Pan-genome Analysis of WOX Gene Family and Function Exploration of CsWOX9 in Cucumber
Abstract
:1. Introduction
2. Results
2.1. Identification of WOX Genes Based on Pan-genome of Cucumber
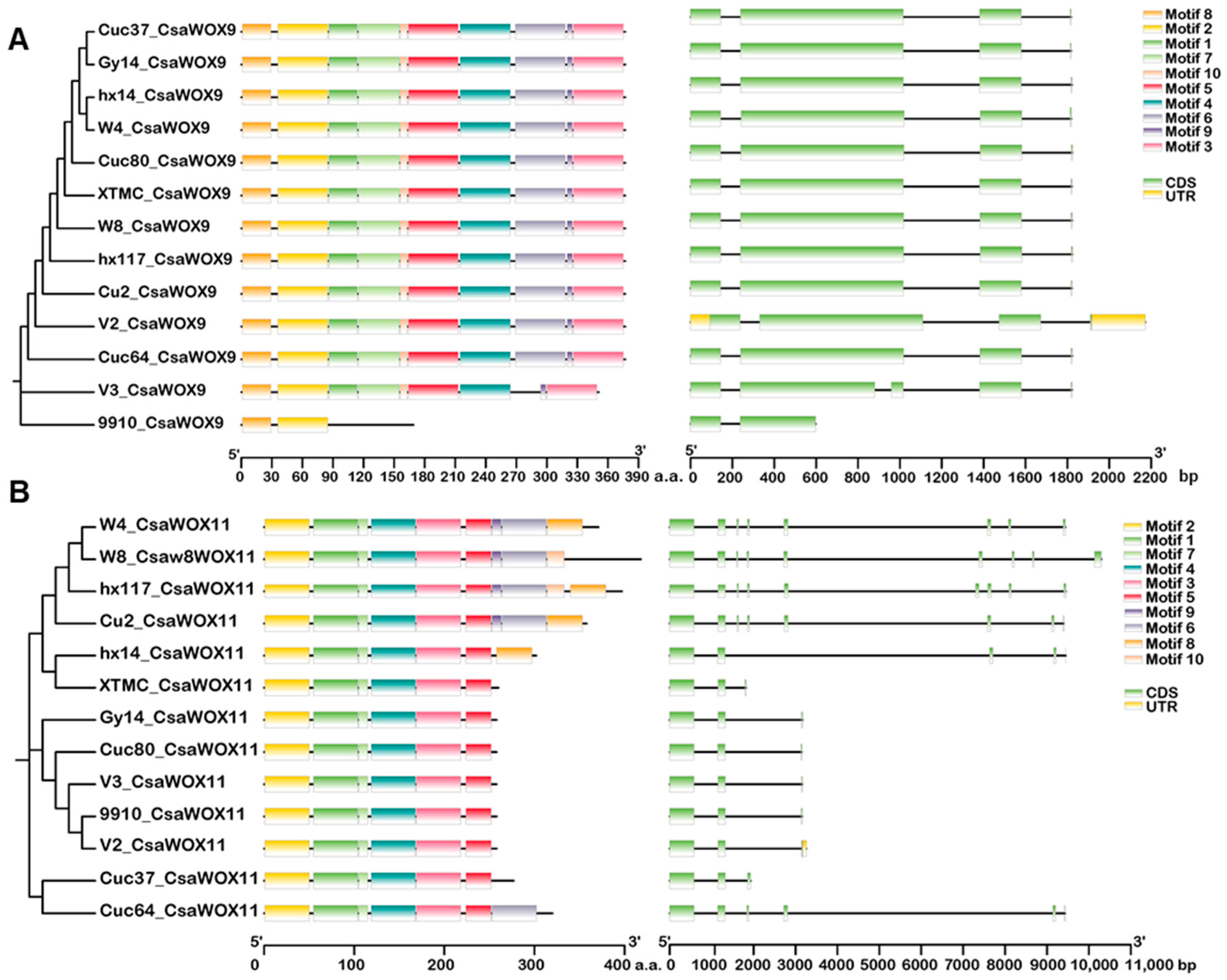
2.2. Gene Structure and Motif Composition of CsWOXs
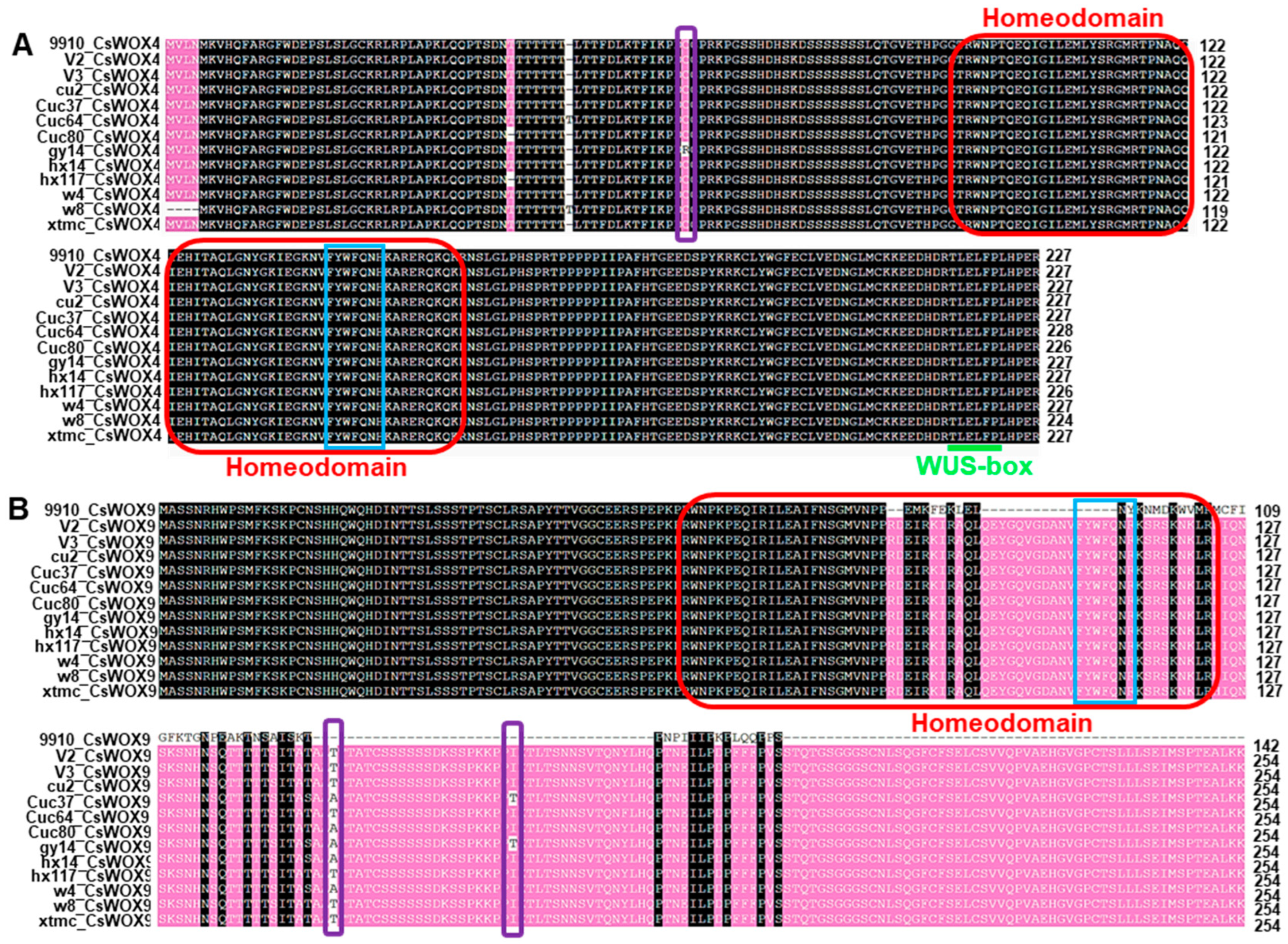
2.3. Multiple Sequence Alignment of CsWOXs
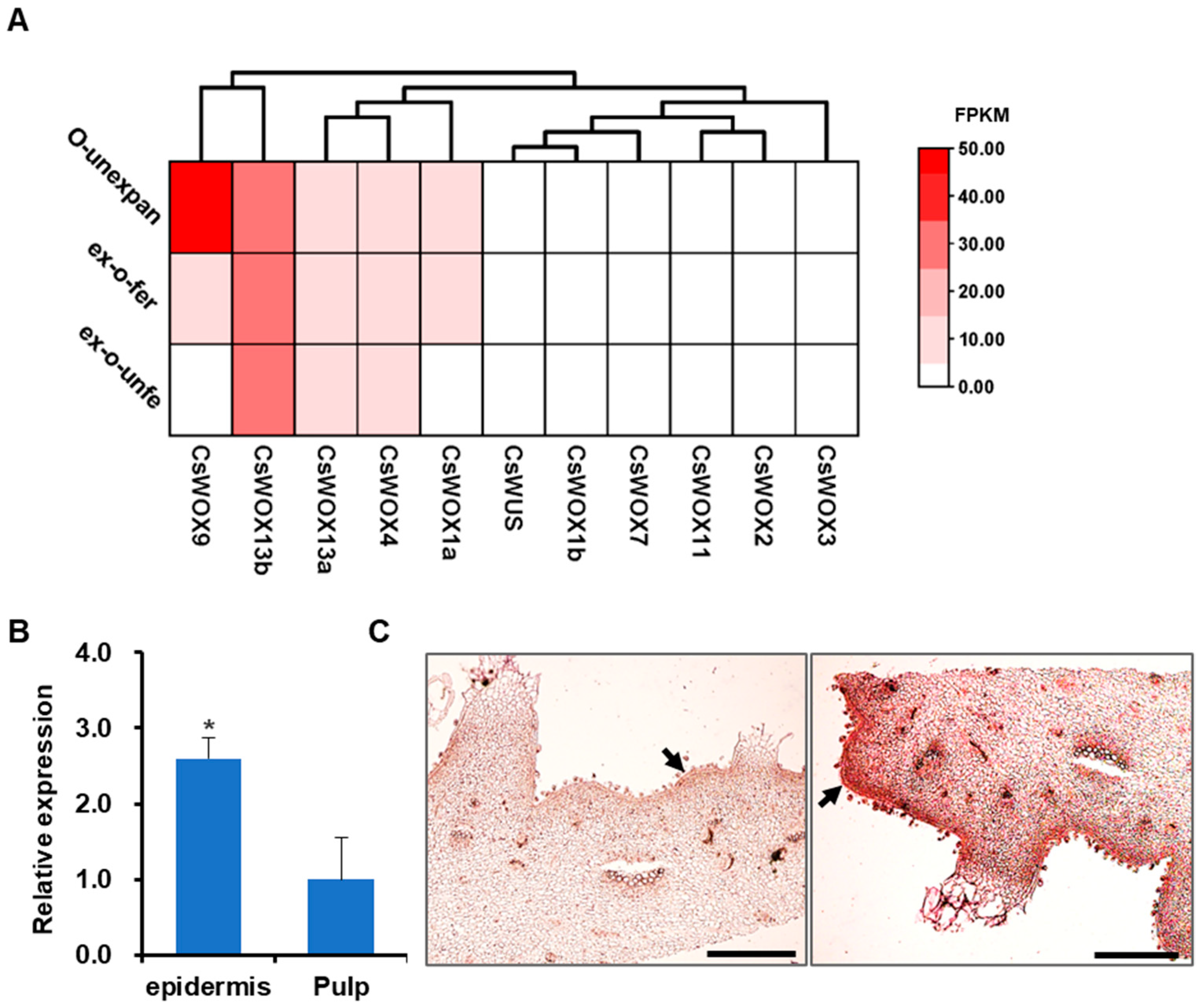
2.4. Identification of Fruit-Specific Expressed Gene CsWOX9
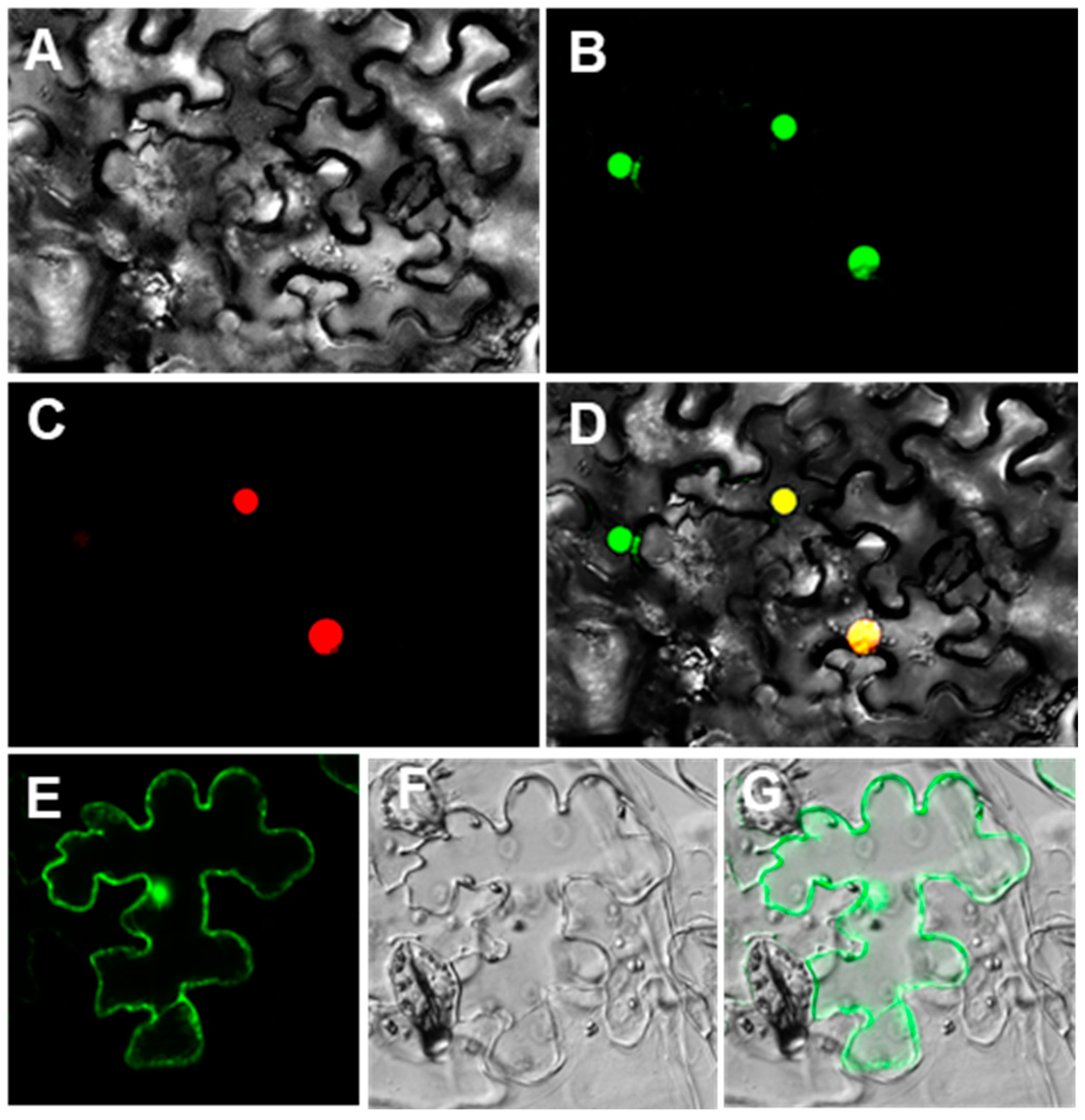
2.5. Subcellular Localization of CsWOX9
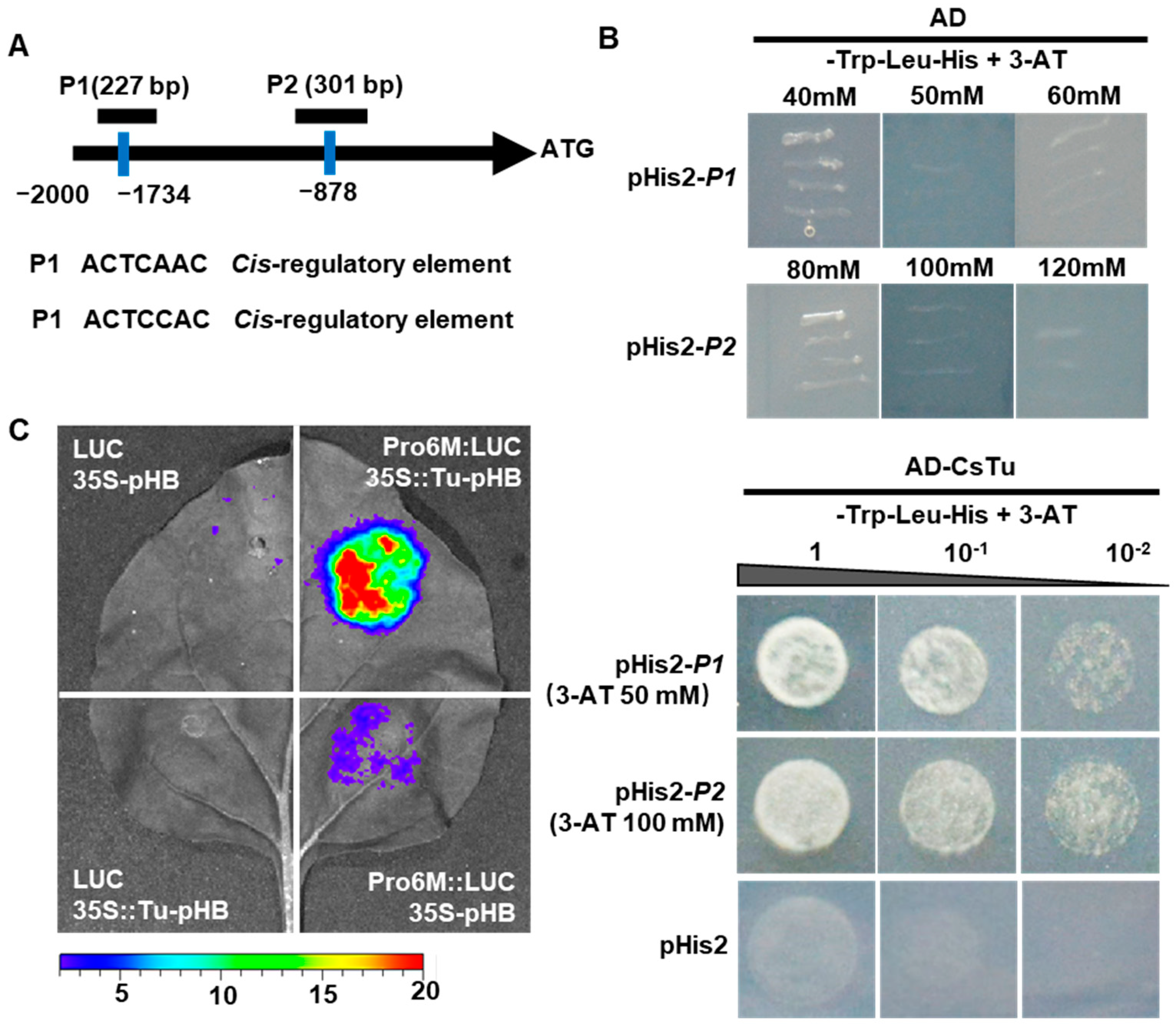
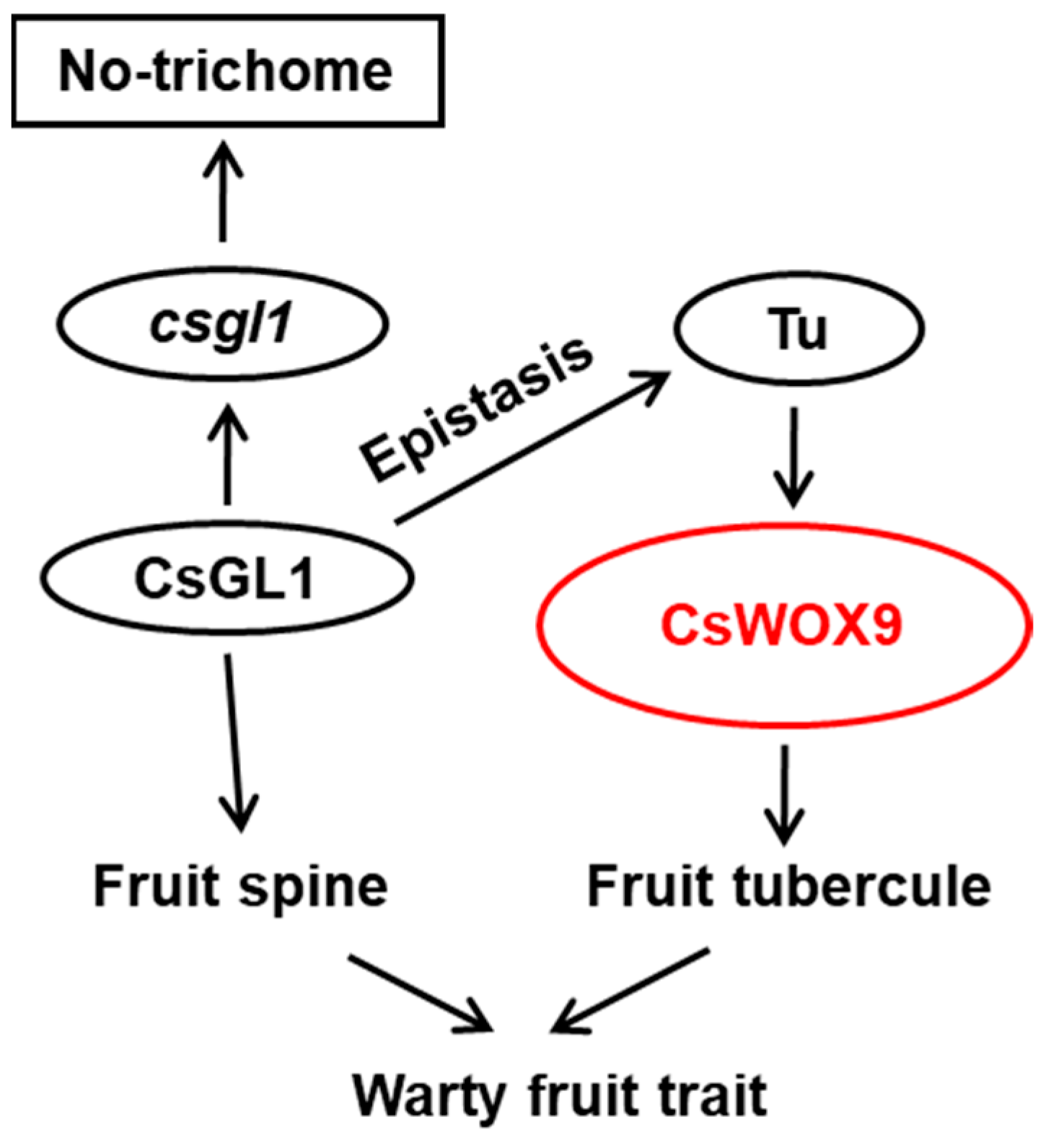
2.6. Tuberculate Fruit Gene Tu Directly Activates CsWOX9 by Binding to Its Promoter
3. Discussion
3.1. Diversification of the CsWOXs in Cucumber Pan-genome
3.2. Identification of Fruit-Specific Expressed CsWOXs in Regulating Fruit Morphogenesis
4. Materials and Methods
4.1. Identification and Phylogenetic Analysis of CsWOX Genes
4.2. Bioinformatics Analysis of CsWOX Genes
4.3. Transcriptome Analysis of WOX Genes in Cucumber Ovaries
4.4. Expression Analysis of CsWOX9 by qRT-PCR
4.5. In Situ Hybridization
4.6. Subcellular Localization of CsWOX9
4.7. Y1H Assay
4.8. Dual-LUC Transaction Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Y1H | yeast one-hybrid assay |
| DLR | dual-luciferase reporter |
| WOX | WUSCHEL-related homeobox |
| HD | homeodomain |
| TFs | transcription factors |
| WUS | WUSCHEL |
| RAMs | root apical meristems |
| EVG | EVERGREEN |
| PFS2 | PRETTY FEW SEEDS2 |
| a.a. | amino acids |
| qRT-PCR | quantitative RT-PCR |
| 3-AT | 3-amino-1,2,4-triazole |
| SGR | STAYGREEN |
| DWT1 | DWARF TILLER1 |
| SD | synthetic dextrose |
| DBA | days before anthesis |
References
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Brocchieri, L.; Burglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Z.; Che, Q.Q.; Cheng, C.X.; Yuan, Y.B.; Wang, Y.Z. Genome-wide identification of WOX gene family in apple and a functional analysis of MdWOX4b during adventitious root formation. J. Integr. Agric. 2022, 21, 1332–1345. [Google Scholar] [CrossRef]
- Hao, Q.N.; Zhang, L.; Yang, Y.Y.; Shan, Z.H.; Zhou, X.A. Genome-Wide Analysis of the WOX Gene Family and Function Exploration of GmWOX18 in Soybean. Plants 2019, 8, 215. [Google Scholar] [CrossRef]
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef]
- Cheng, S.F.; Huang, Y.L.; Zhu, N.; Zhao, Y. The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 2014, 549, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.J.; Affolter, M.; Burglin, T. Homeodomain proteins. Annu. Rev. Biochem. 1994, 63, 487–526. [Google Scholar] [CrossRef]
- Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar]
- Costanzo, E.; Trehin, C.; Vandenbussche, M. The role of WOX genes in flower development. Ann. Bot. 2014, 114, 1545–1553. [Google Scholar] [CrossRef]
- Deveaux, Y.; Toffano-Nioche, C.; Claisse, G.; Thareau, V.; Morin, H.; Laufs, P.; Moreau, H.; Kreis, M.; Lecharny, A. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 2008, 8, 291. [Google Scholar] [CrossRef]
- Rathour, M.; Sharma, A.; Kaur, A.; Upadhyay, S.K. Genome-wide characterization and expression and co-expression analysis suggested diverse functions of WOX genes in bread wheat. Heliyon 2020, 6, e05762. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jurgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.X.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 2010, 22, 2618–2629. [Google Scholar] [CrossRef]
- Pi, L.M.; Aichinger, E.; van der Graaff, E.; Llavata-Peris, C.I.; Weijers, D.; Hennig, L.; Groot, E.; Laux, T. Organizer-Derived WOX5 Signal Maintains Root Columella Stem Cells through Chromatin-Mediated Repression of CDF4 Expression. Dev. Cell 2015, 33, 576–588. [Google Scholar] [CrossRef]
- Osipova, M.A.; Mortier, V.; Demchenko, K.N.; Tsyganov, V.E.; Tikhonovich, I.A.; Lutova, L.A.; Dolgikh, E.A.; Goormachtig, S. Wuschel-related homeobox5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiol. 2012, 158, 1329–1341. [Google Scholar] [CrossRef]
- Zhou, S.L.; Jiang, W.; Long, F.; Cheng, S.F.; Yang, W.J.; Zhao, Y.; Zhou, D.X. Rice Homeodomain Protein WOX11 Recruits a Histone Acetyltransferase Complex to Establish Programs of Cell Proliferation of Crown Root Meristem. Plant Cell 2017, 29, 1088–1104. [Google Scholar] [CrossRef]
- Kong, D.; Hao, Y.; Cui, H. The WUSCHEL Related Homeobox Protein WOX7 Regulates the Sugar Response of Lateral Root Development in Arabidopsis thaliana. Mol. Plant 2016, 9, 261–270. [Google Scholar] [CrossRef]
- Chen, R.; Xu, N.; Yu, B.; Wu, Q.; Li, X.; Wang, G.; Huang, J. The WUSCHEL-related homeobox transcription factor OsWOX4 controls the primary root elongation by activating OsAUX1 in rice. Plant Sci. 2020, 298, 110575. [Google Scholar] [CrossRef]
- Li, J.B.; Jia, H.X.; Sun, P.; Zhang, J.; Xia, Y.X.; Hu, J.J.; Wang, L.J.; Lu, M.Z. The WUSCHELa (PtoWUSa) is involved in developmental plasticity of adventitious root in poplar. Genes 2020, 11, 176. [Google Scholar] [CrossRef] [PubMed]
- Li, J.B.; Zhang, J.; Jia, H.X.; Liu, B.B.; Sun, P.; Hu, J.J.; Wang, L.J.; Lu, M.Z. The WUSCHEL-related homeobox 5a(PtoWOX5a) is involved in adventitious root development in poplar. Tree Physiol. 2018, 38, 139–153. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, J.; Wang, X.; Li, C.; Ye, Z.; Zhang, J. UF, a WOX gene, regulates a novel phenotype of un-fused flower in tomato. Plant Sci. 2020, 297, 110523. [Google Scholar] [CrossRef] [PubMed]
- Park, S.O. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 2005, 132, 841–849. [Google Scholar] [CrossRef]
- Park, S.O.; Hwang, S.; Hauser, B.A. The phenotype of Arabidopsis ovule mutants mimics the morphology of primitive seed plants. Proc. Biol. Sci. 2004, 271, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Gu, R.; Song, X.; Liu, X.; Yan, L.; Zhang, X. Genome-wide analysis of CsWOX transcription factor gene family in cucumber (Cucumis sativus L.). Sci. Rep. 2020, 10, 6216. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, W.; He, H.; Nie, J.; Bie, B.; Zhao, J.; Ren, G.; Li, Y.; Zhang, D.; Pan, J.; et al. Tuberculate fruit gene Tu encodes a C2H2 zinc finger protein that is required for the warty fruit phenotype in cucumber (Cucumis sativus L.). Plant J. 2014, 78, 1034–1046. [Google Scholar] [CrossRef]
- Che, G.; Zhang, X. Molecular basis of cucumber fruit domestication. Curr. Opin. Plant Biol. 2019, 47, 38–46. [Google Scholar] [CrossRef]
- Han, N.; Tang, R.; Chen, X.; Xu, Z.; Ren, Z.; Wang, L. Genome-wide identification and characterization of WOX genes in Cucumis sativus. Genome 2021, 64, 761–776. [Google Scholar] [CrossRef]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Zheng, H.; Luo, R.; Zhu, H.; Li, Q.; Qian, W.; Ren, Y.; Tian, G.; Li, J.; et al. Building the sequence map of the human pan-genome. Nat. Biotechnol. 2010, 28, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Gonda, I.; Sun, H.H.; Ma, Q.Y.; Bao, K.; Tieman, D.M.; Burzynski-Chang, E.A.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L.; et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.F.; Luo, H.; Xu, J.B.; Cruickshank, A.; Zhao, X.R.; Teng, F.; Hathorn, A.; Wu, X.Y.; Liu, Y.M.; Shatte, T.; et al. Extensive variation within the pan-genome of cultivated and wild sorghum. Nat. Plants 2021, 7, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, S.; Chai, S.; Yang, Z.; Zhang, Z. Graph-based pan-genome reveals structural and sequence variations related to agronomic traits and domestication in cucumber. Nat. Commun. 2022, 13, 682. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tan, J.; Wu, Z.; VandenLangenberg, K.; Wehner, T.; Wen, C.; Zheng, X.; Owens, K.; Thornton, A.; Bang, H.; et al. STAYGREEN, STAY HEALTHY: A loss-of-susceptibility mutation in the STAYGREEN gene provides durable, broad-spectrum disease resistances for over 50 years of US cucumber production. New Phytol. 2019, 221, 415–430. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, A.; Zhou, Z.; Zhao, Y.; Yan, A.; Bao, S.; Yu, H.; Gan, Y. GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol. 2015, 206, 220–230. [Google Scholar] [CrossRef]
- Li, Q.; Cao, C.X.; Zhang, C.J.; Zheng, S.S.; Wang, Z.H.; Wang, L.N.; Ren, Z.H. The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J. Exp. Bot. 2015, 66, 2515–2526. [Google Scholar] [CrossRef]
- Wang, W.F.; Li, G.; Zhao, J.; Chu, H.W.; Lin, W.H.; Zhang, D.B.; Wang, Z.Y.; Liang, W.Q. DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice. PLoS Genet. 2014, 10, e1004154. [Google Scholar] [CrossRef]
- Wolabu, T.W.; Wang, H.; Tadesse, D.; Zhang, F.; Behzadirad, M.; Tvorogova, V.E.; Abdelmageed, H.; Liu, Y.; Chen, N.; Chen, J.; et al. WOX9 functions antagonistic to STF and LAM1 to regulate leaf blade expansion in Medicago truncatula and Nicotiana sylvestris. New Phytol. 2021, 229, 1582–1597. [Google Scholar] [CrossRef] [PubMed]
- Rebocho, A.B.; Bliek, M.; Kusters, E.; Castel, R.; Procissi, A.; Roobeek, I.; Souer, E.; Koes, R. Role of EVERGREEN in the development of the cymose petunia inflorescence. Dev. Cell 2008, 15, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011, 12, 540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, Y.; Ding, L.; Wu, Z.; Liu, R.; Meyerowitz, E.M. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 2013, 25, 83–101. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]

| Gene Name | Gene ID 1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9930-V3 * | 9930-V2 * | XTMC | Cu2 | Cuc37 | Cuc64 | Cuc80 | W4 | W8 | Hx14 | Hx117 | Gy14 | 9910gt | |
| CsWOX1a | 1G007120 | 1G042780 | 1G007180 | 1G007360 | 1G007230 | 1G007200 | 1G007190 | 1G007200 | 1G007230 | 1G013410 | 1G010430 | 1G012440 | 1G007480 |
| CsWOX1b | 1G004310 | 1G025040 | 1G004260 | 1G004440 | 1G004350 | 1G004310 | 1G004310 | 1G004370 | 1G004340 | 1G009460 | 1G007470 | 1G008510 | 1G004470 |
| CsWOX2 | 1G034050 | 1G505930 | - | - | - | - | - | - | - | - | - | - | - |
| CsWOX3 | 6G021960 | 6G301060 | 6G032580 | 6G023220 | 6G018920 | 6G021510 | 6G052960 | 6G019330 | 6G022530 | 6G032230 | 6G029450 | 6G030400 | 6G023370 |
| CsWOX4 | 2G026510 | 2G356610 | 2G028100 | 2G027900 | 2G097530 | 2G067930 | 2G100850 | 2G032030 | 2G040170 | 2G036130 | 2G036990 | 2G034990 | 2G028340 |
| CsWOX7 | 6G001670 | 6G010010 | 6G001650 | 6G001610 | 6G001650 | 6G001660 | 6G003640 | 6G001620 | 6G001620 | 6G002610 | 6G001650 | 6G004680 | 6G001650 |
| CsWOX9 | 6G050540 | 6G518270 | 6G062220 | 6G046400 | 6G046950 | 6G045020 | 6G100570 | 6G043880 | 6G044120 | 6G063500 | 6G055780 | 6G056710 | 6G047130 |
| CsWOX11 | 3G039320 | 3G812740 | 3G059960 | 3G046380 | 3G055190 | 3G060870 | 3G051480 | 3G046100 | 3G044550 | 3G064800 | 3G065520 | 3G060130 | 3G046900 |
| CsWOX13a | 3G000330 | 3G002330 | - | 3G000300 | 3G000320 | 3G000320 | 3G000280 | 3G000310 | 3G000300 | 3G000320 | 3G000310 | 3G000330 | 3G000310 |
| CsWOX13b | 4G037370 | 4G663700 | 4G049750 | 4G040360 | 4G101470 | 4G038090 | 4G101290 | 4G033260 | 4G039360 | 4G048450 | 4G045640 | 4G047420 | 4G042180 |
| CsWUS | 6G047050 | 6G505860 | 6G058810 | 6G042980 | 6G043520 | 6G041670 | 6G097310 | 6G040480 | 6G040750 | 6G059150 | 6G052360 | 6G052360 | 6G043720 |
| Protein Name | 9930_V3 * | 9930_V2 * | XTMC | Cu2 | Cuc37 | Cuc64 | Cuc80 | W4 | W8 | Hx14 | Hx117 | Gy14 | 9910gt |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CsWOX1a | 390 | 387 | 390 | 339 | 390 | 389 | 390 | 390 | 388 | 390 | 390 | 390 | 339 |
| CsWOX1b | 334 | 334 | 334 | 334 | 334 | 333 | 333 | 334 | 334 | 334 | 334 | 334 | 334 |
| CsWOX2 | 239 | 239 | - | - | - | - | - | - | - | - | - | - | - |
| CsWOX3 | 193 | 193 | 193 | 193 | 193 | 187 | 193 | 193 | 198 | 193 | 187 | 193 | 193 |
| CsWOX4 | 227 | 227 | 227 | 227 | 227 | 228 | 226 | 227 | 224 | 227 | 226 | 227 | 227 |
| CsWOX7 | 196 | 130 | 116 | 196 | 196 | 196 | 196 | 196 | 196 | 196 | 180 | 168 | 196 |
| CsWOX9 | 350 | 376 | 376 | 376 | 376 | 376 | 376 | 376 | 376 | 376 | 376 | 376 | 168 |
| CsWOX11 | 257 | 257 | 259 | 357 | 276 | 319 | 257 | 370 | 417 | 301 | 396 | 257 | 257 |
| CsWOX13a | 201 | 282 | - | 282 | 282 | 282 | 282 | 282 | 282 | 282 | 282 | 282 | 282 |
| CsWOX13b | 314 | 269 | 269 | 269 | 317 | 269 | 317 | 317 | 317 | 317 | 317 | 269 | 317 |
| CsWUS | 233 | 304 | 233 | 233 | 233 | 233 | 233 | 230 | 233 | 233 | 233 | 233 | 233 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, S.; Zhao, L.; Liu, J.; Sun, Y.; Li, B.; Wang, L.; Ren, Z.; Chen, C. Pan-genome Analysis of WOX Gene Family and Function Exploration of CsWOX9 in Cucumber. Int. J. Mol. Sci. 2023, 24, 17568. https://doi.org/10.3390/ijms242417568
Yin S, Zhao L, Liu J, Sun Y, Li B, Wang L, Ren Z, Chen C. Pan-genome Analysis of WOX Gene Family and Function Exploration of CsWOX9 in Cucumber. International Journal of Molecular Sciences. 2023; 24(24):17568. https://doi.org/10.3390/ijms242417568
Chicago/Turabian StyleYin, Shuai, Lili Zhao, Jiaqi Liu, Yanjie Sun, Bohong Li, Lina Wang, Zhonghai Ren, and Chunhua Chen. 2023. "Pan-genome Analysis of WOX Gene Family and Function Exploration of CsWOX9 in Cucumber" International Journal of Molecular Sciences 24, no. 24: 17568. https://doi.org/10.3390/ijms242417568
APA StyleYin, S., Zhao, L., Liu, J., Sun, Y., Li, B., Wang, L., Ren, Z., & Chen, C. (2023). Pan-genome Analysis of WOX Gene Family and Function Exploration of CsWOX9 in Cucumber. International Journal of Molecular Sciences, 24(24), 17568. https://doi.org/10.3390/ijms242417568





