholar] [CrossRef]
- Pignolo, R.J.; Law, S.F.; Chandra, A. Bone Aging, Cellular Senescence, and Osteoporosis. JBMR Plus 2021, 5, e10488. [Google Scholar] [CrossRef]
- Palmer, A.K.; Tchkonia, T.; LeBrasseur, N.K.; Chini, E.N.; Xu, M.; Kirkland, J.L. Cellular Senescence in Type 2 Diabetes: A Therapeutic Opportunity. Diabetes 2015, 64, 2289–2298. [Google Scholar] [CrossRef]
- Tuttle, C.S.L.; Waaijer, M.E.C.; Slee-Valentijn, M.S.; Stijnen, T.; Westendorp, R.; Maier, A.B. Cellular Senescence and Chronological Age in Various Human Tissues: A Systematic Review and Meta-analysis. Aging Cell 2020, 19, e13083. [Google Scholar] [CrossRef]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; LeBrasseur, N.K.; Childs, B.G.; van de Sluis, B.; Kirkland, J.L.; van Deursen, J.M. Clearance of P16Ink4a-Positive Senescent Cells Delays Ageing-Associated Disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Muthumalage, T.; Rahman, I. Clearance of Senescent Cells Reverts the Cigarette Smoke-induced Lung Senescence and Airspace Enlargement in P16-3MR Mice. Aging Cell 2023, 22, e13850. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Wek, R.C.; Anthony, T.G.; Staschke, K.A. Surviving and Adapting to Stress: Translational Control and the Integrated Stress Response. Antioxid. Redox Signal. 2023, 39, 351–373. [Google Scholar] [CrossRef]
- Huang, W.; Hickson, L.J.; Eirin, A.; Kirkland, J.L.; Lerman, L.O. Cellular Senescence: The Good, the Bad and the Unknown. Nat. Rev. Nephrol. 2022, 18, 611–627. [Google Scholar] [CrossRef]
- Kumari, R.; Jat, P. Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Front. Cell Dev. Biol. 2021, 9, 485. [Google Scholar] [CrossRef]
- Yue, Z.; Nie, L.; Zhao, P.; Ji, N.; Liao, G.; Wang, Q. Senescence-Associated Secretory Phenotype and Its Impact on Oral Immune Homeostasis. Front. Immunol. 2022, 13, 1019313. [Google Scholar] [CrossRef]
- Zhou, R.; Xie, X.; Qin, Z.; Li, X.; Liu, J.; Li, H.; Zheng, Q.; Luo, Y. Cytosolic DsDNA Is a Novel Senescence Marker Associated with Pyroptosis Activation. Tissue Cell 2021, 72, 101554. [Google Scholar] [CrossRef]
- Amor, C.; Feucht, J.; Leibold, J.; Ho, Y.-J.; Zhu, C.; Alonso-Curbelo, D.; Mansilla-Soto, J.; Boyer, J.A.; Li, X.; Giavridis, T.; et al. Senolytic CAR T Cells Reverse Senescence-Associated Pathologies. Nature 2020, 583, 127–132. [Google Scholar] [CrossRef]
- Hall, B.M.; Balan, V.; Gleiberman, A.S.; Strom, E.; Krasnov, P.; Virtuoso, L.P.; Rydkina, E.; Vujcic, S.; Balan, K.; Gitlin, I.I.; et al. P16(Ink4a) and Senescence-Associated β-Galactosidase Can Be Induced in Macrophages as Part of a Reversible Response to Physiological Stimuli. Aging 2017, 9, 1867–1884. [Google Scholar] [CrossRef]
- Piechota, M.; Sunderland, P.; Wysocka, A.; Nalberczak, M.; Sliwinska, M.A.; Radwanska, K.; Sikora, E. Is Senescence-Associated β-Galactosidase a Marker of Neuronal Senescence? Oncotarget 2016, 7, 81099–81109. [Google Scholar] [CrossRef]
- Frippiat, C.; Dewelle, J.; Remacle, J.; Toussaint, O. Signal Transduction in H2O2-Induced Senescence-like Phenotype in Human Diploid Fibroblasts. Free Radic. Biol. Med. 2002, 33, 1334–1346. [Google Scholar] [CrossRef]
- Untergasser, G.; Gander, R.; Rumpold, H.; Heinrich, E.; Plas, E.; Berger, P. TGF-β Cytokines Increase Senescence-Associated Beta-Galactosidase Activity in Human Prostate Basal Cells by Supporting Differentiation Processes, but Not Cellular Senescence. Exp. Gerontol. 2003, 38, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Severino, J.; Allen, R.G.; Balin, S.; Balin, A.; Cristofalo, V.J. Is β-Galactosidase Staining a Marker of Senescence in Vitro and in Vivo? Exp. Cell Res. 2000, 257, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Kurz, D.J.; Decary, S.; Hong, Y.; Erusalimsky, J.D. Senescence-Associated β-Galactosidase Reflects an Increase in Lysosomal Mass during Replicative Ageing of Human Endothelial Cells. J. Cell Sci. 2000, 113, 3613–3622. [Google Scholar] [CrossRef]
- Lee, B.Y.; Han, J.A.; Im, J.S.; Morrone, A.; Johung, K.; Goodwin, E.C.; Kleijer, W.J.; DiMaio, D.; Hwang, E.S. Senescence-associated Β-galactosidase Is Lysosomal Β-galactosidase. Aging Cell 2006, 5, 187–195. [Google Scholar] [CrossRef]
- Wagner, K.-D.; Wagner, N. The Senescence Markers P16INK4A, P14ARF/P19ARF, and P21 in Organ Development and Homeostasis. Cells 2022, 11, 1966. [Google Scholar] [CrossRef]
- Kosar, M.; Bartkova, J.; Hubackova, S.; Hodny, Z.; Lukas, J.; Bartek, J. Senescence-Associated Heterochromatin Foci Are Dispensable for Cellular Senescence, Occur in a Cell Type- and Insult-Dependent Manner and Follow Expression of p16INK4a. Cell Cycle 2011, 10, 457–468. [Google Scholar] [CrossRef]
- Pantoja, C.; Serrano, M. Murine Fibroblasts Lacking P21 Undergo Senescence and Are Resistant to Transformation by Oncogenic Ras. Oncogene 1999, 18, 4974–4982. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Gasek, N.S.; Zhou, Y.; Cohn, R.L.; Martin, D.E.; Zuo, W.; Flynn, W.F.; Guo, C.; Jellison, E.R.; et al. Targeting P21Cip1 Highly Expressing Cells in Adipose Tissue Alleviates Insulin Resistance in Obesity. Cell Metab. 2022, 34, 75–89.e8. [Google Scholar] [CrossRef]
- Grosse, L.; Wagner, N.; Emelyanov, A.; Molina, C.; Lacas-Gervais, S.; Wagner, K.-D.; Bulavin, D.V. Defined P16High Senescent Cell Types Are Indispensable for Mouse Healthspan. Cell Metab. 2020, 32, 87–99.e6. [Google Scholar] [CrossRef]
- Natarajan, E.; Omobono, J.D.; Jones, J.C.; Rheinwald, J.G. Co-Expression of P16INK4A and Laminin 5 by Keratinocytes: A Wound-Healing Response Coupling Hypermotility with Growth Arrest That Goes Awry During Epithelial Neoplastic Progression. J. Investig. Dermatol. Symp. Proc. 2005, 10, 72–85. [Google Scholar] [CrossRef]
- Demaria, M.; Ohtani, N.; Youssef, S.A.; Rodier, F.; Toussaint, W.; Mitchell, J.R.; Laberge, R.-M.; Vijg, J.; Van Steeg, H.; Dollé, M.E.T.; et al. An Essential Role for Senescent Cells in Optimal Wound Healing through Secretion of PDGF-AA. Dev. Cell 2014, 31, 722–733. [Google Scholar] [CrossRef]
- Cohn, R.L.; Gasek, N.S.; Kuchel, G.A.; Xu, M. The Heterogeneity of Cellular Senescence: Insights at the Single-Cell Level. Trends Cell Biol. 2023, 33, 9–17. [Google Scholar] [CrossRef]
- Muñoz-Espín, D.; Cañamero, M.; Maraver, A.; Gómez-López, G.; Contreras, J.; Murillo-Cuesta, S.; Rodríguez-Baeza, A.; Varela-Nieto, I.; Ruberte, J.; Collado, M.; et al. Programmed Cell Senescence during Mammalian Embryonic Development. Cell 2013, 155, 1104–1118. [Google Scholar] [CrossRef]
- Antelo-Iglesias, L.; Picallos-Rabina, P.; Estévez-Souto, V.; Da Silva-Álvarez, S.; Collado, M. The Role of Cellular Senescence in Tissue Repair and Regeneration. Mech. Ageing Dev. 2021, 198, 111528. [Google Scholar] [CrossRef]
- Fulop, T.; Larbi, A.; Dupuis, G.; Le Page, A.; Frost, E.H.; Cohen, A.A.; Witkowski, J.M.; Franceschi, C. Immunosenescence and Inflamm-Aging as Two Sides of the Same Coin: Friends or Foes? Front. Immunol. 2018, 8, 1960. [Google Scholar] [CrossRef]
- Roger, L.; Tomas, F.; Gire, V. Mechanisms and Regulation of Cellular Senescence. Int. J. Mol. Sci. 2021, 22, 13173. [Google Scholar] [CrossRef]
- Aiello, A.; Farzaneh, F.; Candore, G.; Caruso, C.; Davinelli, S.; Gambino, C.M.; Ligotti, M.E.; Zareian, N.; Accardi, G. Immunosenescence and Its Hallmarks: How to Oppose Aging Strategically? A Review of Potential Options for Therapeutic Intervention. Front. Immunol. 2019, 10, 2247. [Google Scholar] [CrossRef]
- Lagoumtzi, S.M.; Chondrogianni, N. Senolytics and Senomorphics: Natural and Synthetic Therapeutics in the Treatment of Aging and Chronic Diseases. Free Radic. Biol. Med. 2021, 171, 169–190. [Google Scholar] [CrossRef]
- Le Pelletier, L.; Mantecon, M.; Gorwood, J.; Auclair, M.; Foresti, R.; Motterlini, R.; Laforge, M.; Atlan, M.; Fève, B.; Capeau, J.; et al. Metformin Alleviates Stress-Induced Cellular Senescence of Aging Human Adipose Stromal Cells and the Ensuing Adipocyte Dysfunction. elife 2021, 10, e62635. [Google Scholar] [CrossRef]
- Chen, D.; Xia, D.; Pan, Z.; Xu, D.; Zhou, Y.; Wu, Y.; Cai, N.; Tang, Q.; Wang, C.; Yan, M.; et al. Metformin Protects against Apoptosis and Senescence in Nucleus Pulposus Cells and Ameliorates Disc Degeneration in Vivo. Cell Death Dis. 2016, 7, e2441. [Google Scholar] [CrossRef]
- Xu, M.; Tchkonia, T.; Ding, H.; Ogrodnik, M.; Lubbers, E.R.; Pirtskhalava, T.; White, T.A.; Johnson, K.O.; Stout, M.B.; Mezera, V.; et al. JAK Inhibition Alleviates the Cellular Senescence-Associated Secretory Phenotype and Frailty in Old Age. Proc. Natl. Acad. Sci. USA 2015, 112, E6301–E6310. [Google Scholar] [CrossRef]
- Yi, G.; He, Z.; Zhou, X.; Xian, L.; Yuan, T.; Jia, X.; Hong, J.; He, L.; Liu, J. Low Concentration of Metformin Induces a P53-Dependent Senescence in Hepatoma Cells via Activation of the AMPK Pathway. Int. J. Oncol. 2013, 43, 1503–1510. [Google Scholar] [CrossRef]
- Lussana, F.; Cattaneo, M.; Rambaldi, A.; Squizzato, A. Ruxolitinib-Associated Infections: A Systematic Review and Meta-Analysis. Am. J. Hematol. 2018, 93, 339–347. [Google Scholar] [CrossRef]
- Wysham, N.G.; Sullivan, D.R.; Allada, G. An Opportunistic Infection Associated With Ruxolitinib, a Novel Janus Kinase 1,2 Inhibitor. Chest 2013, 143, 1478–1479. [Google Scholar] [CrossRef]
- Cai, Y.; Zhou, H.; Zhu, Y.; Sun, Q.; Ji, Y.; Xue, A.; Wang, Y.; Chen, W.; Yu, X.; Wang, L.; et al. Elimination of Senescent Cells by β-Galactosidase-Targeted Prodrug Attenuates Inflammation and Restores Physical Function in Aged Mice. Cell Res. 2020, 30, 574–589. [Google Scholar] [CrossRef]
- Saul, D.; Kosinsky, R.L.; Atkinson, E.J.; Doolittle, M.L.; Zhang, X.; LeBrasseur, N.K.; Pignolo, R.J.; Robbins, P.D.; Niedernhofer, L.J.; Ikeno, Y.; et al. A New Gene Set Identifies Senescent Cells and Predicts Senescence-Associated Pathways across Tissues. Nat. Commun. 2022, 13, 4827. [Google Scholar] [CrossRef]
- Song, Q.; Hou, Y.; Zhang, Y.; Liu, J.; Wang, Y.; Fu, J.; Zhang, C.; Cao, M.; Cui, Y.; Zhang, X.; et al. Integrated Multi-Omics Approach Revealed Cellular Senescence Landscape. Nucleic Acids Res. 2022, 50, 10947–10963. [Google Scholar] [CrossRef]
- Gao, Y.; Chi, Y.; Chen, Y.; Wang, W.; Li, H.; Zheng, W.; Zhu, P.; An, J.; Duan, Y.; Sun, T.; et al. Multi-Omics Analysis of Human Mesenchymal Stem Cells Shows Cell Aging That Alters Immunomodulatory Activity through the Downregulation of PD-L1. Nat. Commun. 2023, 14, 4373. [Google Scholar] [CrossRef]
- Lu, P.D.; Jousse, C.; Marciniak, S.J.; Zhang, Y.; Novoa, I.; Scheuner, D.; Kaufman, R.J.; Ron, D.; Harding, H.P. Cytoprotection by Pre-Emptive Conditional Phosphorylation of Translation Initiation Factor 2. EMBO J. 2004, 23, 169–179. [Google Scholar] [CrossRef]
- Andreev, D.E.; O’Connor, P.B.; Fahey, C.; Kenny, E.M.; Terenin, I.M.; Dmitriev, S.E.; Cormican, P.; Morris, D.W.; Shatsky, I.N.; Baranov, P.V. Translation of 5′ Leaders Is Pervasive in Genes Resistant to EIF2 Repression. elife 2015, 4, e03971. [Google Scholar] [CrossRef]
- Taniuchi, S.; Miyake, M.; Tsugawa, K.; Oyadomari, M.; Oyadomari, S. Integrated Stress Response of Vertebrates Is Regulated by Four EIF2α Kinases. Sci. Rep. 2016, 6, 32886. [Google Scholar] [CrossRef]
- Guo, X.; Aviles, G.; Liu, Y.; Tian, R.; Unger, B.A.; Lin, Y.-H.T.; Wiita, A.P.; Xu, K.; Correia, M.A.; Kampmann, M. Mitochondrial Stress Is Relayed to the Cytosol by an OMA1–DELE1–HRI Pathway. Nature 2020, 579, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-J.; Zhang, S. Heme-Regulated EIF2α Kinase in Erythropoiesis and Hemoglobinopathies. Blood 2019, 134, 1697–1707. [Google Scholar] [CrossRef]
- Fessler, E.; Eckl, E.-M.; Schmitt, S.; Mancilla, I.A.; Meyer-Bender, M.F.; Hanf, M.; Philippou-Massier, J.; Krebs, S.; Zischka, H.; Jae, L.T. A Pathway Coordinated by DELE1 Relays Mitochondrial Stress to the Cytosol. Nature 2020, 579, 433–437. [Google Scholar] [CrossRef]
- Hinnebusch, A.G.; Lorsch, J.R. The Mechanism of Eukaryotic Translation Initiation: New Insights and Challenges. Cold Spring Harb. Perspect. Biol. 2012, 4, a011544. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.T.; Pestova, T.V. The Mechanism of Eukaryotic Translation Initiation and Principles of Its Regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef]
- Adomavicius, T.; Guaita, M.; Zhou, Y.; Jennings, M.D.; Latif, Z.; Roseman, A.M.; Pavitt, G.D. The Structural Basis of Translational Control by EIF2 Phosphorylation. Nat. Commun. 2019, 10, 2136. [Google Scholar] [CrossRef]
- Starck, S.R.; Tsai, J.C.; Chen, K.; Shodiya, M.; Wang, L.; Yahiro, K.; Martins-Green, M.; Shastri, N.; Walter, P. Translation from the 5′ Untranslated Region Shapes the Integrated Stress Response. Science 2016, 351, aad3867. [Google Scholar] [CrossRef] [PubMed]
- Calvo, S.E.; Pagliarini, D.J.; Mootha, V.K. Upstream Open Reading Frames Cause Widespread Reduction of Protein Expression and Are Polymorphic among Humans. Proc. Natl. Acad. Sci. USA 2009, 106, 7507–7512. [Google Scholar] [CrossRef]
- Torrence, M.E.; MacArthur, M.R.; Hosios, A.M.; Valvezan, A.J.; Asara, J.M.; Mitchell, J.R.; Manning, B.D. The MTORC1-Mediated Activation of ATF4 Promotes Protein and Glutathione Synthesis Downstream of Growth Signals. elife 2021, 10, e63326. [Google Scholar] [CrossRef]
- Young, S.K.; Wek, R.C. Upstream Open Reading Frames Differentially Regulate Gene-Specific Translation in the Integrated Stress Response. J. Biol. Chem. 2016, 291, 16927–16935. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.-Y.; Wek, S.A.; McGrath, B.C.; Lu, D.; Hai, T.; Harding, H.P.; Wang, X.; Ron, D.; Cavener, D.R.; Wek, R.C. Activating Transcription Factor 3 Is Integral to the Eukaryotic Initiation Factor 2 Kinase Stress Response. Mol. Cell. Biol. 2004, 24, 1365–1377. [Google Scholar] [CrossRef]
- Reid, D.W.; Tay, A.S.L.; Sundaram, J.R.; Lee, I.C.J.; Chen, Q.; George, S.E.; Nicchitta, C.V.; Shenolikar, S. Complementary Roles of GADD34- and CReP-Containing Eukaryotic Initiation Factor 2α Phosphatases during the Unfolded Protein Response. Mol. Cell. Biol. 2016, 36, 1868–1880. [Google Scholar] [CrossRef]
- Jousse, C.; Oyadomari, S.; Novoa, I.; Lu, P.; Zhang, Y.; Harding, H.P.; Ron, D. Inhibition of a Constitutive Translation Initiation Factor 2α Phosphatase, CReP, Promotes Survival of Stressed Cells. J. Cell Biol. 2003, 163, 767–775. [Google Scholar] [CrossRef]
- Kojima, E.; Takeuchi, A.; Haneda, M.; Yagi, F.; Hasegawa, T.; Yamaki, K.-I.; Takeda, K.; Akira, S.; Shimokata, K.; Isobe, K.-I. The Function of GADD34 Is a Recovery from a Shutoff of Protein Synthesis Induced by ER Stress—Elucidation by GADD34-deficient Mice. FASEB J. 2003, 17, 1–18. [Google Scholar] [CrossRef]
- Ma, Y.; Hendershot, L.M. Delineation of a Negative Feedback Regulatory Loop That Controls Protein Translation during Endoplasmic Reticulum Stress. J. Biol. Chem. 2003, 278, 34864–34873. [Google Scholar] [CrossRef]
- Clemens, M.J. Initiation Factor EIF2α Phosphorylation in Stress Responses and Apoptosis. In Progress in Molecular and Subcellular Biology; Springer: Berlin/Heidelberg, Germany, 2001; pp. 57–89. [Google Scholar]
- Li, T.; Su, L.; Lei, Y.; Liu, X.; Zhang, Y.; Liu, X. DDIT3 and KAT2A Proteins Regulate TNFRSF10A and TNFRSF10B Expression in Endoplasmic Reticulum Stress-Mediated Apoptosis in Human Lung Cancer Cells. J. Biol. Chem. 2015, 290, 11108–11118. [Google Scholar] [CrossRef]
- Liu, K.; Shi, Y.; Guo, X.; Wang, S.; Ouyang, Y.; Hao, M.; Liu, D.; Qiao, L.; Li, N.; Zheng, J.; et al. CHOP Mediates ASPP2-Induced Autophagic Apoptosis in Hepatoma Cells by Releasing Beclin-1 from Bcl-2 and Inducing Nuclear Translocation of Bcl-2. Cell Death Dis. 2014, 5, e1323. [Google Scholar] [CrossRef]
- Ghosh, A.P.; Klocke, B.J.; Ballestas, M.E.; Roth, K.A. CHOP Potentially Co-Operates with FOXO3a in Neuronal Cells to Regulate PUMA and BIM Expression in Response to ER Stress. PLoS ONE 2012, 7, e39586. [Google Scholar] [CrossRef]
- Tsukano, H.; Gotoh, T.; Endo, M.; Miyata, K.; Tazume, H.; Kadomatsu, T.; Yano, M.; Iwawaki, T.; Kohno, K.; Araki, K.; et al. The Endoplasmic Reticulum Stress-C/EBP Homologous Protein Pathway-Mediated Apoptosis in Macrophages Contributes to the Instability of Atherosclerotic Plaques. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1925–1932. [Google Scholar] [CrossRef]
- Teske, B.F.; Fusakio, M.E.; Zhou, D.; Shan, J.; McClintick, J.N.; Kilberg, M.S.; Wek, R.C. CHOP Induces Activating Transcription Factor 5 (ATF5) to Trigger Apoptosis in Response to Perturbations in Protein Homeostasis. Mol. Biol. Cell 2013, 24, 2477–2490. [Google Scholar] [CrossRef]
- Syed, V.; Mukherjee, K.; Lyons-Weiler, J.; Lau, K.-M.; Mashima, T.; Tsuruo, T.; Ho, S. Identification of ATF-3, Caveolin-1, DLC-1, and NM23-H2 as Putative Antitumorigenic, Progesterone-Regulated Genes for Ovarian Cancer Cells by Gene Profiling. Oncogene 2005, 24, 1774–1787. [Google Scholar] [CrossRef]
- Nobori, K.; Ito, H.; Tamamori-Adachi, M.; Adachi, S.; Ono, Y.; Kawauchi, J.; Kitajima, S.; Marumo, F.; Isobe, M. ATF3 Inhibits Doxorubicin-Induced Apoptosis in Cardiac Myocytes: A Novel Cardioprotective Role of ATF3. J. Mol. Cell. Cardiol. 2002, 34, 1387–1397. [Google Scholar] [CrossRef]
- Wang, X.; Yu, X.; Li, Y.; Liu, F.; Du, L.; Xie, N.; Wang, C. ATF5 Attenuates Apoptosis in Hippocampal Neurons with Seizures Evoked by Mg2+-Free Medium via Regulating Mitochondrial Unfolded Protein Response. Neurochem. Res. 2023, 48, 62–71. [Google Scholar] [CrossRef]
- Peng, X.; Yang, T.; He, L.; Chen, X.; Jiang, Y.; Zhang, H.; Liu, H.; Peng, Y. Impact of GADD34 on Apoptosis of Tonsillar Mononuclear Cells from IgA Nephropathy Patients by Regulating Eif2α Phosphorylation. Cell. Physiol. Biochem. 2018, 50, 2203–2215. [Google Scholar] [CrossRef]
- Song, P.; Yang, S.; Hua, H.; Zhang, H.; Kong, Q.; Wang, J.; Luo, T.; Jiang, Y. The Regulatory Protein GADD34 Inhibits TRAIL-Induced Apoptosis via TRAF6/ERK-Dependent Stabilization of Myeloid Cell Leukemia 1 in Liver Cancer Cells. J. Biol. Chem. 2019, 294, 5945–5955. [Google Scholar] [CrossRef]
- Ito, S.; Tanaka, Y.; Oshino, R.; Aiba, K.; Thanasegaran, S.; Nishio, N.; Isobe, K. GADD34 Inhibits Activation-Induced Apoptosis of Macrophages through Enhancement of Autophagy. Sci. Rep. 2015, 5, 8327. [Google Scholar] [CrossRef]
- Li, S.; Guo, L.; Qian, P.; Zhao, Y.; Liu, A.; Ji, F.; Chen, L.; Wu, X.; Qian, G. Lipopolysaccharide Induces Autophagic Cell Death through the PERK-Dependent Branch of the Unfolded Protein Response in Human Alveolar Epithelial A549 Cells. Cell. Physiol. Biochem. 2015, 36, 2403–2417. [Google Scholar] [CrossRef]
- Verfaillie, T.; Rubio, N.; Garg, A.D.; Bultynck, G.; Rizzuto, R.; Decuypere, J.-P.; Piette, J.; Linehan, C.; Gupta, S.; Samali, A.; et al. PERK Is Required at the ER-Mitochondrial Contact Sites to Convey Apoptosis after ROS-Based ER Stress. Cell Death Differ. 2012, 19, 1880–1891. [Google Scholar] [CrossRef]
- Burwick, N.; Zhang, M.Y.; de la Puente, P.; Azab, A.K.; Hyun, T.S.; Ruiz-Gutierrez, M.; Sanchez-Bonilla, M.; Nakamura, T.; Delrow, J.J.; MacKay, V.L.; et al. The EIF2-Alpha Kinase HRI Is a Novel Therapeutic Target in Multiple Myeloma. Leuk. Res. 2017, 55, 23–32. [Google Scholar] [CrossRef]
- Wei, C.; Lin, M.; Jinjun, B.; Su, F.; Dan, C.; Yan, C.; Jie, Y.; Jin, Z.; Zi-Chun, H.; Wu, Y. Involvement of General Control Nonderepressible Kinase 2 in Cancer Cell Apoptosis by Posttranslational Mechanisms. Mol. Biol. Cell 2015, 26, 1044–1057. [Google Scholar] [CrossRef]
- Xu, C.; Gamil, A.; Munang’andu, H.; Evensen, Ø. Apoptosis Induction by DsRNA-Dependent Protein Kinase R (PKR) in EPC Cells via Caspase 8 and 9 Pathways. Viruses 2018, 10, 526. [Google Scholar] [CrossRef]
- D’Aniello, C.; Fico, A.; Casalino, L.; Guardiola, O.; Di Napoli, G.; Cermola, F.; De Cesare, D.; Tatè, R.; Cobellis, G.; Patriarca, E.J.; et al. A Novel Autoregulatory Loop between the Gcn2-Atf4 Pathway and L-Proline Metabolism Controls Stem Cell Identity. Cell Death Differ. 2015, 22, 1094–1105. [Google Scholar] [CrossRef]
- Mielke, N.; Schwarzer, R.; Calkhoven, C.F.; Kaufman, R.J.; Dorken, B.; Leutz, A.; Jundt, F. Eukaryotic Initiation Factor 2 Phosphorylation Is Required for B-Cell Maturation and Function in Mice. Haematologica 2011, 96, 1261–1268. [Google Scholar] [CrossRef][Green Version]
- Zismanov, V.; Chichkov, V.; Colangelo, V.; Jamet, S.; Wang, S.; Syme, A.; Koromilas, A.E.; Crist, C. Phosphorylation of EIF2α Is a Translational Control Mechanism Regulating Muscle Stem Cell Quiescence and Self-Renewal. Cell Stem Cell 2016, 18, 79–90. [Google Scholar] [CrossRef]
- Wang, W.; Lian, N.; Li, L.; Moss, H.E.; Wang, W.; Perrien, D.S.; Elefteriou, F.; Yang, X. Atf4 Regulates Chondrocyte Proliferation and Differentiation during Endochondral Ossification by Activating Ihh Transcription. Development 2009, 136, 4143–4153. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, J.; Liu, D.; Dong, F.; Cheng, H.; Wang, W.; Pang, Y.; Wang, Y.; Mu, X.; Ni, Y.; et al. ATF4 Plays a Pivotal Role in the Development of Functional Hematopoietic Stem Cells in Mouse Fetal Liver. Blood 2015, 126, 2383–2391. [Google Scholar] [CrossRef]
- Fischer, C.; Johnson, J.; Stillwell, B.; Conner, J.; Cerovac, Z.; Wilson-Rawls, J.; Rawls, A. Activating Transcription Factor 4 Is Required for the Differentiation of the Lamina Propria Layer of the Vas Deferens1. Biol. Reprod. 2004, 70, 371–378. [Google Scholar] [CrossRef]
- Yu, S.; Zhu, K.; Lai, Y.; Zhao, Z.; Fan, J.; Im, H.-J.; Chen, D.; Xiao, G. ATF4 Promotes β-Catenin Expression and Osteoblastic Differentiation of Bone Marrow Mesenchymal Stem Cells. Int. J. Biol. Sci. 2013, 9, 256–266. [Google Scholar] [CrossRef]
- Neill, G.; Masson, G.R. A Stay of Execution: ATF4 Regulation and Potential Outcomes for the Integrated Stress Response. Front. Mol. Neurosci. 2023, 16, 1112253. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and Medical Implications of Mammalian Autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- B’chir, W.; Maurin, A.-C.; Carraro, V.; Averous, J.; Jousse, C.; Muranishi, Y.; Parry, L.; Stepien, G.; Fafournoux, P.; Bruhat, A. The EIF2α/ATF4 Pathway Is Essential for Stress-Induced Autophagy Gene Expression. Nucleic Acids Res. 2013, 41, 7683–7699. [Google Scholar] [CrossRef]
- Yang, Y.; Gatica, D.; Liu, X.; Wu, R.; Kang, R.; Tang, D.; Klionsky, D.J. Upstream Open Reading Frames Mediate Autophagy-Related Protein Translation. Autophagy 2023, 19, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.M. Role of the Transcription Factor ATF4 in the Anabolic Actions of Insulin and the Anti-Anabolic Actions of Glucocorticoids. J. Biol. Chem. 2007, 282, 16744–16753. [Google Scholar] [CrossRef]
- Selvarajah, B.; Azuelos, I.; Platé, M.; Guillotin, D.; Forty, E.J.; Contento, G.; Woodcock, H.V.; Redding, M.; Taylor, A.; Brunori, G.; et al. MTORC1 Amplifies the ATF4-Dependent De Novo Serine-Glycine Pathway to Supply Glycine during TGF-β 1 –Induced Collagen Biosynthesis. Sci. Signal. 2019, 12, eaav3048. [Google Scholar] [CrossRef]
- Park, Y.; Reyna-Neyra, A.; Philippe, L.; Thoreen, C.C. MTORC1 Balances Cellular Amino Acid Supply with Demand for Protein Synthesis through Post-Transcriptional Control of ATF4. Cell Rep. 2017, 19, 1083–1090. [Google Scholar] [CrossRef]
- He, L.; Zhang, J.; Zhao, J.; Ma, N.; Kim, S.W.; Qiao, S.; Ma, X. Autophagy: The Last Defense against Cellular Nutritional Stress. Adv. Nutr. 2018, 9, 493–504. [Google Scholar] [CrossRef]
- Averous, J.; Lambert-Langlais, S.; Mesclon, F.; Carraro, V.; Parry, L.; Jousse, C.; Bruhat, A.; Maurin, A.-C.; Pierre, P.; Proud, C.G.; et al. GCN2 Contributes to MTORC1 Inhibition by Leucine Deprivation through an ATF4 Independent Mechanism. Sci. Rep. 2016, 6, 27698. [Google Scholar] [CrossRef] [PubMed]
- Maurin, A.-C.; Parry, L.; B’chir, W.; Carraro, V.; Coudy-Gandilhon, C.; Chaouki, G.; Chaveroux, C.; Mordier, S.; Martinie, B.; Reinhardt, V.; et al. GCN2 Upregulates Autophagy in Response to Short-Term Deprivation of a Single Essential Amino Acid. Autophagy Rep. 2022, 1, 119–142. [Google Scholar] [CrossRef]
- Ye, J.; Palm, W.; Peng, M.; King, B.; Lindsten, T.; Li, M.O.; Koumenis, C.; Thompson, C.B. GCN2 Sustains MTORC1 Suppression upon Amino Acid Deprivation by Inducing Sestrin2. Genes Dev. 2015, 29, 2331–2336. [Google Scholar] [CrossRef]
- Uddin, M.N.; Ito, S.; Nishio, N.; Suganya, T.; Isobe, K. Gadd34 Induces Autophagy through the Suppression of the MTOR Pathway during Starvation. Biochem. Biophys. Res. Commun. 2011, 407, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Kapuy, O.; Holczer, M.; Márton, M.; Korcsmáros, T. Autophagy-Dependent Survival Is Controlled with a Unique Regulatory Network upon Various Cellular Stress Events. Cell Death Dis. 2021, 12, 309. [Google Scholar] [CrossRef]
- Spinelli, J.B.; Haigis, M.C. The Multifaceted Contributions of Mitochondria to Cellular Metabolism. Nat. Cell Biol. 2018, 20, 745–754. [Google Scholar] [CrossRef]
- HARMAN, D. The Biologic Clock: The Mitochondria? J. Am. Geriatr. Soc. 1972, 20, 145–147. [Google Scholar] [CrossRef]
- Wiley, C.D.; Velarde, M.C.; Lecot, P.; Liu, S.; Sarnoski, E.A.; Freund, A.; Shirakawa, K.; Lim, H.W.; Davis, S.S.; Ramanathan, A.; et al. Mitochondrial Dysfunction Induces Senescence with a Distinct Secretory Phenotype. Cell Metab. 2016, 23, 303–314. [Google Scholar] [CrossRef]
- Quirós, P.M.; Prado, M.A.; Zamboni, N.; D’Amico, D.; Williams, R.W.; Finley, D.; Gygi, S.P.; Auwerx, J. Multi-Omics Analysis Identifies ATF4 as a Key Regulator of the Mitochondrial Stress Response in Mammals. J. Cell Biol. 2017, 216, 2027–2045. [Google Scholar] [CrossRef]
- Krall, A.S.; Mullen, P.J.; Surjono, F.; Momcilovic, M.; Schmid, E.W.; Halbrook, C.J.; Thambundit, A.; Mittelman, S.D.; Lyssiotis, C.A.; Shackelford, D.B.; et al. Asparagine Couples Mitochondrial Respiration to ATF4 Activity and Tumor Growth. Cell Metab. 2021, 33, 1013–1026.e6. [Google Scholar] [CrossRef]
- Bao, X.R.; Ong, S.-E.; Goldberger, O.; Peng, J.; Sharma, R.; Thompson, D.A.; Vafai, S.B.; Cox, A.G.; Marutani, E.; Ichinose, F.; et al. Mitochondrial Dysfunction Remodels One-Carbon Metabolism in Human Cells. elife 2016, 5, e10575. [Google Scholar] [CrossRef]
- Palikaras, K.; Lionaki, E.; Tavernarakis, N. Mechanisms of Mitophagy in Cellular Homeostasis, Physiology and Pathology. Nat. Cell Biol. 2018, 20, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Shpilka, T.; Haynes, C.M. The Mitochondrial UPR: Mechanisms, Physiological Functions and Implications in Ageing. Nat. Rev. Mol. Cell Biol. 2018, 19, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Condon, K.J.; Orozco, J.M.; Adelmann, C.H.; Spinelli, J.B.; van der Helm, P.W.; Roberts, J.M.; Kunchok, T.; Sabatini, D.M. Genome-Wide CRISPR Screens Reveal Multitiered Mechanisms through Which MTORC1 Senses Mitochondrial Dysfunction. Proc. Natl. Acad. Sci. USA 2021, 118, e2022120118. [Google Scholar] [CrossRef] [PubMed]
- Whitney, M.L.; Jefferson, L.S.; Kimball, S.R. ATF4 Is Necessary and Sufficient for ER Stress-Induced Upregulation of REDD1 Expression. Biochem. Biophys. Res. Commun. 2009, 379, 451–455. [Google Scholar] [CrossRef]
- Zhu, S.; Nguyen, A.; Pang, J.; Zhao, J.; Chen, Z.; Liang, Z.; Gu, Y.; Huynh, H.; Bao, Y.; Lee, S.; et al. Mitochondrial Stress Induces an HRI-EIF2α Pathway Protective for Cardiomyopathy. Circulation 2022, 146, 1028–1031. [Google Scholar] [CrossRef]
- Memme, J.M.; Sanfrancesco, V.C.; Hood, D.A. Activating Transcription Factor 4 Regulates Mitochondrial Content, Morphology, and Function in Differentiating Skeletal Muscle Myotubes. Am. J. Physiol. Physiol. 2023, 325, C224–C242. [Google Scholar] [CrossRef]
- Diwan, B.; Sharma, R. Nutritional Components as Mitigators of Cellular Senescence in Organismal Aging: A Comprehensive Review. Food Sci. Biotechnol. 2022, 31, 1089–1109. [Google Scholar] [CrossRef]
- Canfield, C.A.; Bradshaw, P.C. Amino Acids in the Regulation of Aging and Aging-Related Diseases. Transl. Med. Aging 2019, 3, 70–89. [Google Scholar] [CrossRef]
- Lehman, S.L.; Cerniglia, G.J.; Johannes, G.J.; Ye, J.; Ryeom, S.; Koumenis, C. Translational Upregulation of an Individual P21Cip1 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress. PLoS Genet. 2015, 11, e1005212. [Google Scholar] [CrossRef]
- Missiaen, R.; Anderson, N.M.; Kim, L.C.; Nance, B.; Burrows, M.; Skuli, N.; Carens, M.; Riscal, R.; Steensels, A.; Li, F.; et al. GCN2 Inhibition Sensitizes Arginine-Deprived Hepatocellular Carcinoma Cells to Senolytic Treatment. Cell Metab. 2022, 34, 1151–1167.e7. [Google Scholar] [CrossRef]
- Cordova, R.A.; Misra, J.; Amin, P.H.; Klunk, A.J.; Damayanti, N.P.; Carlson, K.R.; Elmendorf, A.J.; Kim, H.G.; Mirek, E.T.; Elzey, B.D.; et al. GCN2 EIF2 Kinase Promotes Prostate Cancer by Maintaining Amino Acid Homeostasis. elife 2022, 11, e81083. [Google Scholar] [CrossRef]
- Ye, J.; Kumanova, M.; Hart, L.S.; Sloane, K.; Zhang, H.; De Panis, D.N.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Ron, D.; Koumenis, C. The GCN2-ATF4 Pathway Is Critical for Tumour Cell Survival and Proliferation in Response to Nutrient Deprivation. EMBO J. 2010, 29, 2082–2096. [Google Scholar] [CrossRef]
- Rajesh, K.; Papadakis, A.I.; Kazimierczak, U.; Peidis, P.; Wang, S.; Ferbeyre, G.; Kaufman, R.J.; Koromilas, A.E. EIF2α Phosphorylation Bypasses Premature Senescence Caused by Oxidative Stress and Pro-Oxidant Antitumor Therapies. Aging 2013, 5, 884–901. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, Y.; Zhu, Y.; Zhang, Q.; Guan, H.; Liu, S.; Chen, S.; Mei, C.; Chen, C.; Liao, Z.; et al. A Non-Canonical CGAS–STING–PERK Pathway Facilitates the Translational Program Critical for Senescence and Organ Fibrosis. Nat. Cell Biol. 2022, 24, 766–782. [Google Scholar] [CrossRef]
- Yoon, C.H.; Lee, E.S.; Lim, D.S.; Bae, Y.S. PKR, a P53 Target Gene, Plays a Crucial Role in the Tumor-Suppressor Function of P53. Proc. Natl. Acad. Sci. USA 2009, 106, 7852–7857. [Google Scholar] [CrossRef]
- Gal-Ben-Ari, S.; Barrera, I.; Ehrlich, M.; Rosenblum, K. PKR: A Kinase to Remember. Front. Mol. Neurosci. 2019, 11, 480. [Google Scholar] [CrossRef]
- Li, Y.; Peng, Z.; Wang, C.; Li, L.; Leng, Y.; Chen, R.; Yuan, H.; Zhou, S.; Zhang, Z.; Chen, A.F. Novel Role of PKR in Palmitate-Induced Sirt1 Inactivation and Endothelial Cell Senescence. Am. J. Physiol. Circ. Physiol. 2018, 315, H571–H580. [Google Scholar] [CrossRef]
- Lee, J.-J.; Lee, J.-H.; Ko, Y.-G.; Hong, S.I.; Lee, J.-S. Prevention of Premature Senescence Requires JNK Regulation of Bcl-2 and Reactive Oxygen Species. Oncogene 2010, 29, 561–575. [Google Scholar] [CrossRef]
- Spallarossa, P.; Altieri, P.; Barisione, C.; Passalacqua, M.; Aloi, C.; Fugazza, G.; Frassoni, F.; Podestà, M.; Canepa, M.; Ghigliotti, G.; et al. P38 MAPK and JNK Antagonistically Control Senescence and Cytoplasmic P16INK4A Expression in Doxorubicin-Treated Endothelial Progenitor Cells. PLoS ONE 2010, 5, e15583. [Google Scholar] [CrossRef]
- Das, M.; Jiang, F.; Sluss, H.K.; Zhang, C.; Shokat, K.M.; Flavell, R.A.; Davis, R.J. Suppression of P53-Dependent Senescence by the JNK Signal Transduction Pathway. Proc. Natl. Acad. Sci. USA 2007, 104, 15759–15764. [Google Scholar] [CrossRef]
- Baltzis, D.; Pluquet, O.; Papadakis, A.I.; Kazemi, S.; Qu, L.K.; Koromilas, A.E. The EIF2α Kinases PERK and PKR Activate Glycogen Synthase Kinase 3 to Promote the Proteasomal Degradation of P53. J. Biol. Chem. 2007, 282, 31675–31687. [Google Scholar] [CrossRef]
- Tomiyoshi, G.; Co, F.K.; Nakamura, R.; Co, F.K.; Shinmen, N.; Co, F.K.; Yoshida, Y.; Mine, S.; Machida, T.; Kitamura, K.; et al. GADD34 Activates P53 and May Have Utility as a Marker of Atherosclerosis. Front. Med. 2023, 10, 1128921. [Google Scholar] [CrossRef]
- Yang, T.; He, R.; Li, G.; Liang, J.; Zhao, L.; Zhao, X.; Li, L.; Wang, P. Growth Arrest and DNA Damage-Inducible Protein 34 (GADD34) Contributes to Cerebral Ischemic Injury and Can Be Detected in Plasma Exosomes. Neurosci. Lett. 2021, 758, 136004. [Google Scholar] [CrossRef]
- Minami, K.; Inoue, H.; Terashita, T.; Kawakami, T.; Watanabe, R.; Haneda, M.; Isobe, K.I.; Okabe, H.; Chano, T. GADD34 Induces P21 Expression and Cellular Senescence. Oncol. Rep. 2007, 17, 1481–1485. [Google Scholar] [CrossRef][Green Version]
- Otsuka, R.; Harada, N.; Aoki, S.; Shirai, K.; Nishitsuji, K.; Nozaki, A.; Hatakeyama, A.; Shono, M.; Mizusawa, N.; Yoshimoto, K.; et al. C-Terminal Region of GADD34 Regulates EIF2α Dephosphorylation and Cell Proliferation in CHO-K1 Cells. Cell Stress Chaperones 2016, 21, 29–40. [Google Scholar] [CrossRef]
- Yagi, A.; Hasegawa, Y.; Xiao, H.; Haneda, M.; Kojima, E.; Nishikimi, A.; Hasegawa, T.; Shimokata, K.; Isobe, K.I. GADD34 Induces P53 Phosphorylation and P21/WAF1 Transcription. J. Cell. Biochem. 2003, 90, 1242–1249. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, X.; Huang, L.; Guan, Y.; Huang, X.; Tian, X.; Zhang, L.; Tao, W. ATF3 Drives Senescence by Reconstructing Accessible Chromatin Profiles. Aging Cell 2021, 20, e13315. [Google Scholar] [CrossRef]
- Zhao, Q.; Luo, Y.-F.; Tian, M.; Xiao, Y.-L.; Cai, H.-R.; Li, H. Activating Transcription Factor 3 Involved in Pseudomonas Aeruginosa PAO1-Induced Macrophage Senescence. Mol. Immunol. 2021, 133, 122–127. [Google Scholar] [CrossRef]
- Kim, K.-H.; Park, B.; Rhee, D.-K.; Pyo, S. Acrylamide Induces Senescence in Macrophages through a Process Involving ATF3, ROS, P38/JNK, and a Telomerase-Independent Pathway. Chem. Res. Toxicol. 2015, 28, 71–86. [Google Scholar] [CrossRef]
- Yan, C.; Lu, D.; Hai, T.; Boyd, D.D. Activating Transcription Factor 3, a Stress Sensor, Activates P53 by Blocking Its Ubiquitination. EMBO J. 2005, 24, 2425–2435. [Google Scholar] [CrossRef]
- Taketani, K.; Kawauchi, J.; Tanaka-Okamoto, M.; Ishizaki, H.; Tanaka, Y.; Sakai, T.; Miyoshi, J.; Maehara, Y.; Kitajima, S. Key Role of ATF3 in P53-Dependent DR5 Induction upon DNA Damage of Human Colon Cancer Cells. Oncogene 2012, 31, 2210–2221. [Google Scholar] [CrossRef]
- Tanaka, Y.; Nakamura, A.; Morioka, M.S.; Inoue, S.; Tamamori-Adachi, M.; Yamada, K.; Taketani, K.; Kawauchi, J.; Tanaka-Okamoto, M.; Miyoshi, J.; et al. Systems Analysis of ATF3 in Stress Response and Cancer Reveals Opposing Effects on Pro-Apoptotic Genes in P53 Pathway. PLoS ONE 2011, 6, e26848. [Google Scholar] [CrossRef]
- Sun, Y.; Lin, X.; Liu, B.; Zhang, Y.; Li, W.; Zhang, S.; He, F.; Tian, H.; Zhu, X.; Liu, X.; et al. Loss of ATF4 Leads to Functional Aging-like Attrition of Adult Hematopoietic Stem Cells. Sci. Adv. 2021, 7, eabj6877. [Google Scholar] [CrossRef]
- Liu, J.; Yang, J.-R.; Chen, X.-M.; Cai, G.-Y.; Lin, L.-R.; He, Y.-N. Impact of ER Stress-Regulated ATF4/P16 Signaling on the Premature Senescence of Renal Tubular Epithelial Cells in Diabetic Nephropathy. Am. J. Physiol. Physiol. 2015, 308, C621–C630. [Google Scholar] [CrossRef]
- Sakai, T.; Kurokawa, R.; Hirano, S.; Imai, J. Hydrogen Indirectly Suppresses Increases in Hydrogen Peroxide in Cytoplasmic Hydroxyl Radical-Induced Cells and Suppresses Cellular Senescence. Int. J. Mol. Sci. 2019, 20, 456. [Google Scholar] [CrossRef]
- Salminen, A.; Kaarniranta, K.; Hiltunen, M.; Kauppinen, A. Histone Demethylase Jumonji D3 (JMJD3/KDM6B) at the Nexus of Epigenetic Regulation of Inflammation and the Aging Process. J. Mol. Med. 2014, 92, 1035–1043. [Google Scholar] [CrossRef]
- Moskalev, A.A.; Smit-McBride, Z.; Shaposhnikov, M.V.; Plyusnina, E.N.; Zhavoronkov, A.; Budovsky, A.; Tacutu, R.; Fraifeld, V.E. Gadd45 Proteins: Relevance to Aging, Longevity and Age-Related Pathologies. Ageing Res. Rev. 2012, 11, 51–66. [Google Scholar] [CrossRef]
- Fernández-Hernando, C.; Suárez, Y. ANGPTL4. Curr. Opin. Hematol. 2020, 27, 206–213. [Google Scholar] [CrossRef]
- Wischhusen, J.; Melero, I.; Fridman, W.H. Growth/Differentiation Factor-15 (GDF-15): From Biomarker to Novel Targetable Immune Checkpoint. Front. Immunol. 2020, 11, 951. [Google Scholar] [CrossRef]
- Schmitt, C.A.; Wang, B.; Demaria, M. Senescence and Cancer—Role and Therapeutic Opportunities. Nat. Rev. Clin. Oncol. 2022, 19, 619–636. [Google Scholar] [CrossRef]
- Podust, L.M.; Krezel, A.M.; Kim, Y. Crystal Structure of the CCAAT Box/Enhancer-Binding Protein β Activating Transcription Factor-4 Basic Leucine Zipper Heterodimer in the Absence of DNA. J. Biol. Chem. 2001, 276, 505–513. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.A.; Reinke, A.W.; Bhimsaria, D.; Keating, A.E.; Ansari, A.Z. Combinatorial BZIP Dimers Display Complex DNA-Binding Specificity Landscapes. elife 2017, 6, e19272. [Google Scholar] [CrossRef]
- Juliana, C.A.; Yang, J.; Cannon, C.E.; Good, A.L.; Haemmerle, M.W.; Stoffers, D.A. A PDX1-ATF Transcriptional Complex Governs β Cell Survival during Stress. Mol. Metab. 2018, 17, 39–48. [Google Scholar] [CrossRef]
- Ebrahim, N.; Shakirova, K.; Dashinimaev, E. PDX1 Is the Cornerstone of Pancreatic β-Cell Functions and Identity. Front. Mol. Biosci. 2022, 9, 1091757. [Google Scholar] [CrossRef]
- RAMJI, D.P.; FOKA, P. CCAAT/Enhancer-Binding Proteins: Structure, Function and Regulation. Biochem. J. 2002, 365, 561–575. [Google Scholar] [CrossRef]
- Horiguchi, M.; Koyanagi, S.; Okamoto, A.; Suzuki, S.O.; Matsunaga, N.; Ohdo, S. Stress-Regulated Transcription Factor ATF4 Promotes Neoplastic Transformation by Suppressing Expression of the INK4a/ARF Cell Senescence Factors. Cancer Res. 2012, 72, 395–401. [Google Scholar] [CrossRef]
- Huggins, C.J.; Mayekar, M.K.; Martin, N.; Saylor, K.L.; Gonit, M.; Jailwala, P.; Kasoji, M.; Haines, D.C.; Quiñones, O.A.; Johnson, P.F. C/EBPγ Is a Critical Regulator of Cellular Stress Response Networks through Heterodimerization with ATF4. Mol. Cell. Biol. 2016, 36, 693–713. [Google Scholar] [CrossRef]
- Huggins, C.J.; Malik, R.; Lee, S.; Salotti, J.; Thomas, S.; Martin, N.; Quiñones, O.A.; Alvord, W.G.; Olanich, M.E.; Keller, J.R.; et al. C/EBPγ Suppresses Senescence and Inflammatory Gene Expression by Heterodimerizing with C/EBPβ. Mol. Cell. Biol. 2013, 33, 3242–3258. [Google Scholar] [CrossRef]
- Dey, S.; Savant, S.; Teske, B.F.; Hatzoglou, M.; Calkhoven, C.F.; Wek, R.C. Transcriptional Repression of ATF4 Gene by CCAAT/Enhancer-Binding Protein β (C/EBPβ) Differentially Regulates Integrated Stress Response. J. Biol. Chem. 2012, 287, 21936–21949. [Google Scholar] [CrossRef]
- Guan, Y.; Zhang, C.; Lyu, G.; Huang, X.; Zhang, X.; Zhuang, T.; Jia, L.; Zhang, L.; Zhang, C.; Li, C.; et al. Senescence-Activated Enhancer Landscape Orchestrates the Senescence-Associated Secretory Phenotype in Murine Fibroblasts. Nucleic Acids Res. 2020, 48, 10909–10923. [Google Scholar] [CrossRef]
- Oyadomari, S.; Mori, M. Roles of CHOP/GADD153 in Endoplasmic Reticulum Stress. Cell Death Differ. 2004, 11, 381–389. [Google Scholar] [CrossRef]
- Kaspar, S.; Oertlin, C.; Szczepanowska, K.; Kukat, A.; Senft, K.; Lucas, C.; Brodesser, S.; Hatzoglou, M.; Larsson, O.; Topisirovic, I.; et al. Adaptation to Mitochondrial Stress Requires CHOP-Directed Tuning of ISR. Sci. Adv. 2021, 7, eabf0971. [Google Scholar] [CrossRef]
- Inoue, Y.; Kawachi, S.; Ohkubo, T.; Nagasaka, M.; Ito, S.; Fukuura, K.; Itoh, Y.; Ohoka, N.; Morishita, D.; Hayashi, H. The CDK Inhibitor P21 Is a Novel Target Gene of ATF4 and Contributes to Cell Survival under ER Stress. FEBS Lett. 2017, 591, 3682–3691. [Google Scholar] [CrossRef]
- Jing, X.; Sun, W.; Yang, X.; Huang, H.; Wang, P.; Luo, Q.; Xia, S.; Fang, C.; Zhang, Q.; Guo, J.; et al. CCAAT/Enhancer-Binding Protein (C/EBP) Homologous Protein Promotes Alveolar Epithelial Cell Senescence via the Nuclear Factor-Kappa B Pathway in Pulmonary Fibrosis. Int. J. Biochem. Cell Biol. 2022, 143, 106142. [Google Scholar] [CrossRef]
- Ahmed, S.M.U.; Luo, L.; Namani, A.; Wang, X.J.; Tang, X. Nrf2 Signaling Pathway: Pivotal Roles in Inflammation. Biochim. Biophys. Acta—Mol. Basis Dis. 2017, 1863, 585–597. [Google Scholar] [CrossRef]
- Yuan, H.; Xu, Y.; Luo, Y.; Wang, N.-X.; Xiao, J.-H. Role of Nrf2 in Cell Senescence Regulation. Mol. Cell. Biochem. 2021, 476, 247–259. [Google Scholar] [CrossRef]
- Malavolta, M.; Bracci, M.; Santarelli, L.; Sayeed, M.A.; Pierpaoli, E.; Giacconi, R.; Costarelli, L.; Piacenza, F.; Basso, A.; Cardelli, M.; et al. Inducers of Senescence, Toxic Compounds, and Senolytics: The Multiple Faces of Nrf2-Activating Phytochemicals in Cancer Adjuvant Therapy. Mediat. Inflamm. 2018, 2018, 4159013. [Google Scholar] [CrossRef]
- He, C.H.; Gong, P.; Hu, B.; Stewart, D.; Choi, M.E.; Choi, A.M.K.; Alam, J. Identification of Activating Transcription Factor 4 (ATF4) as an Nrf2-Interacting Protein. J. Biol. Chem. 2001, 276, 20858–20865. [Google Scholar] [CrossRef]
- Reinke, A.W.; Baek, J.; Ashenberg, O.; Keating, A.E. Networks of BZIP Protein-Protein Interactions Diversified Over a Billion Years of Evolution. Science 2013, 340, 730–734. [Google Scholar] [CrossRef]
- Poh, J.; Ponsford, A.H.; Boyd, J.; Woodsmith, J.; Stelzl, U.; Wanker, E.; Harper, N.; MacEwan, D.; Sanderson, C.M. A Functionally Defined High-Density NRF2 Interactome Reveals New Conditional Regulators of ARE Transactivation. Redox Biol. 2020, 37, 101686. [Google Scholar] [CrossRef]
- Su, N.; Kilberg, M.S. C/EBP Homology Protein (CHOP) Interacts with Activating Transcription Factor 4 (ATF4) and Negatively Regulates the Stress-Dependent Induction of the Asparagine Synthetase Gene. J. Biol. Chem. 2008, 283, 35106–35117. [Google Scholar] [CrossRef]
- Cullinan, S.B.; Zhang, D.; Hannink, M.; Arvisais, E.; Kaufman, R.J.; Diehl, J.A. Nrf2 Is a Direct PERK Substrate and Effector of PERK-Dependent Cell Survival. Mol. Cell. Biol. 2003, 23, 7198–7209. [Google Scholar] [CrossRef]
- Sarcinelli, C.; Dragic, H.; Piecyk, M.; Barbet, V.; Duret, C.; Barthelaix, A.; Ferraro-Peyret, C.; Fauvre, J.; Renno, T.; Chaveroux, C.; et al. ATF4-Dependent NRF2 Transcriptional Regulation Promotes Antioxidant Protection during Endoplasmic Reticulum Stress. Cancers 2020, 12, 569. [Google Scholar] [CrossRef]
- Kreß, J.K.C.; Jessen, C.; Hufnagel, A.; Schmitz, W.; Xavier da Silva, T.N.; Ferreira dos Santos, A.; Mosteo, L.; Goding, C.R.; Friedmann Angeli, J.P.; Meierjohann, S. The Integrated Stress Response Effector ATF4 Is an Obligatory Metabolic Activator of NRF2. Cell Rep. 2023, 42, 112724. [Google Scholar] [CrossRef]
- Huang, X.; Ordemann, J.; Müller, J.M.; Dubiel, W. The COP9 Signalosome, Cullin 3 and Keap1 Supercomplex Regulates CHOP Stability and Adipogenesis. Biol. Open 2012, 1, 705–710. [Google Scholar] [CrossRef]
- Nakade, K.; Pan, J.; Yamasaki, T.; Murata, T.; Wasylyk, B.; Yokoyama, K.K. JDP2 (Jun Dimerization Protein 2)-Deficient Mouse Embryonic Fibroblasts Are Resistant to Replicative Senescence. J. Biol. Chem. 2009, 284, 10808–10817. [Google Scholar] [CrossRef]
- Milde-Langosch, K. The Fos Family of Transcription Factors and Their Role in Tumourigenesis. Eur. J. Cancer 2005, 41, 2449–2461. [Google Scholar] [CrossRef]
- Seshadri, T.; Campisi, J. Repression of C-Fos Transcription and an Altered Genetic Program in Senescent Human Fibroblasts. Science 1990, 247, 205–209. [Google Scholar] [CrossRef]
- Delpoux, A.; Marcel, N.; Hess Michelini, R.; Katayama, C.D.; Allison, K.A.; Glass, C.K.; Quiñones-Parra, S.M.; Murre, C.; Loh, L.; Kedzierska, K.; et al. FOXO1 Constrains Activation and Regulates Senescence in CD8 T Cells. Cell Rep. 2021, 34, 108674. [Google Scholar] [CrossRef]
- Chen, P.-M.; Lin, C.-H.; Li, N.-T.; Wu, Y.-M.; Lin, M.-T.; Hung, S.-C.; Yen, M.-L. C-Maf Regulates Pluripotency Genes, Proliferation/Self-Renewal, and Lineage Commitment in ROS-Mediated Senescence of Human Mesenchymal Stem Cells. Oncotarget 2015, 6, 35404–35418. [Google Scholar] [CrossRef]
- Yang, D.; Xiao, C.; Long, F.; Wu, W.; Huang, M.; Qu, L.; Liu, X.; Zhu, Y. Fra-1 Plays a Critical Role in Angiotensin II—Induced Vascular Senescence. FASEB J. 2019, 33, 7603–7614. [Google Scholar] [CrossRef]
- Eyers, P.A.; Keeshan, K.; Kannan, N. Tribbles in the 21st Century: The Evolving Roles of Tribbles Pseudokinases in Biology and Disease. Trends Cell Biol. 2017, 27, 284–298. [Google Scholar] [CrossRef]
- Dobens, L.L.; Bouyain, S. Developmental Roles of Tribbles Protein Family Members. Dev. Dyn. 2012, 241, 1239–1248. [Google Scholar] [CrossRef]
- Lohan, F.; Keeshan, K. The Functionally Diverse Roles of Tribbles. Biochem. Soc. Trans. 2013, 41, 1096–1100. [Google Scholar] [CrossRef]
- Bowers, A.J.; Scully, S.; Boylan, J.F. SKIP3, a Novel Drosophila Tribbles Ortholog, Is Overexpressed in Human Tumors and Is Regulated by Hypoxia. Oncogene 2003, 22, 2823–2835. [Google Scholar] [CrossRef]
- Örd, T.; Örd, D.; Kaikkonen, M.U.; Örd, T. Pharmacological or TRIB3-Mediated Suppression of ATF4 Transcriptional Activity Promotes Hepatoma Cell Resistance to Proteasome Inhibitor Bortezomib. Cancers 2021, 13, 2341. [Google Scholar] [CrossRef]
- Ohoka, N.; Yoshii, S.; Hattori, T.; Onozaki, K.; Hayashi, H. TRB3, a Novel ER Stress-Inducible Gene, Is Induced via ATF4–CHOP Pathway and Is Involved in Cell Death. EMBO J. 2005, 24, 1243–1255. [Google Scholar] [CrossRef]
- Örd, D.; Örd, T. Characterization of Human NIPK (TRB3, SKIP3) Gene Activation in Stressful Conditions. Biochem. Biophys. Res. Commun. 2005, 330, 210–218. [Google Scholar] [CrossRef]
- Jousse, C.; Deval, C.; Maurin, A.-C.; Parry, L.; Chérasse, Y.; Chaveroux, C.; Lefloch, R.; Lenormand, P.; Bruhat, A.; Fafournoux, P. TRB3 Inhibits the Transcriptional Activation of Stress-Regulated Genes by a Negative Feedback on the ATF4 Pathway. J. Biol. Chem. 2007, 282, 15851–15861. [Google Scholar] [CrossRef]
- Örd, D.; Meerits, K.; Örd, T. TRB3 Protects Cells against the Growth Inhibitory and Cytotoxic Effect of ATF4. Exp. Cell Res. 2007, 313, 3556–3567. [Google Scholar] [CrossRef]
- Selim, E.; Frkanec, J.T.; Cunard, R. Fibrates Upregulate TRB3 in Lymphocytes Independent of PPARα by Augmenting CCAAT/Enhancer-Binding Proteinβ (C/EBPβ) Expression. Mol. Immunol. 2007, 44, 1218–1229. [Google Scholar] [CrossRef]
- Bezy, O.; Vernochet, C.; Gesta, S.; Farmer, S.R.; Kahn, C.R. TRB3 Blocks Adipocyte Differentiation through the Inhibition of C/EBPβ Transcriptional Activity. Mol. Cell. Biol. 2007, 27, 6818–6831. [Google Scholar] [CrossRef]
- Li, K.; Wang, F.; Cao, W.-B.; Lv, X.-X.; Hua, F.; Cui, B.; Yu, J.-J.; Zhang, X.-W.; Shang, S.; Liu, S.-S.; et al. TRIB3 Promotes APL Progression through Stabilization of the Oncoprotein PML-RARα and Inhibition of P53-Mediated Senescence. Cancer Cell 2017, 31, 697–710.e7. [Google Scholar] [CrossRef]
- Gu, Y.; Yan, R.; Wang, Y.; Zeng, Y.; Yao, Q. High TRB3 Expression Induces Chondrocyte Autophagy and Senescence in Osteoarthritis Cartilage. Aging 2022, 14, 5366–5375. [Google Scholar] [CrossRef]
- Corcoran, C.A.; Luo, X.; He, Q.; Jiang, C.; Huang, Y.; Sheikh, M.S. Genotoxic and Endoplasmic Reticulum Stresses Differentially Regulate TRB3 Expression. Cancer Biol. Ther. 2005, 4, 1063–1067. [Google Scholar] [CrossRef][Green Version]
- Wang, L.; Zhao, W.; Xia, C.; Li, Z.; Zhao, W.; Xu, K.; Wang, N.; Lian, H.; Rosas, I.O.; Yu, G. TRIB3 Mediates Fibroblast Activation and Fibrosis Though Interaction with ATF4 in IPF. Int. J. Mol. Sci. 2022, 23, 15705. [Google Scholar] [CrossRef]
- Rabouw, H.H.; Langereis, M.A.; Anand, A.A.; Visser, L.J.; de Groot, R.J.; Walter, P.; van Kuppeveld, F.J.M. Small Molecule ISRIB Suppresses the Integrated Stress Response within a Defined Window of Activation. Proc. Natl. Acad. Sci. USA 2019, 116, 2097–2102. [Google Scholar] [CrossRef]
- Li, Y.-Q.; An, X.-L.; Jin, F.-Y.; Bai, Y.-F.; Li, T.; Yang, X.-Y.; Liu, S.-P.; Gao, X.-M.; Mao, N.; Xu, H.; et al. ISRIB Inhibits the Senescence of Type II Pulmonary Epithelial Cells to Alleviate Pulmonary Fibrosis Induced by Silica in Mice. Ecotoxicol. Environ. Saf. 2023, 264, 115410. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, H.; Fan, J.; Li, Q.; Guo, R.; Pan, J.; Liu, Y.; Peng, J.; Zhu, Q.; Feng, Y.; et al. Inhibition of Integrated Stress Response Protects against Lipid-Induced Senescence in Hypothalamic Neural Stem Cells in Adamantinomatous Craniopharyngioma. Neuro. Oncol. 2023, 25, 720–732. [Google Scholar] [CrossRef]
- Rojas-Rivera, D.; Delvaeye, T.; Roelandt, R.; Nerinckx, W.; Augustyns, K.; Vandenabeele, P.; Bertrand, M.J.M. When PERK Inhibitors Turn out to Be New Potent RIPK1 Inhibitors: Critical Issues on the Specificity and Use of GSK2606414 and GSK2656157. Cell Death Differ. 2017, 24, 1100–1110. [Google Scholar] [CrossRef]
- Watanabe, T.; Ninomiya, H.; Saitou, T.; Takanezawa, S.; Yamamoto, S.; Imai, Y.; Yoshida, O.; Kawakami, R.; Hirooka, M.; Abe, M.; et al. Therapeutic Effects of the PKR Inhibitor C16 Suppressing Tumor Proliferation and Angiogenesis in Hepatocellular Carcinoma in Vitro and in Vivo. Sci. Rep. 2020, 10, 5133. [Google Scholar] [CrossRef]
- Boyce, M.; Bryant, K.F.; Jousse, C.; Long, K.; Harding, H.P.; Scheuner, D.; Kaufman, R.J.; Ma, D.; Coen, D.M.; Ron, D.; et al. A Selective Inhibitor of EIF2α Dephosphorylation Protects Cells from ER Stress. Science 2005, 307, 935–939. [Google Scholar] [CrossRef]
- Kessel, D. Protection of Bcl-2 by Salubrinal. Biochem. Biophys. Res. Commun. 2006, 346, 1320–1323. [Google Scholar] [CrossRef]
- Li, L.; Hu, G.; Xie, R.; Yang, J.; Shi, X.; Jia, Z.; Qu, X.; Wang, M.; Wu, Y. Salubrinal-Mediated Activation of EIF2α Signaling Improves Oxidative Stress-Induced BMSCs Senescence and Senile Osteoporosis. Biochem. Biophys. Res. Commun. 2022, 610, 70–76. [Google Scholar] [CrossRef]
- Keller, T.L.; Zocco, D.; Sundrud, M.S.; Hendrick, M.; Edenius, M.; Yum, J.; Kim, Y.-J.; Lee, H.-K.; Cortese, J.F.; Wirth, D.F.; et al. Halofuginone and Other Febrifugine Derivatives Inhibit Prolyl-TRNA Synthetase. Nat. Chem. Biol. 2012, 8, 311–317. [Google Scholar] [CrossRef]
- Pitera, A.P.; Szaruga, M.; Peak-Chew, S.; Wingett, S.W.; Bertolotti, A. Cellular Responses to Halofuginone Reveal a Vulnerability of the GCN2 Branch of the Integrated Stress Response. EMBO J. 2022, 41, e109985. [Google Scholar] [CrossRef]
- Tsuchida, K.; Tsujita, T.; Hayashi, M.; Ojima, A.; Keleku-Lukwete, N.; Katsuoka, F.; Otsuki, A.; Kikuchi, H.; Oshima, Y.; Suzuki, M.; et al. Halofuginone Enhances the Chemo-Sensitivity of Cancer Cells by Suppressing NRF2 Accumulation. Free Radic. Biol. Med. 2017, 103, 236–247. [Google Scholar] [CrossRef]
- Chen, T.; Ozel, D.; Qiao, Y.; Harbinski, F.; Chen, L.; Denoyelle, S.; He, X.; Zvereva, N.; Supko, J.G.; Chorev, M.; et al. Chemical Genetics Identify EIF2α Kinase Heme-Regulated Inhibitor as an Anticancer Target. Nat. Chem. Biol. 2011, 7, 610–616. [Google Scholar] [CrossRef]
- Xu, Y.; Yuan, H.; Luo, Y.; Zhao, Y.-J.; Xiao, J.-H. Ganoderic Acid D Protects Human Amniotic Mesenchymal Stem Cells against Oxidative Stress-Induced Senescence through the PERK/NRF2 Signaling Pathway. Oxid. Med. Cell. Longev. 2020, 2020, 8291413. [Google Scholar] [CrossRef]

| Transcription Factor | Interacting Partner | Effect on Senescence | References |
|---|---|---|---|
| ATF4 | CEBPA, CEBPG | Senescence alleviation through regulation of CARE-containing genes | Horiguchi, M. et al. (2012) [158]; Huggins, C.J. et al. (2016) [159]; Huggins, C.J. et al. (2013) [160] |
| CHOP | Stimulation of pro-senescence protein p21 expression | Inoue, Y. et al. (2017) [165] | |
| NRF2 | Senescence alleviation presumably through the expression of NRF2-target genes, which exert an anti-senescence effect | He, C.H. et al. (2001) [170] | |
| ATF3 | Not stated | Stimulation of replicative senescence through upregulation of pro-senescence proteins like p16 and p21 | Zhang, C. et al. (2021) [139] |
| ATF3 knockdown results in aggravated senescence in macrophages exposed to Pseudomonas aeruginosa | Zhao, Q. et al. (2021) [140] | ||
| Aggravation of acrylamide-induced senescence by upregulation of p53 and p21 pro-senescent proteins in macrophages | Kim, K.-H. et al. (2015) [141] | ||
| ATF4 | Not stated | Upregulation of pro-senescence protein p16 in senescent renal tubular epithelial cells | Liu, J. et al. (2015) [146] |
| Upregulation of pro-senescence protein p16 in oxidative stress induces senescence in MEFs | Sakai, T. et al. (2019) [147] | ||
| CHOP | Not stated | Senescence aggravation in alveolar epithelial cells from patients with idiopathic pulmonary fibrosis through enhancement of ROS generation and activation of the NF-κB pathway—factors promoting senescence | Jing, X. et al. (2022) [166] |
| CEBPA | Not stated | Senescence aggravation by upregulation pro-senescence factors: CXCL1, CXCL5 and CXCL15 | Guan, Y. et al. (2020) [162] |
| JDP2 | Not stated | Aggravation of MEFs replicative senescence by upregulation of p16 and p19 proteins | Nakade, K. et al. (2009) [178] |
| FOS | Not stated | FOS expression is significantly increased during fibroblast replicative senescence | Seshadri, T. et al. (1990) [180] |
| Not stated | FOS is upregulated in senescent T-cells, whether FOXO1 ensures the naïve state of T-cells through downregulation of FOS and FOSB | Delpoux, A. et al. (2021) [181] | |
| MAF | Not stated | MAF disappears in senescent hADMSCs, leading to reduced osteogenic differentiation capacity | Chen P.-M., et al. (2015) [182] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalinin, A.; Zubkova, E.; Menshikov, M. Integrated Stress Response (ISR) Pathway: Unraveling Its Role in Cellular Senescence. Int. J. Mol. Sci. 2023, 24, 17423. https://doi.org/10.3390/ijms242417423
Kalinin A, Zubkova E, Menshikov M. Integrated Stress Response (ISR) Pathway: Unraveling Its Role in Cellular Senescence. International Journal of Molecular Sciences. 2023; 24(24):17423. https://doi.org/10.3390/ijms242417423
Chicago/Turabian StyleKalinin, Alexander, Ekaterina Zubkova, and Mikhail Menshikov. 2023. "Integrated Stress Response (ISR) Pathway: Unraveling Its Role in Cellular Senescence" International Journal of Molecular Sciences 24, no. 24: 17423. https://doi.org/10.3390/ijms242417423
APA StyleKalinin, A., Zubkova, E., & Menshikov, M. (2023). Integrated Stress Response (ISR) Pathway: Unraveling Its Role in Cellular Senescence. International Journal of Molecular Sciences, 24(24), 17423. https://doi.org/10.3390/ijms242417423







