A Comprehensive Look at In Vitro Angiogenesis Image Analysis Software
Abstract
1. Introduction
2. Vascular Tissue
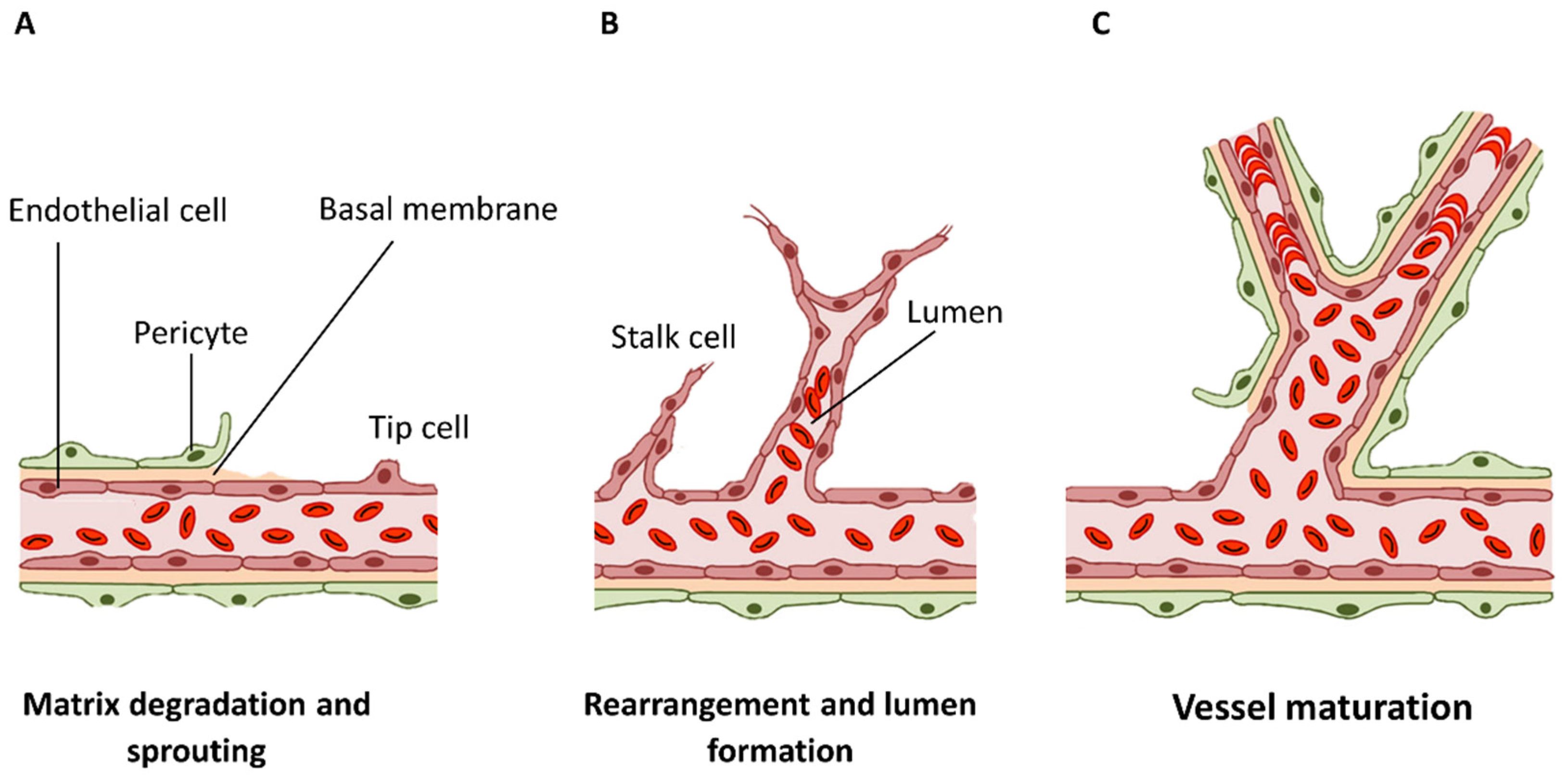
2.1. Blood Vessel Development
Angiogenesis Mechanism for Adaptation and Stabilization
3. Hydrogels Applied in Pre-Vascularization
3.1. Hydrogel Structure and Properties
3.2. Hydrogels for Angiogenesis and In Vitro Analysis
4. Imaging and Analyzing Assays in Angiogenesis
Microscopy Techniques: Optical and Fluorescence Microscopy
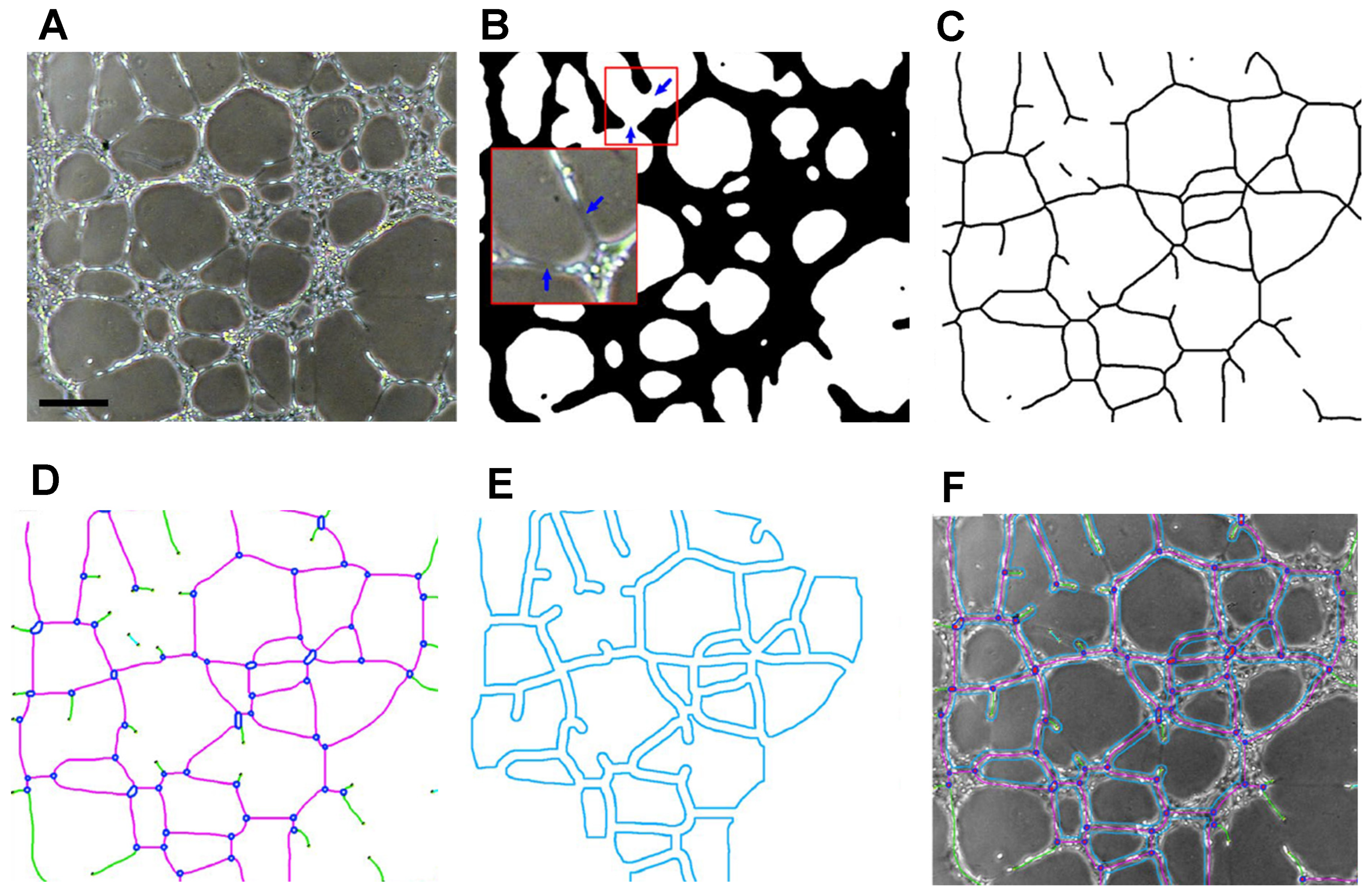
5. Image Analysis
5.1. ImageJ
5.2. Alternatives to ImageJ
5.3. Future Perspectives
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wahid, F.; Khan, T.; Hussain, Z.; Ullah, H. Nanocomposite scaffolds for tissue engineering; properties, preparation and applications. In Applications of Nanocomposite Materials in Drug Delivery; Woodhead Publishing: Duxford, UK, 2018; pp. 701–735. [Google Scholar]
- O’Brien, F.J. Biomaterials & scaffolds for tissue engineering. Mater. Today 2011, 14, 88–95. [Google Scholar] [CrossRef]
- Ahmed, S.; Chauhan, V.M.; Ghaemmaghami, A.M.; Aylott, J.W. New generation of bioreactors that advance extracellular matrix modelling and tissue engineering. Biotechnol. Lett. 2019, 41, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Kottamasu, P.; Herman, I. Engineering a microcirculation for perfusion control of ex vivo-assembled organ systems: Challenges and opportunities. J. Tissue Eng. 2018, 9, 2041731418772949. [Google Scholar] [CrossRef]
- Feijao, T.; Neves, M.I.; Sousa, A.; Torres, A.L.; Bidarra, S.J.; Orge, I.D.; Carvalho, D.T.O.; Barrias, C.C. Engineering injectable vascularized tissues from the bottom-up: Dynamics of in-gel extra-spheroid dermal tissue assembly. Biomaterials 2021, 279, 121222. [Google Scholar] [CrossRef] [PubMed]
- Jamalpoor, Z.; Taromi, N. Pre-vascularization of biomimetic 3-D scaffolds via direct co-culture of human umbilical cord derived osteogenic and angiogenic progenitor cells. J. Drug Deliv. Sci. Technol. 2021, 65, 102703. [Google Scholar] [CrossRef]
- Ren, Y.; Senarathna, J.; Grayson, W.L.; Pathak, A.P. State-of-the-art techniques for imaging the vascular microenvironment in craniofacial bone tissue engineering applications. Am. J. Physiol. Cell Physiol. 2022, 323, C1524–C1538. [Google Scholar] [CrossRef] [PubMed]
- Corliss, B.A.; Doty, R.W.; Mathews, C.; Yates, P.A.; Zhang, T.; Peirce, S.M. REAVER: A program for improved analysis of high-resolution vascular network images. Microcirculation 2020, 27, e12618. [Google Scholar] [CrossRef] [PubMed]
- Spangenberg, P.; Hagemann, N.; Squire, A.; Förster, N.; Krauß, S.D.; Qi, Y.; Mohamud Yusuf, A.; Wang, J.; Grüneboom, A.; Kowitz, L.; et al. Rapid and fully automated blood vasculature analysis in 3D light-sheet image volumes of different organs. Cell Rep. Methods 2023, 3, 100436. [Google Scholar] [CrossRef]
- Rahman, H.S.; Tan, B.L.; Othman, H.H.; Chartrand, M.S.; Pathak, Y.; Mohan, S.; Abdullah, R.; Alitheen, N.B. An Overview of in Vitro, in Vivo, and Computational Techniques for Cancer-Associated Angiogenesis Studies. BioMed Res. Int. 2020, 2020, 8857428. [Google Scholar] [CrossRef]
- Claesson-Welsh, L. Vascular permeability—The essentials. Ups. J. Med. Sci. 2015, 120, 135–143. [Google Scholar] [CrossRef]
- Mazurek, R.; Dave, J.M.; Chandran, R.R.; Misra, A.; Sheikh, A.Q.; Greif, D.M. Vascular Cells in Blood Vessel Wall Development and Disease. Adv. Pharmacol. 2017, 78, 323–350. [Google Scholar] [CrossRef]
- Shiu, Y.-T.; Weiss, J.A.; Hoying, J.B.; Iwamoto, M.N.; Joung, I.S.; Quam, C.T. The Role of Mechanical Stresses in Angiogenesis. Crit. Rev. Biomed. Eng. 2005, 33, 431–510. [Google Scholar] [CrossRef] [PubMed]
- Esser, T.U.; Roshanbinfar, K.; Engel, F.B. Promoting vascularization for tissue engineering constructs: Current strategies focusing on HIF-regulating scaffolds. Expert Opin. Biol. Ther. 2019, 19, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Mastrullo, V.; Cathery, W.; Velliou, E.; Madeddu, P.; Campagnolo, P. Angiogenesis in Tissue Engineering: As Nature Intended? Front. Bioeng. Biotechnol. 2020, 8, 188. [Google Scholar] [CrossRef] [PubMed]
- Laschke, M.W.; Gu, Y.; Menger, M.D. Replacement in angiogenesis research: Studying mechanisms of blood vessel development by animal-free in vitro, in vivo and in silico approaches. Front. Physiol. 2022, 13, 981161. [Google Scholar] [CrossRef] [PubMed]
- Irvin, M.W.; Zijlstra, A.; Wikswo, J.P.; Pozzi, A. Techniques and assays for the study of angiogenesis. Exp. Biol. Med. 2014, 239, 1476–1488. [Google Scholar] [CrossRef] [PubMed]
- Ardakani, M.R.; Hormozi, A.K.; Ardakani, J.R.; Davarpanahjazi, A.H.; Moghadam, A.S. Introduction of a potent single-donor fibrin glue for vascular anastomosis: An animal study. J. Res. Med. Sci. 2012, 17, 461–465. [Google Scholar]
- Hunt, J.A.; Chen, R.; van Veen, T.; Bryan, N. Hydrogels for tissue engineering and regenerative medicine. J. Mater. Chem. B 2014, 2, 5319–5338. [Google Scholar] [CrossRef]
- Montalbano, G.; Toumpaniari, S.; Popov, A.; Duan, P.; Chen, J.; Dalgarno, K.; Scott, W.E., 3rd; Ferreira, A.M. Synthesis of bioinspired collagen/alginate/fibrin based hydrogels for soft tissue engineering. Mater. Sci. Eng. C Mater. Biol. Appl. 2018, 91, 236–246. [Google Scholar] [CrossRef]
- O’Dwyer, J.; Murphy, R.; Dolan, E.B.; Kovarova, L.; Pravda, M.; Velebny, V.; Heise, A.; Duffy, G.P.; Cryan, S.A. Development of a nanomedicine-loaded hydrogel for sustained delivery of an angiogenic growth factor to the ischaemic myocardium. Drug Deliv. Transl. Res. 2020, 10, 440–454. [Google Scholar] [CrossRef]
- Thakur, S.; Thakur, V.K.; Arotiba, O.A. History, Classification, Properties and Application of Hydrogels: An Overview. In Hydrogels: Recent Advances; Thakur, V.K., Thakur, M.K., Eds.; Springer: Singapore, 2018; pp. 29–50. [Google Scholar]
- Chai, Q.; Jiao, Y.; Yu, X. Hydrogels for Biomedical Applications: Their Characteristics and the Mechanisms behind Them. Gels 2017, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Oyen, M.L. Mechanical characterisation of hydrogel materials. Int. Mater. Rev. 2014, 59, 44–59. [Google Scholar] [CrossRef]
- Roh, H.S.; Lee, C.M.; Hwang, Y.H.; Kook, M.S.; Yang, S.W.; Lee, D.; Kim, B.H. Addition of MgO nanoparticles and plasma surface treatment of three-dimensional printed polycaprolactone/hydroxyapatite scaffolds for improving bone regeneration. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 74, 525–535. [Google Scholar] [CrossRef] [PubMed]
- de Mendonça Narciso, M.L.B.T. Optimization of Nanoindentation Protocols for Hydrogel Mechanical Characterization. Master’s Thesis, Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal, 2019. [Google Scholar]
- Yan, C.; Pochan, D.J. Rheological properties of peptide-based hydrogels for biomedical and other applications. Chem. Soc. Rev. 2010, 39, 3528–3540. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Szczesny, S.E.; Caliari, S.R.; Charrier, E.E.; Chaudhuri, O.; Cao, X.; Lin, Y.; Mauck, R.L.; Janmey, P.A.; Burdick, J.A.; et al. Matching material and cellular timescales maximizes cell spreading on viscoelastic substrates. Proc. Natl. Acad. Sci. USA 2018, 115, E2686–E2695. [Google Scholar] [CrossRef] [PubMed]
- Ullah, F.; Othman, M.B.; Javed, F.; Ahmad, Z.; Md Akil, H. Classification, processing and application of hydrogels: A review. Mater. Sci. Eng. C Mater. Biol. Appl. 2015, 57, 414–433. [Google Scholar] [CrossRef] [PubMed]
- Alpesh, P.; Kibret, M. Hydrogel Biomaterials. In Biomedical Engineering; Reza, F.-R., Ed.; IntechOpen: Rijeka, Croatia, Chapter 14; 2011. [Google Scholar]
- Nsiah, B.A.; Moore, E.M.; Roudsari, L.C.; Virdone, N.K.; West, J.L. Angiogenesis in hydrogel biomaterials. In Biosynthetic Polymers for Medical Applications; Woodhead Publishing: Cambridge, UK, 2016; pp. 189–203. [Google Scholar]
- Caliari, S.R.; Burdick, J.A. A practical guide to hydrogels for cell culture. Nat. Methods 2016, 13, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Nicosia, A.; Salamone, M.; Costa, S.; Ragusa, M.A.; Ghersi, G. Mimicking Molecular Pathways in the Design of Smart Hydrogels for the Design of Vascularized Engineered Tissues. Int. J. Mol. Sci. 2023, 24, 12314. [Google Scholar] [CrossRef]
- Tae, G.; Scatena, M.; Stayton, P.S.; Hoffman, A.S. PEG-cross-linked heparin is an affinity hydrogel for sustained release of vascular endothelial growth factor. J. Biomater. Sci. Polym. Ed. 2006, 17, 187–197. [Google Scholar] [CrossRef]
- Vallmajo-Martin, Q.; Broguiere, N.; Millan, C.; Zenobi-Wong, M.; Ehrbar, M. PEG/HA Hybrid Hydrogels for Biologically and Mechanically Tailorable Bone Marrow Organoids. Adv. Funct. Mater. 2020, 30, 1910282. [Google Scholar] [CrossRef]
- Singh, R.K.; Seliktar, D.; Putnam, A.J. Capillary morphogenesis in PEG-collagen hydrogels. Biomaterials 2013, 34, 9331–9340. [Google Scholar] [CrossRef] [PubMed]
- Ara, C.; Jabeen, S.; Afshan, G.; Farooq, A.; Akram, M.S.; Asmatullah; Islam, A.; Ziafat, S.; Nawaz, B.; Khan, R.U. Angiogenic potential and wound healing efficacy of chitosan derived hydrogels at varied concentrations of APTES in chick and mouse models. Int. J. Biol. Macromol. 2022, 202, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Hwang, M.R.; Kim, J.O.; Lee, J.H.; Kim, Y.I.; Kim, J.H.; Chang, S.W.; Jin, S.G.; Kim, J.A.; Lyoo, W.S.; Han, S.S.; et al. Gentamicin-loaded wound dressing with polyvinyl alcohol/dextran hydrogel: Gel characterization and in vivo healing evaluation. AAPS PharmSciTech 2010, 11, 1092–1103. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Liu, S.; Feng, W. PVA hydrogel properties for biomedical application. J. Mech. Behav. Biomed. Mater. 2011, 4, 1228–1233. [Google Scholar] [CrossRef] [PubMed]
- Clavane, E.M.; Taylor, H.A.; Cubbon, R.M.; Meakin, P.J. Endothelial Cell FibrinFibrin Gel Angiogenesis Bead Assay. In Angiogenesis: Methods and Protocols; Benest, A.V., Ed.; Springer: New York, NY, USA, 2022; pp. 321–327. [Google Scholar]
- Morin, K.T.; Tranquillo, R.T. In vitro models of angiogenesis and vasculogenesis in fibrin gel. Exp. Cell Res. 2013, 319, 2409–2417. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Meng, H.; Liu, Y.; Lee, B.P. Fibrin Gel as an Injectable Biodegradable Scaffold and Cell Carrier for Tissue Engineering. Sci. World J. 2015, 2015, 685690. [Google Scholar] [CrossRef] [PubMed]
- Giraudo, M.V.; Di Francesco, D.; Catoira, M.C.; Cotella, D.; Fusaro, L.; Boccafoschi, F. Angiogenic Potential in Biological Hydrogels. Biomedicines 2020, 8, 436. [Google Scholar] [CrossRef]
- Bao, W.; Li, M.; Yang, Y.; Wan, Y.; Wang, X.; Bi, N.; Li, C. Advancements and Frontiers in the High Performance of Natural Hydrogels for Cartilage Tissue Engineering. Front. Chem. 2020, 8, 53. [Google Scholar] [CrossRef]
- Catoira, M.C.; Fusaro, L.; Di Francesco, D.; Ramella, M.; Boccafoschi, F. Overview of natural hydrogels for regenerative medicine applications. J. Mater. Sci. Mater. Med. 2019, 30, 115. [Google Scholar] [CrossRef]
- Salisbury, V.A. High Resolution Imaging and Analysis of Endothelial Tubulogenesis and Blood Vessel Formation. Ph.D. Thesis, Uiversity of Birmingham, Birmingham, UK, 2017. [Google Scholar]
- Dokland, T.; Hutmacher, D.W.; Ng, M.M.-L.; Schantz, J.-T. Techniques in Microscopy for Biomedical Applications; World Scientific: Singapore, 2006; Volume 2, p. 420. [Google Scholar]
- Rasband, W.S. ImageJ. Available online: http://imagej.nih.gov/ij/ (accessed on 21 August 2023).
- Niemisto, A.; Dunmire, V.; Yli-Harja, O.; Wei, Z.; Shmulevich, I. Robust quantification of in vitro angiogenesis through image analysis. IEEE Trans. Med. Imaging 2005, 24, 549–553. [Google Scholar] [CrossRef]
- Tu, F.; Pang, Q.; Chen, X.; Huang, T.; Liu, M.; Zhai, Q. Angiogenic effects of apigenin on endothelial cells after hypoxia-reoxygenation via the caveolin-1 pathway. Int. J. Mol. Med. 2017, 40, 1639–1648. [Google Scholar] [CrossRef] [PubMed]
- Gershovich, J.G.; Dahlin, R.L.; Kasper, F.K.; Mikos, A.G. Enhanced Osteogenesis in Cocultures with Human Mesenchymal Stem Cells and Endothelial Cells on Polymeric Microfiber Scaffolds. Tissue Eng. Part A 2013, 19, 2565–2576. [Google Scholar] [CrossRef] [PubMed]
- Skrzypek, K.; Nibbelink, M.G.; Karbaat, L.P.; Karperien, M.; van Apeldoorn, A.; Stamatialis, D. An important step towards a prevascularized islet macroencapsulation device—Effect of micropatterned membranes on development of endothelial cell network. J. Mater. Sci. Mater. Med. 2018, 29, 91. [Google Scholar] [CrossRef] [PubMed]
- Yavvari, P.; Laporte, A.; Elomaa, L.; Schraufstetter, F.; Pacharzina, I.; Daberkow, A.D.; Hoppensack, A.; Weinhart, M. 3D-Cultured Vascular-like Networks Enable Validation of Vascular Disruption Properties of Drugs in Vitro. Front. Bioeng. Biotechnol. 2022, 10, 888492. [Google Scholar] [CrossRef] [PubMed]
- Mulder, W.J.M.; Castermans, K.; van Beijnum, J.R.; Egbrink, O.M.G.A.; Chin, P.T.K.; Fayad, Z.A.; Löwik, C.W.G.M.; Kaijzel, E.L.; Que, I.; Storm, G.; et al. Molecular imaging of tumor angiogenesis using αvβ3-integrin targeted multimodal quantum dots. Angiogenesis 2009, 12, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Sample Preparation for Fluorescence Microscopy: An Introduction Concepts and Tips for Better Fixed Sample Imaging Results. 2015. Available online: https://health.uconn.edu/cell-analysis-modeling/wp-content/uploads/sites/149/2023/04/cell-fixation-5994-2778EN-agilent.pdf (accessed on 6 December 2023).
- Lakowicz, J. Principles of Fluorescence Spectroscopy; Springer: New York, NY, USA, 2006; Volume 1. [Google Scholar]
- Yuste, R. Fluorescence microscopy today. Nat. Methods 2005, 2, 902–904. [Google Scholar] [CrossRef] [PubMed]
- Shotton, D.; White, N. Confocal scanning microscopy: Three-dimensional biological imaging. Trends Biochem. Sci. 1989, 14, 435–439. [Google Scholar] [CrossRef]
- Hoffman, R.M. Imaging tumor angiogenesis with fluorescent proteins. APMIS J. 2004, 112, 441–449. [Google Scholar] [CrossRef]
- Pian, Q.; Wang, C.; Chen, X.; Liang, J.; Zhao, L.; Wang, G.; Intes, X. Multimodal Biomedical Optical Imaging Review: Towards Comprehensive Investigation of Biological Tissues. Curr. Mol. Imaging 2015, 3, 72–87. [Google Scholar] [CrossRef]
- Upputuri, P.K.; Sivasubramanian, K.; Mark, C.S.K.; Pramanik, M. Recent Developments in Vascular Imaging Techniques in Tissue Engineering and Regenerative Medicine. BioMed Res. Int. 2015, 2015, 783983. [Google Scholar] [CrossRef]
- Provenzale, J.M. Imaging of Angiogenesis: Clinical Techniques and Novel Imaging Methods. Am. J. Roentgenol. 2007, 188, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Corliss, B.A.; Mathews, C.; Doty, R.; Rohde, G.; Peirce, S.M. Methods to label, image, and analyze the complex structural architectures of microvascular networks. Microcirculation 2019, 26, e12520. [Google Scholar] [CrossRef] [PubMed]
- Medina-Leyte, D.J.; Domínguez-Pérez, M.; Mercado, I.; Villarreal-Molina, M.T.; Jacobo-Albavera, L. Use of Human Umbilical Vein Endothelial Cells (HUVEC) as a Model to Study Cardiovascular Disease: A Review. Appl. Sci. 2020, 10, 938. [Google Scholar] [CrossRef]
- Kumar, A.; D’Souza, S.S.; Moskvin, O.V.; Toh, H.; Wang, B.; Zhang, J.; Swanson, S.; Guo, L.W.; Thomson, J.A.; Slukvin, I.I. Specification and Diversification of Pericytes and Smooth Muscle Cells from Mesenchymoangioblasts. Cell Rep. 2017, 19, 1902–1916. [Google Scholar] [CrossRef] [PubMed]
- Bachetti, T.; Morbidelli, L. Endothelial cells in culture: A model for studying vascular functions. Pharmacol. Res. 2000, 42, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Cenni, E.; Perut, F.; Baldini, N. In vitro models for the evaluation of angiogenic potential in bone engineering. Acta Pharmacol. Sin. 2011, 32, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Strauss, O.; Phillips, A.; Ruggiero, K.; Bartlett, A.; Dunbar, P.R. Immunofluorescence identifies distinct subsets of endothelial cells in the human liver. Sci. Rep. 2017, 7, 44356. [Google Scholar] [CrossRef]
- Keiper, T.; Santoso, S.; Nawroth, P.P.; Orlova, V.; Chavakis, T. The role of junctional adhesion molecules in cell-cell interactions. Histol. Histopathol. 2005, 20, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Jaff, N.; Grankvist, R.; Muhl, L.; Chireh, A.; Sandell, M.; Jonsson, S.; Arnberg, F.; Eriksson, U.; Holmin, S. Transcriptomic analysis of the harvested endothelial cells in a swine model of mechanical thrombectomy. Neuroradiology 2018, 60, 759–768. [Google Scholar] [CrossRef]
- Zudaire, E.; Gambardella, L.; Kurcz, C.; Vermeren, S. A computational tool for quantitative analysis of vascular networks. PLoS ONE 2011, 6, e27385. [Google Scholar] [CrossRef]
- Gurevich, D.B.; Severn, C.E.; Twomey, C.; Greenhough, A.; Cash, J.; Toye, A.M.; Mellor, H.; Martin, P. Live imaging of wound angiogenesis reveals macrophage orchestrated vessel sprouting and regression. EMBO J. 2018, 37, e97786. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gao, L.; Han, Y.; Xing, M.; Zhao, C.; Peng, J.; Chang, J. Silicon-Enhanced Adipogenesis and Angiogenesis for Vascularized Adipose Tissue Engineering. Adv. Sci. 2018, 5, 1800776. [Google Scholar] [CrossRef] [PubMed]
- Gholobova, D.; Decroix, L.; Van Muylder, V.; Desender, L.; Gerard, M.; Carpentier, G.; Vandenburgh, H.; Thorrez, L. Endothelial Network Formation within Human Tissue-Engineered Skeletal Muscle. Tissue Eng. Part A 2015, 21, 2548–2558. [Google Scholar] [CrossRef] [PubMed]
- Seaman, M.E.; Peirce, S.M.; Kelly, K. Rapid analysis of vessel elements (RAVE): A tool for studying physiologic, pathologic and tumor angiogenesis. PLoS ONE 2011, 6, e20807. [Google Scholar] [CrossRef][Green Version]
- Chong, D.C.; Yu, Z.; Brighton, H.E.; Bear, J.E.; Bautch, V.L. Tortuous Microvessels Contribute to Wound Healing via Sprouting Angiogenesis. Arter. Thromb. Vasc. Biol. 2017, 37, 1903–1912. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Abramoff, M.; Magalhães, P.; Ram, S.J. Image Processing with ImageJ. Biophotonics Int. 2003, 11, 36–42. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Dashnyam, K.; Buitrago, J.O.; Bold, T.; Mandakhbayar, N.; Perez, R.A.; Knowles, J.C.; Lee, J.H.; Kim, H.W. Angiogenesis-promoted bone repair with silicate-shelled hydrogel fiber scaffolds. Biomater. Sci. 2019, 7, 5221–5231. [Google Scholar] [CrossRef]
- Eglinger, J.; Karsjens, H.; Lammert, E. Quantitative assessment of angiogenesis and pericyte coverage in human cell-derived vascular sprouts. Inflamm. Regen. 2017, 37, 2. [Google Scholar] [CrossRef]
- Freudenberg, U.; Zieris, A.; Chwalek, K.; Tsurkan, M.V.; Maitz, M.F.; Atallah, P.; Levental, K.R.; Eming, S.A.; Werner, C. Heparin desulfation modulates VEGF release and angiogenesis in diabetic wounds. J. Control. Release 2015, 220, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.R.; Clark, A.Y.; Garcia, A.J. Integrin-specific hydrogels functionalized with VEGF for vascularization and bone regeneration of critical-size bone defects. J. Biomed. Mater. Res. A 2016, 104, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Pan, C.C.; Yang, Y.P. Development of a Dual Hydrogel Model System for Vascularization. Macromol. Biosci. 2020, 20, e2000204. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Cui, J.; Huang, H.; Yue, Z.; Chang, Y.; Li, N.; Han, Z.; Han, Z.-C.; Guo, Z.; Li, Z. IGF-1C domain-modified chitosan hydrogel accelerates cutaneous wound healing by promoting angiogenesis. Future Med. Chem. 2020, 12, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Yu, Y.; Wang, J.; Liu, H.; Pan, H.; Wang, G.; Liu, C. Enhancement and orchestration of osteogenesis and angiogenesis by a dual-modular design of growth factors delivery scaffolds and 26SCS decoration. Biomaterials 2020, 232, 119645. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Yu, Y.; Dai, K.; Zhang, T.; Ji, L.; Wang, X.; Wang, J.; Liu, C. Engineering Neutrophil Immunomodulatory Hydrogels Promoted Angiogenesis. ACS Appl. Mater Interfaces 2022, 14, 39746–39758. [Google Scholar] [CrossRef]
- Flora, T.; de Torre, I.G.; Alonso, M.; Rodriguez-Cabello, J.C. Tethering QK peptide to enhance angiogenesis in elastin-like recombinamer (ELR) hydrogels. J. Mater. Sci. Mater. Med. 2019, 30, 30. [Google Scholar] [CrossRef]
- Hao, Z.; Ren, L.; Zhang, Z.; Yang, Z.; Wu, S.; Liu, G.; Cheng, B.; Wu, J.; Xia, J. A multifunctional neuromodulation platform utilizing Schwann cell-derived exosomes orchestrates bone microenvironment via immunomodulation, angiogenesis and osteogenesis. Bioact. Mater. 2023, 23, 206–222. [Google Scholar] [CrossRef]
- Pal, A.; Smith, C.I.; Palade, J.; Nagaraju, S.; Alarcon-Benedetto, B.A.; Kilbourne, J.; Rawls, A.; Wilson-Rawls, J.; Vernon, B.L.; Nikkhah, M. Poly(N-isopropylacrylamide)-based dual-crosslinking biohybrid injectable hydrogels for vascularization. Acta Biomater. 2020, 107, 138–151. [Google Scholar] [CrossRef]
- Williams, P.A.; Campbell, K.T.; Silva, E.A. Alginate hydrogels of varied molecular weight distribution enable sustained release of sphingosine-1-phosphate and promote angiogenesis. J. Biomed. Mater. Res. A 2018, 106, 138–146. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, J.; Sun, H.; Wang, F.; Wang, Y.; Zhang, Z.; Teng, W.; Ye, Y.; Huang, D.; Zhang, W.; et al. Spatiotemporal regulation of angiogenesis/osteogenesis emulating natural bone healing cascade for vascularized bone formation. J. Nanobiotechnol. 2021, 19, 420. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, J.; Hao, H.; Chen, D.; Zhizhong, L.; Li, M.; Song, H.; Xiang, R.; Jiang, C.; Fu, X.; et al. Preferred M2 Polarization by ASC-Based Hydrogel Accelerated Angiogenesis and Myogenesis in Volumetric Muscle Loss Rats. Stem Cells Int. 2017, 2017, 2896874. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Guan, F.; Cui, F.; Sun, X.; Zhao, L.; Wang, Y.; Wang, X. Enhanced angiogenesis by the hyaluronic acid hydrogels immobilized with a VEGF mimetic peptide in a traumatic brain injury model in rats. Regen. Biomater. 2019, 6, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Mizuguchi, Y.; Mashimo, Y.; Mie, M.; Kobatake, E. Temperature-Responsive Multifunctional Protein Hydrogels with Elastin-like Polypeptides for 3-D Angiogenesis. Biomacromolecules 2020, 21, 1126–1135. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Fan, Y.; Akay, Y.M.; Akay, M. TNP-470 Reduces Glioblastoma Angiogenesis in Three Dimensional GelMA Microwell Platform. IEEE Trans. Nanobiosci. 2016, 15, 683–688. [Google Scholar] [CrossRef]
- Li, M.N.; Yu, H.P.; Ke, Q.F.; Zhang, C.Q.; Gao, Y.S.; Guo, Y.P. Gelatin methacryloyl hydrogels functionalized with endothelin-1 for angiogenesis and full-thickness wound healing. J. Mater. Chem. B 2021, 9, 4700–4709. [Google Scholar] [CrossRef]
- Liu, H.; Wu, B.; Shi, X.; Cao, Y.; Zhao, X.; Liang, D.; Qin, Q.; Liang, X.; Lu, W.; Wang, D.; et al. Aerobic exercise-induced circulating extracellular vesicle combined decellularized dermal matrix hydrogel facilitates diabetic wound healing by promoting angiogenesis. Front. Bioeng. Biotechnol. 2022, 10, 903779. [Google Scholar] [CrossRef]
- Paul, A.; Hasan, A.; Kindi, H.A.; Gaharwar, A.K.; Rao, V.T.S.; Nikkhah, M.; Shin, S.R.; Krafft, D.; Dokmeci, M.R.; Shum-Tim, D.; et al. Injectable Graphene Oxide/Hydrogel-Based Angiogenic Gene Delivery System for Vasculogenesis and Cardiac Repair. ACS Nano 2014, 8, 8050–8062. [Google Scholar] [CrossRef]
- Peterson, A.W.; Caldwell, D.J.; Rioja, A.Y.; Rao, R.R.; Putnam, A.J.; Stegemann, J.P. Vasculogenesis and Angiogenesis in Modular Collagen-Fibrin Microtissues. Biomater. Sci. 2014, 2, 1497–1508. [Google Scholar] [CrossRef]
- Tonello, S.; Moore, M.C.; Sharma, B.; Dobson, J.; McFetridge, P.S. Controlled release of a heterogeneous human placental matrix from PLGA microparticles to modulate angiogenesis. Drug Deliv. Transl. Res. 2016, 6, 174–183. [Google Scholar] [CrossRef]
- Carpentier, G.; Berndt, S.; Ferratge, S.; Rasband, W.; Cuendet, M.; Uzan, G.; Albanese, P. Angiogenesis Analyzer for ImageJ—A comparative morphometric analysis of “Endothelial Tube Formation Assay” and “Fibrin Bead Assay”. Sci. Rep. 2020, 10, 11568. [Google Scholar] [CrossRef] [PubMed]
- Huuskes, B.M.; DeBuque, R.J.; Kerr, P.G.; Samuel, C.S.; Ricardo, S.D. The Use of Live Cell Imaging and Automated Image Analysis to Assist with Determining Optimal Parameters for Angiogenic Assay in vitro. Front. Cell Dev. Biol. 2019, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Médina, C.; Stachowicz, M.-L.; Paiva dos Santos, B.; Chagot, L.; Dusserre, N.; Fricain, J.-C. Extracellular matrix (ECM)-derived bioinks designed to foster vasculogenesis and neurite outgrowth: Characterization and bioprinting. Bioprinting 2021, 22, e00134. [Google Scholar] [CrossRef]
- Page, D.J.; Clarkin, C.E.; Mani, R.; Khan, N.A.; Dawson, J.I.; Evans, N.D. Injectable nanoclay gels for angiogenesis. Acta Biomater. 2019, 100, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Schaly, S.; Islam, P.; Abosalha, A.; Boyajian, J.L.; Shum-Tim, D.; Prakash, S. Alginate-Chitosan Hydrogel Formulations Sustain Baculovirus Delivery and VEGFA Expression Which Promotes Angiogenesis for Wound Dressing Applications. Pharmaceuticals 2022, 15, 1382. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, G.; Martinelli, M.; Courty, J.; Cascone, I. Angiogenesis Analyzer for ImageJ. In Proceedings of the 4th ImageJ User and Developer Conference, Mondorf Domaine Thermal, Luxembourg, 24–26 October 2012; pp. 198–201. [Google Scholar]
- Issa, M.E.; Berndt, S.; Carpentier, G.; Pezzuto, J.M.; Cuendet, M. Bruceantin inhibits multiple myeloma cancer stem cell proliferation. Cancer Biol. Ther. 2016, 17, 966–975. [Google Scholar] [CrossRef]
- Sakr, O.S.; Berndt, S.; Carpentier, G.; Cuendet, M.; Jordan, O.; Borchard, G. Arming embolic beads with anti-VEGF antibodies and controlling their release using LbL technology. J. Control. Release 2016, 224, 199–207. [Google Scholar] [CrossRef]
- Berndt, S.; Carpentier, G.; Turzi, A.; Borlat, F.; Cuendet, M.; Modarressi, A. Angiogenesis Is Differentially Modulated by Platelet-Derived Products. Biomedicines 2021, 9, 251. [Google Scholar] [CrossRef]
- Beter, M.; Abdollahzadeh, A.; Pulkkinen, H.H.; Huang, H.; Orsenigo, F.; Magnusson, P.U.; Yla-Herttuala, S.; Tohka, J.; Laakkonen, J.P. SproutAngio: An open-source bioimage informatics tool for quantitative analysis of sprouting angiogenesis and lumen space. Sci. Rep. 2023, 13, 7279. [Google Scholar] [CrossRef]
- Kempers, L.; van der Bijl, I.; van Stalborch, A.D.; Ponsioen, B.; Margadant, C. Fast in vitro protocol for the visualization and quantitative high-throughput analysis of sprouting angiogenesis by confocal microscopy. STAR Protoc. 2021, 2, 100690. [Google Scholar] [CrossRef]
- Nunes, J.P.S.; Dias, A.A.M. ImageJ macros for the user-friendly analysis of soft-agar and wound-healing assays. Biotechniques 2017, 62, 175–179. [Google Scholar] [CrossRef]
- Shin, Y.; Moriya, A.; Tohnishi, Y.; Watanabe, T.; Imamura, Y. Basement membrane-like structures containing NTH alpha1(IV) are formed around the endothelial cell network in a novel in vitro angiogenesis model. Am. J. Physiol.-Cell Physiol. 2019, 317, C314–C325. [Google Scholar] [CrossRef]
- Blinder, Y.J.; Freiman, A.; Raindel, N.; Mooney, D.J.; Levenberg, S. Vasculogenic dynamics in 3D engineered tissue constructs. Sci. Rep. 2015, 5, 17840. [Google Scholar] [CrossRef]
- Siller, I.G.; Epping, N.M.; Lavrentieva, A.; Scheper, T.; Bahnemann, J. Customizable 3D-Printed (Co-)Cultivation Systems for in Vitro Study of Angiogenesis. Materials 2020, 13, 4290. [Google Scholar] [CrossRef]
- Perry, L.; Flugelman, M.Y.; Levenberg, S. Elderly Patient-Derived Endothelial Cells for Vascularization of Engineered Muscle. Mol. Ther. 2017, 25, 935–948. [Google Scholar] [CrossRef]
- Zippusch, S.; Böer, U.; Behrens, P. Towards the Development of a Pre-Vascularized Attachable Tissue Construct: Impact of Chemically-Induced Hypoxia, Growth Factors and Fluid Dynamics on Vascular Tube Formation in Fibrin Hydrogels; Bibliothek der Medizinischen Hochschule Hannover: Hannover, Germany, 2021. [Google Scholar]
- Stalling, D.; Westerhoff, M.; Hege, H.-C. 38—Amira: A Highly Interactive System for Visual Data Analysis. In Visualization Handbook; Hansen, C.D., Johnson, C.R., Eds.; Butterworth-Heinemann: Burlington, 2005; pp. 749–767. [Google Scholar]
- Demene, C.; Payen, T.; Dizeux, A.; Barrois, G.; Gennisson, J.L.; Bridal, L.; Tanter, M. 3-D Longitudinal Imaging of Tumor Angiogenesis in Mice in Vivo Using Ultrafast Doppler Tomography. Ultrasound Med. Biol. 2019, 45, 1284–1296. [Google Scholar] [CrossRef]
- Boyd, N.L.; Nunes, S.S.; Krishnan, L.; Jokinen, J.D.; Ramakrishnan, V.M.; Bugg, A.R.; Hoying, J.B. Dissecting the role of human embryonic stem cell-derived mesenchymal cells in human umbilical vein endothelial cell network stabilization in three-dimensional environments. Tissue Eng. Part A 2013, 19, 211–223. [Google Scholar] [CrossRef]
- Davern, J.W. Development of a Bioengineered Microvascular In Vitro Model. Master’s Thesis, Queensland University of Technology, Brisbane, QLD, Australia, 2020. [Google Scholar]
- WinFiber3D User’s Manual; University of Utah: Salt Lake City, UT, USA, 2010.
- Barnes, D. In Vitro Bioengineering Applications of Melt Electrowritten and Hydrogel Composite Scaffolds. Master’s Thesis, Queensland University of Technology, Brisane City, QLD, Australia, 2021. [Google Scholar]
- Koch, M.K.; Jaeschke, A.; Murekatete, B.; Ravichandran, A.; Tsurkan, M.; Werner, C.; Soon, P.; Hutmacher, D.W.; Haupt, L.M.; Bray, L.J. Stromal fibroblasts regulate microvascular-like network architecture in a bioengineered breast tumour angiogenesis model. Acta Biomater. 2020, 114, 256–269. [Google Scholar] [CrossRef]
- Hsu, C.W.; Poche, R.A.; Saik, J.E.; Ali, S.; Wang, S.; Yosef, N.; Calderon, G.A.; Scott, L., Jr.; Vadakkan, T.J.; Larina, I.V.; et al. Improved Angiogenesis in Response to Localized Delivery of Macrophage-Recruiting Molecules. PLoS ONE 2015, 10, e0131643. [Google Scholar] [CrossRef]
- Verbridge, S.S.; Chakrabarti, A.; DelNero, P.; Kwee, B.; Varner, J.D.; Stroock, A.D.; Fischbach, C. Physicochemical regulation of endothelial sprouting in a 3D microfluidic angiogenesis model. J. Biomed. Mater. Res. A 2013, 101, 2948–2956. [Google Scholar] [CrossRef]
- Bhanushali, U.; Rajendran, S.; Sarma, K.; Kulkarni, P.; Chatti, K.; Chatterjee, S.; Ramaa, C.S. 5-Benzylidene-2,4-thiazolidenedione derivatives: Design, synthesis and evaluation as inhibitors of angiogenesis targeting VEGR-2. Bioorg. Chem. 2016, 67, 139–147. [Google Scholar] [CrossRef]
- Rema, R.B.; Rajendran, K.; Ragunathan, M. Angiogenic efficacy of Heparin on chick chorioallantoic membrane. Vasc. Cell 2012, 4, 8. [Google Scholar] [CrossRef]
- Vimalraj, S.; Subramanian, R.; Dhanasekaran, A. LncRNA MALAT1 Promotes Tumor Angiogenesis by Regulating MicroRNA-150-5p/VEGFA Signaling in Osteosarcoma: In-Vitro and in-Vivo Analyses. Front. Oncol. 2021, 11, 742789. [Google Scholar] [CrossRef]
- Zhang, B.; Liya, A.; Li, S.; Xu, Z. AngioIQ: A Novel Automated Analysis Approach for Angiogenesis Image Quantification. In Proceedings of the 2009 2nd International Conference on Biomedical Engineering and Informatics, Tianjin, China, 17–19 October 2009; pp. 1–5. [Google Scholar]
- Gutknecht, M.F.; Seaman, M.E.; Ning, B.; Cornejo, D.A.; Mugler, E.; Antkowiak, P.F.; Moskaluk, C.A.; Hu, S.; Epstein, F.H.; Kelly, K.A. Identification of the S100 fused-type protein hornerin as a regulator of tumor vascularity. Nat. Commun. 2017, 8, 552. [Google Scholar] [CrossRef]
- James, A.C.; Szot, J.O.; Iyer, K.; Major, J.A.; Pursglove, S.E.; Chapman, G.; Dunwoodie, S.L. Notch4 reveals a novel mechanism regulating Notch signal transduction. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2014, 1843, 1272–1284. [Google Scholar] [CrossRef]
- Marquez-Curtis, L.A.; Sultani, A.B.; McGann, L.E.; Elliott, J.A.W. Beyond membrane integrity: Assessing the functionality of human umbilical vein endothelial cells after cryopreservation. Cryobiology 2016, 72, 183–190. [Google Scholar] [CrossRef]
- Edgar, L.T.; Maas, S.A.; Guilkey, J.E.; Weiss, J.A. A coupled model of neovessel growth and matrix mechanics describes and predicts angiogenesis in vitro. Biomech. Model. Mechanobiol. 2015, 14, 767–782. [Google Scholar] [CrossRef]
- Pulkkinen, H.H.; Kiema, M.; Lappalainen, J.P.; Toropainen, A.; Beter, M.; Tirronen, A.; Holappa, L.; Niskanen, H.; Kaikkonen, M.U.; Yla-Herttuala, S.; et al. BMP6/TAZ-Hippo signaling modulates angiogenesis and endothelial cell response to VEGF. Angiogenesis 2021, 24, 129–144. [Google Scholar] [CrossRef]
- Prentasic, P.; Heisler, M.; Mammo, Z.; Lee, S.; Merkur, A.; Navajas, E.; Beg, M.F.; Sarunic, M.; Loncaric, S. Segmentation of the foveal microvasculature using deep learning networks. J. Biomed. Opt. 2016, 21, 75008. [Google Scholar] [CrossRef]
- Fu, H.; Xu, Y.; Lin, S.; Kee Wong, D.W.; Liu, J. DeepVessel: Retinal Vessel Segmentation via Deep Learning and Conditional Random Field. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016, Athens, Greece, 17–21 October 2016; pp. 132–139. [Google Scholar]
- Gallardo, M.; Munk, M.R.; Kurmann, T.; De Zanet, S.; Mosinska, A.; Karagoz, I.K.; Zinkernagel, M.S.; Wolf, S.; Sznitman, R. Machine Learning Can Predict Anti-VEGF Treatment Demand in a Treat-and-Extend Regimen for Patients with Neovascular AMD, DME, and RVO Associated Macular Edema. Ophthalmol. Retin. 2021, 5, 604–624. [Google Scholar] [CrossRef]
- Kuri, P.M.; Pion, E.; Mahl, L.; Kainz, P.; Schwarz, S.; Brochhausen, C.; Aung, T.; Haerteis, S. Deep Learning-Based Image Analysis for the Quantification of Tumor-Induced Angiogenesis in the 3D in Vivo Tumor Model-Establishment and Addition to Laser Speckle Contrast Imaging (LSCI). Cells 2022, 11, 2321. [Google Scholar] [CrossRef]
- Ing, N.; Huang, F.; Conley, A.; You, S.; Ma, Z.; Klimov, S.; Ohe, C.; Yuan, X.; Amin, M.B.; Figlin, R.; et al. A novel machine learning approach reveals latent vascular phenotypes predictive of renal cancer outcome. Sci. Rep. 2017, 7, 13190. [Google Scholar] [CrossRef]
- Chen, J.; Yang, Y.; Luo, B.; Wen, Y.; Chen, Q.; Ma, R.; Huang, Z.; Zhu, H.; Li, Y.; Chen, Y.; et al. Further predictive value of lymphovascular invasion explored via supervised deep learning for lymph node metastases in breast cancer. Hum. Pathol. 2023, 131, 26–37. [Google Scholar] [CrossRef]
- Vu, Q.D.; Graham, S.; Kurc, T.; To, M.N.N.; Shaban, M.; Qaiser, T.; Koohbanani, N.A.; Khurram, S.A.; Kalpathy-Cramer, J.; Zhao, T.; et al. Methods for Segmentation and Classification of Digital Microscopy Tissue Images. Front. Bioeng. Biotechnol. 2019, 7, 53. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Schonmehl, R.; Artinger, A.; Winter, L.; Bock, H.; Schreml, S.; Gurtler, F.; Daza, J.; Schmitt, V.H.; Mamilos, A.; et al. 3D Visualization, Skeletonization and Branching Analysis of Blood Vessels in Angiogenesis. Int. J. Mol. Sci. 2023, 24, 7714. [Google Scholar] [CrossRef]
- Timakova, A.; Ananev, V.; Fayzullin, A.; Makarov, V.; Ivanova, E.; Shekhter, A.; Timashev, P. Artificial Intelligence Assists in the Detection of Blood Vessels in Whole Slide Images: Practical Benefits for Oncological Pathology. Biomolecules 2023, 13, 1327. [Google Scholar] [CrossRef]
- Deng, R.; Liu, Q.; Cui, C.; Yao, T.; Long, J.; Asad, Z.; Womick, R.M.; Zhu, Z.; Fogo, A.B.; Zhao, S.; et al. Omni-Seg: A Scale-Aware Dynamic Network for Renal Pathological Image Segmentation. IEEE Trans. Biomed. Eng. 2023, 70, 2636–2644. [Google Scholar] [CrossRef]
- Edgar, L.T.; Hoying, J.B.; Weiss, J.A. In Silico Investigation of Angiogenesis with Growth and Stress Generation Coupled to Local Extracellular Matrix Density. Ann. Biomed. Eng. 2015, 43, 1531–1542. [Google Scholar] [CrossRef][Green Version]
- Hira, Z.M.; Gillies, D.F. A Review of Feature Selection and Feature Extraction Methods Applied on Microarray Data. Adv. Bioinform. 2015, 2015, 198363. [Google Scholar] [CrossRef]



| Software | Input | Output | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| ImageJ (“Angiogenenis Analyser” and “Automated Sprout Analysis”) | Phase-contrast or fluorescence 2D images | Area covered by the cells, total network length, number of meshes, nodes, extremities, and isolated elements, length of segments and branching interval | Runs on any operating system; Code can be customized according to the users’ objectives; Automatic process | Requires large amounts of RAM; Some level of computer programming may be required, depending on the objective; Mostly dependent on custom plugins and macros. Some plugins may involve image manipulation to fully function | [48,78,79,102,113,134] |
| AngioTool | 2D fluorescence images | Explant area, vessel density, branching index, number of endpoints, lacunarity, and total and average vessel length | Parameters can be adjusted to better define the vessels; Automatic process with a lower chance of human error | Results are dependent on how the initial parameters are set; Wrongful detection of vessels | [71] |
| Amira | 2D and 3D images | Vessel volume fraction, vessel length density, fractal dimension, and mean vessel radius | 3D reconstruction, noise removal, quantification of 3D networks | Not freely available | [119,120,121] |
| WinFiber3D | 3D fluorescence images | Segment orientation, average and total vessel length, even in a defined range, number of vessels and diameter | Can analyze segment orientation | - | [121,122,124,135] |
| AngioQuant | 2D brightfield images | Vessel length, segment area, branchpoints, segment count | Designed for co-culture assays; Can be used for CAM assays | Long running time | [63,131] |
| RAVE | 2D fluorescent images | Vessel volume fraction, vessel length density, fractal dimension, mean vessel radius | Rapid analysis; Detects differences between healthy and tumor-associated vasculature | Cannot be modified to perform 3D vessel analysis | [75,132] |
| REAVER | 2D fluorescent images | Vessel length density, vessel area fraction, branchpoint count, mean vessel diameter | High pixel-by-pixel accuracy for vessel segmentation | Automated segmentation is considered inaccurate, and it is recommended to combine it with manual assistance | [8] |
| VesselExpress | LSFM 3D data of blood vessels | Microvascular length, branching, diameter, tortuosity | Fast image analysis, processing, and graph composition; High-volume analysis | Hollow tubes show up if endothelial-specific antibodies are used | [9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, M.; Pinto, J.; Arteaga, B.; Guerra, A.; Jorge, R.N.; Monteiro, F.J.; Salgado, C.L. A Comprehensive Look at In Vitro Angiogenesis Image Analysis Software. Int. J. Mol. Sci. 2023, 24, 17625. https://doi.org/10.3390/ijms242417625
Pereira M, Pinto J, Arteaga B, Guerra A, Jorge RN, Monteiro FJ, Salgado CL. A Comprehensive Look at In Vitro Angiogenesis Image Analysis Software. International Journal of Molecular Sciences. 2023; 24(24):17625. https://doi.org/10.3390/ijms242417625
Chicago/Turabian StylePereira, Mariana, Jéssica Pinto, Belén Arteaga, Ana Guerra, Renato Natal Jorge, Fernando Jorge Monteiro, and Christiane Laranjo Salgado. 2023. "A Comprehensive Look at In Vitro Angiogenesis Image Analysis Software" International Journal of Molecular Sciences 24, no. 24: 17625. https://doi.org/10.3390/ijms242417625
APA StylePereira, M., Pinto, J., Arteaga, B., Guerra, A., Jorge, R. N., Monteiro, F. J., & Salgado, C. L. (2023). A Comprehensive Look at In Vitro Angiogenesis Image Analysis Software. International Journal of Molecular Sciences, 24(24), 17625. https://doi.org/10.3390/ijms242417625










