Constructed Risk Prognosis Model Associated with Disulfidptosis lncRNAs in HCC
Abstract
:1. Introduction
2. Results
2.1. Construction of the Prognostic Model Associated with Disulfidptosis lncRNAs for HCC
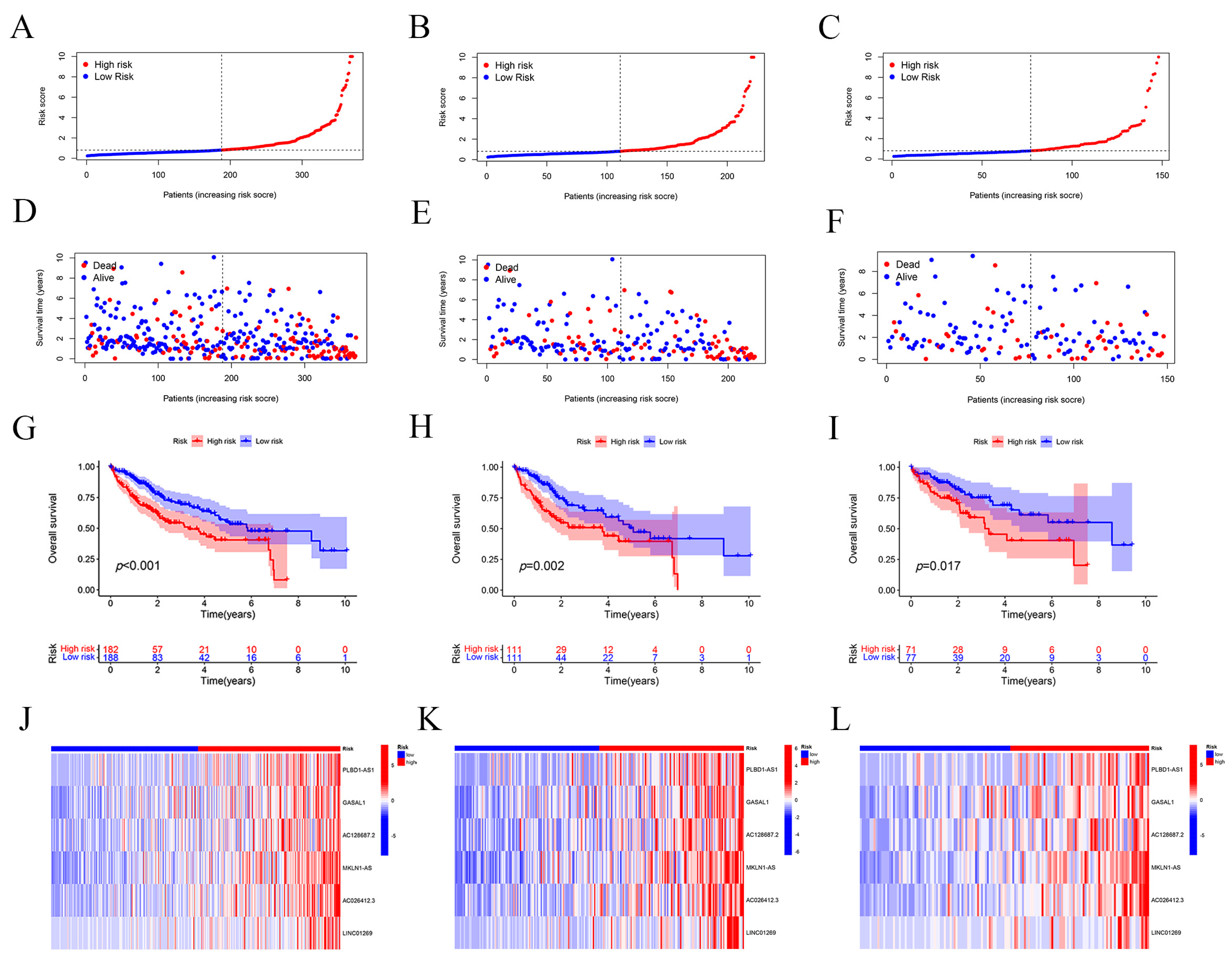
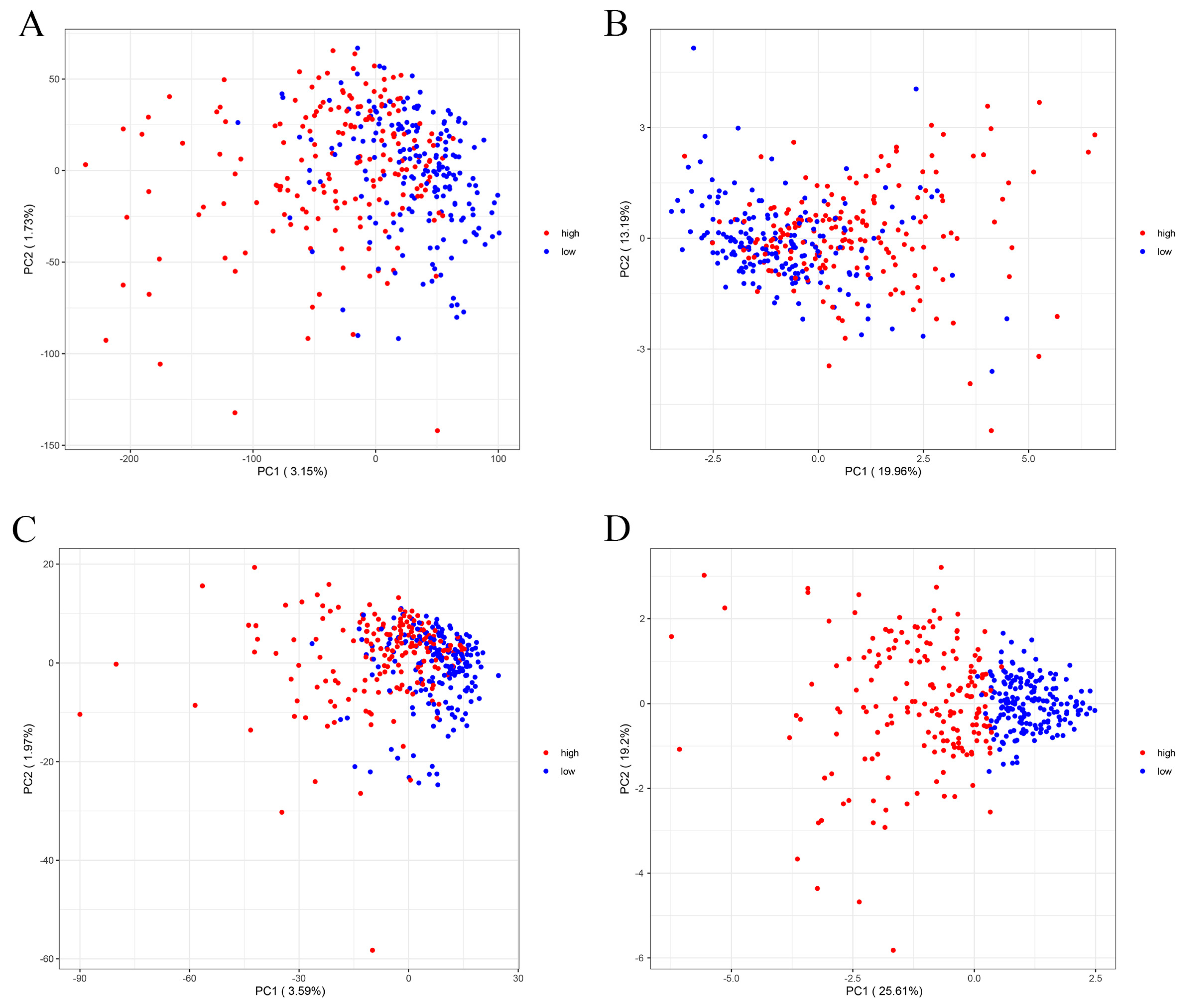
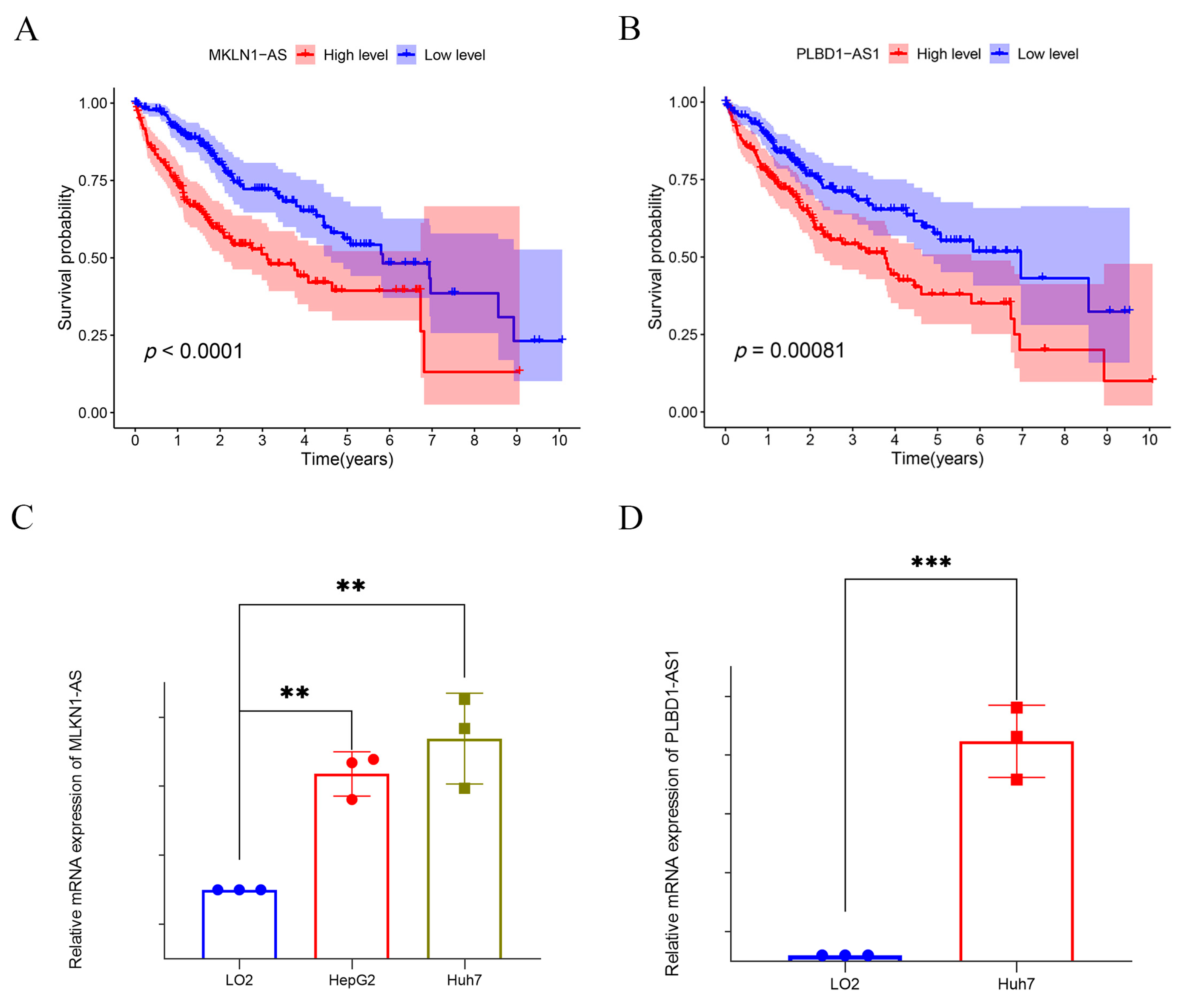
2.2. Assessment and Validation of the Risk Prognostic Models for Hepatocellular Carcinoma Associated with Disulfidptosis lncRNAs
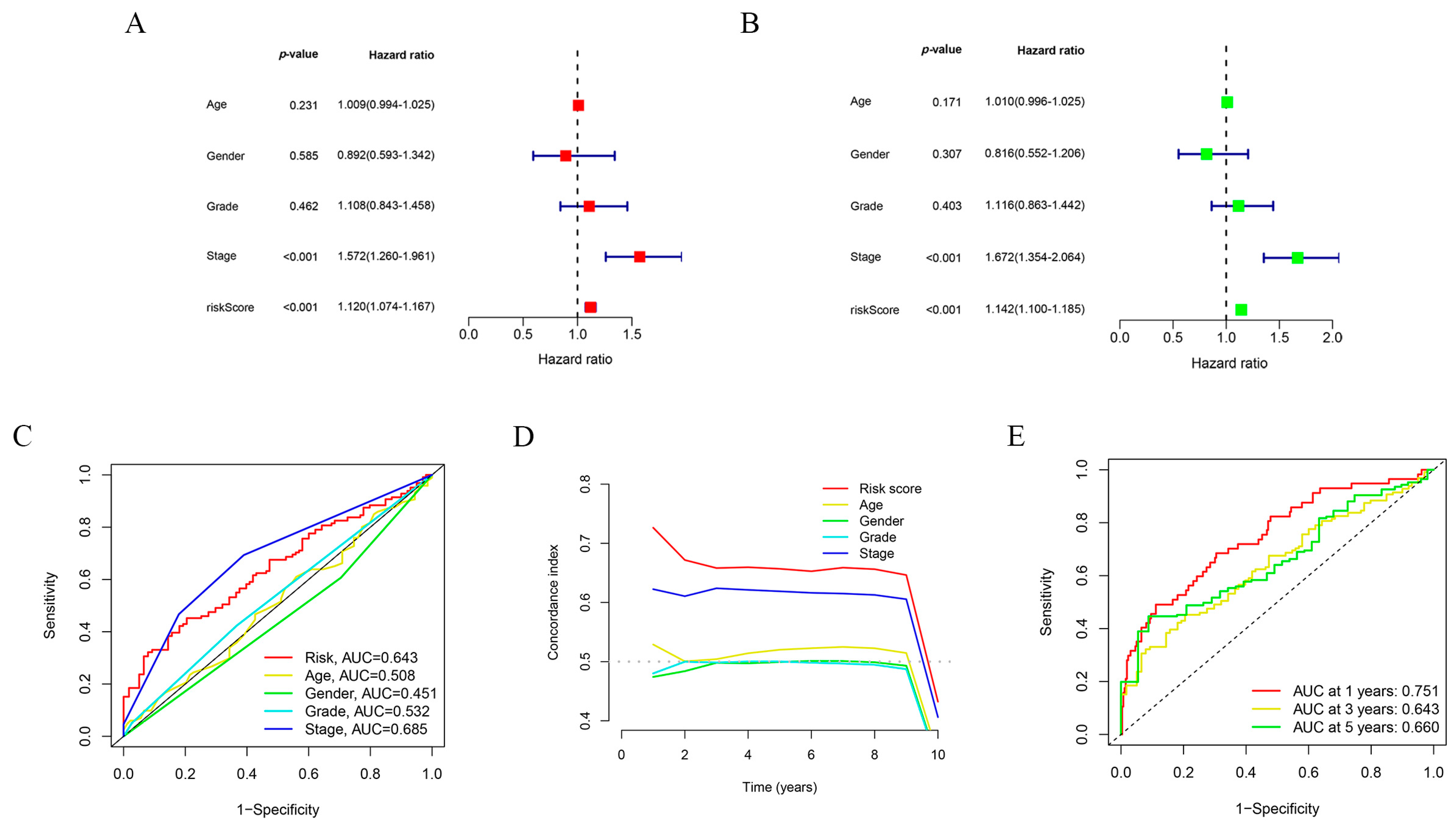
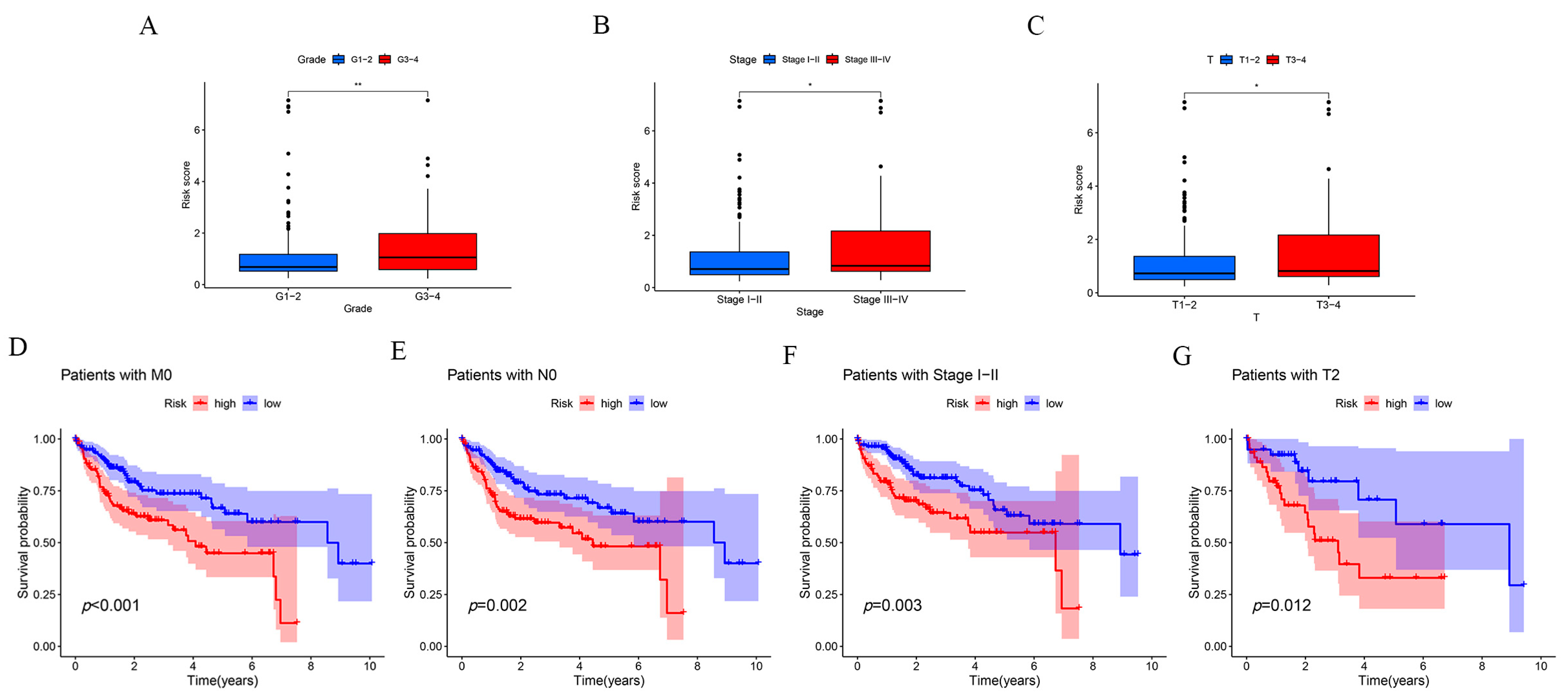
2.3. Clinical Subgroup Validation of the Risk Prognostic Model for Hepatocellular Carcinoma Associated with Disulfidptosis lncRNAs
2.4. Functional Enrichment Analysis of the Risk Prognostic Model Associated with Disulfidptosis lncRNAs in Hepatocellular Carcinoma
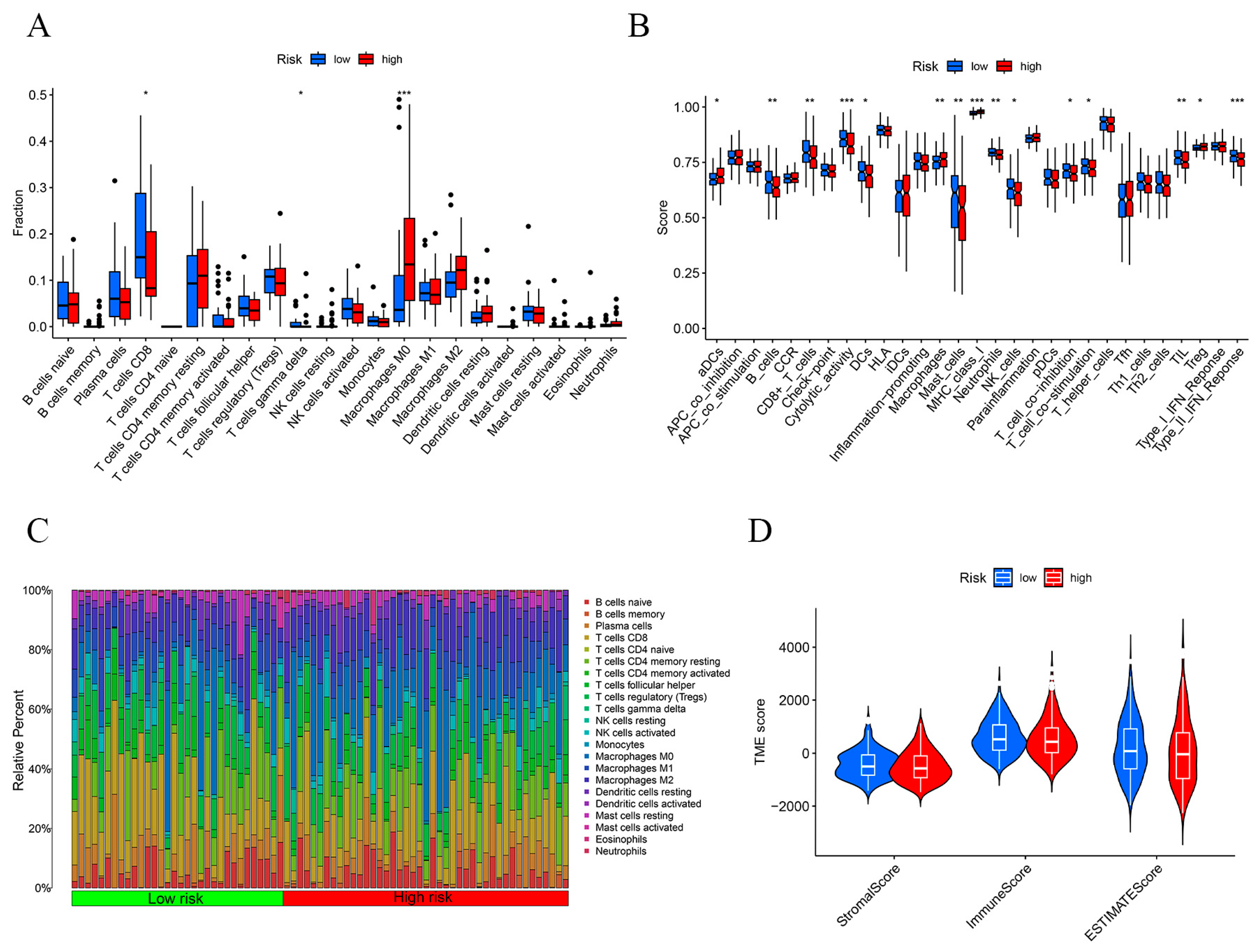
2.5. Immunosignature Characterization of the Risk Prognostic Model Associated with Disulfidptosis lncRNAs in Hepatocellular Carcinoma
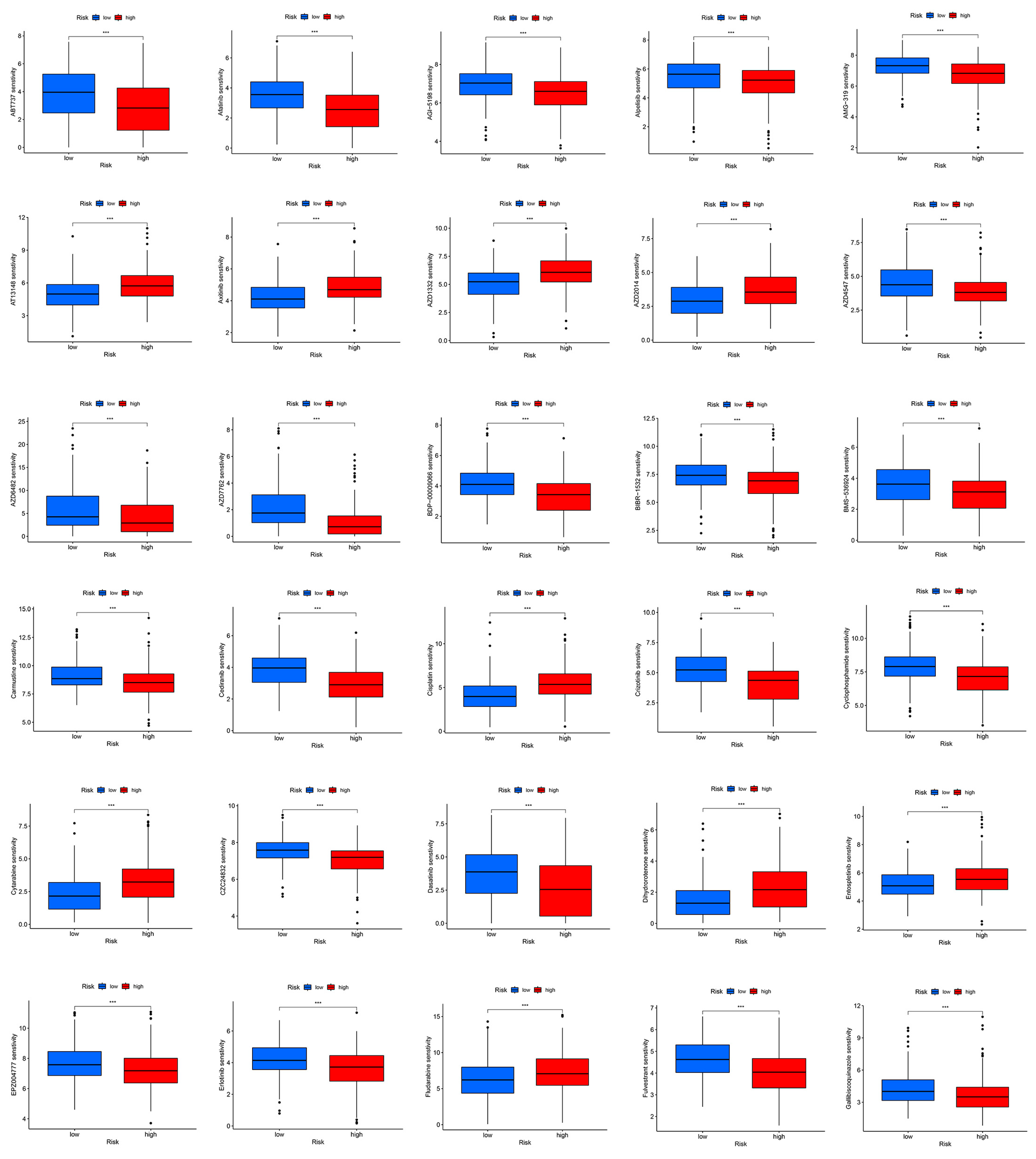
2.6. Potential Therapeutic Agents in Hepatocellular Carcinoma
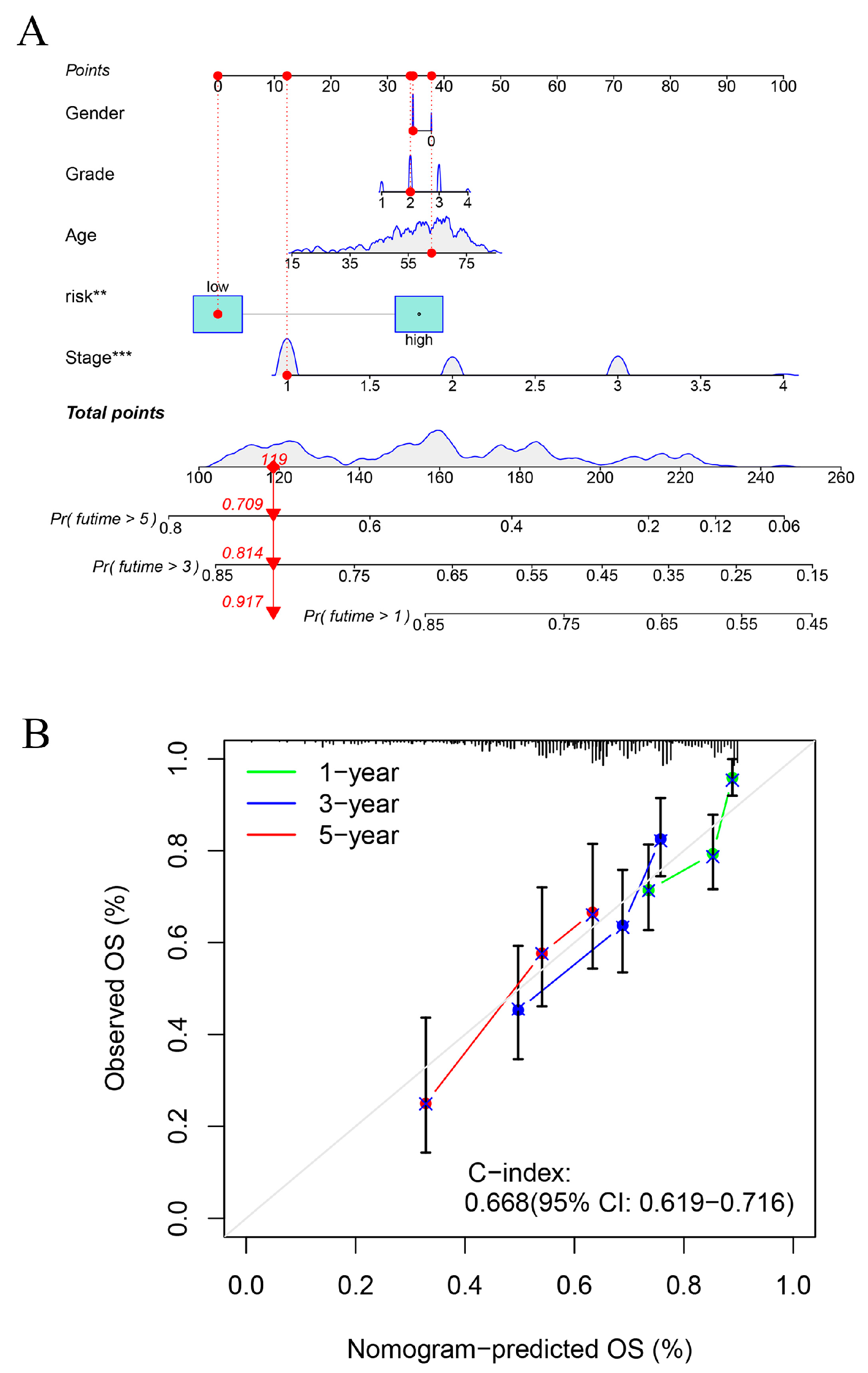
2.7. Construction of Line Graphs for the Risk Prognostic Modelling
2.8. Exploring Potential Prognostic Therapeutic Targets in Hepatocellular Carcinoma
3. Discussion
4. Materials and Methods
4.1. Data Acquisition
4.2. Identification of lncRNAs Associated with Disulfidptosis in HCC
4.3. Construction of a Predictive Model for HCC Based on Disulfidptosis-Related lncRNAs
4.4. Validation of the Risk Prognostic Models and Construction of Column-Line Diagrams
4.5. Immune Infiltration and Functional Analysis
4.6. Biological Function Enrichment Analysis
4.7. Drug Sensitivity Analysis
4.8. Cell Culture
4.9. Quantitative Reverse Transcription PCR (RT-qPCR)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A Global View of Hepatocellular Carcinoma: Trends, Risk, Prevention and Management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular Carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of Liver Diseases in the World. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular Carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Kim, E.; Viatour, P. Viatour. Hepatocellular Carcinoma: Old Friends and New Tricks. Exp. Mol. Med. 2020, 52, 1898–1907. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Liu, Q.; Xing, F.; Zeng, C.; Wang, W. Disulfidptosis: A New Form of Programmed Cell Death. J. Exp. Clin. Cancer Res. 2023, 42, 137. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Nie, L.; Zhang, Y.; Yan, Y.; Wang, C.; Colic, M.; Olszewski, K.; Horbath, A.; Chen, X.; Lei, G.; et al. Actin Cytoskeleton Vulnerability to Disulfide Stress Mediates Disulfidptosis. Nat. Cell. Biol. 2023, 25, 404–414. [Google Scholar] [CrossRef]
- Meng, Y.; Chen, X.; Deng, G. Disulfidptosis: A New Form of Regulated Cell Death for Cancer Treatment. Mol. Biomed. 2023, 4, 18. [Google Scholar] [CrossRef]
- Tan, Y.T.; Lin, J.F.; Li, T.; Li, J.J.; Xu, R.H.; Ju, H.Q. Lncrna-Mediated Posttranslational Modifications and Reprogramming of Energy Metabolism in Cancer. Cancer Commun. 2021, 41, 109–120. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, J.-K.; Peng, Y.; He, W.; Huang, C. The Role of Long Noncoding Rnas in Hepatocellular Carcinoma. Mol. Cancer 2020, 19, 77. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Zhang, Y.; Zhan, Z.; Ye, F.; Liang, Y.; Huang, J.; Chen, K.; Chen, L.; Ding, Y. A Novel Lncrna Uc.134 Represses Hepatocellular Carcinoma Progression by Inhibiting Cul4a-Mediated Ubiquitination of Lats1. J. Hematol. Oncol. 2017, 10, 91. [Google Scholar] [CrossRef]
- Lv, E.; Sheng, J.; Yu, C.; Rao, D.; Huang, W. Lncrna Influence Sequential Steps of Hepatocellular Carcinoma Metastasis. Biomed. Pharmacother. 2021, 136, 111224. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, B.; Zhang, M.; Guo, W.; Wu, Z.; Wang, Y.; Jia, L.; Li, S.; Xie, W.; Yang, D.; et al. Lncrna Epigenetic Landscape Analysis Identifies Epic1 as an Oncogenic Lncrna That Interacts with Myc and Promotes Cell-Cycle Progression in Cancer. Cancer Cell. 2018, 33, 706–720.e9. [Google Scholar] [CrossRef] [PubMed]
- Kamel, M.M.; Matboli, M.; Sallam, M.; Montasser, I.F.; Saad, A.S.; El-Tawdi, A.H. Investigation of Long Noncoding Rnas Expression Profile as Potential Serum Biomarkers in Patients with Hepatocellular Carcinoma. Transl. Res. 2016, 168, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Zhou, C.; Ding, Y.; Duan, S. Disulfidptosis: A New Target for Metabolic Cancer Therapy. J. Exp. Clin. Cancer Res. 2023, 42, 103. [Google Scholar] [CrossRef]
- Luo, L.; Hu, X.; Huang, A.; Liu, X.; Wang, L.; Du, T.; Liu, L.; Li, M. A Noval Established Cuproptosis-Associated Lncrna Signature for Prognosis Prediction in Primary Hepatic Carcinoma. Evid. Based Complement. Alternat. Med. 2022, 2022, 2075638. [Google Scholar] [CrossRef]
- Deng, X.; Bi, Q.; Chen, S.; Chen, X.; Li, S.; Zhong, Z.; Guo, W.; Li, X.; Deng, Y.; Yang, Y. Identification of a Five-Autophagy-Related-Lncrna Signature as a Novel Prognostic Biomarker for Hepatocellular Carcinoma. Front Mol. Biosci. 2020, 7, 611626. [Google Scholar] [CrossRef]
- Shen, C.; Li, J.; Zhang, Q.; Tao, Y.; Li, R.; Ma, Z.; Wang, Z. Lncrna Gasal1 Promotes Hepatocellular Carcinoma Progression by up-Regulating Usp10-Stabilized Pcna. Exp. Cell Res. 2022, 415, 112973. [Google Scholar] [CrossRef]
- Sun, R.; Wang, X.; Chen, J.; Teng, D.; Chan, S.; Tu, X.; Wang, Z.; Zuo, X.; Wei, X.; Lin, L.; et al. Development and Validation of a Novel Cellular Senescence-Related Prognostic Signature for Predicting the Survival and Immune Landscape in Hepatocellular Carcinoma. Front. Genet. 2022, 13, 949110. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Chen, X.; Chi, W.; Xue, M. Long Non-Coding Rna Mkln1-as Aggravates Hepatocellular Carcinoma Progression by Functioning as a Molecular Sponge for Mir-654-3p, Thereby Promoting Hepatoma-Derived Growth Factor Expression. Int. J. Mol. Med. 2020, 46, 1743–1754. [Google Scholar] [CrossRef]
- Guo, C.; Zhou, S.; Yi, W.; Yang, P.; Li, O.; Liu, J.; Peng, C. Long Non-Coding Rna Muskelin 1 Antisense Rna (Mkln1-as) Is a Potential Diagnostic and Prognostic Biomarker and Therapeutic Target for Hepatocellular Carcinoma. Exp. Mol. Pathol. 2021, 120, 104638. [Google Scholar] [CrossRef] [PubMed]
- Hikita, H.; Takehara, T.; Shimizu, S.; Kodama, T.; Shigekawa, M.; Iwase, K.; Hosui, A.; Miyagi, T.; Tatsumi, T.; Ishida, H.; et al. The Bcl-Xl Inhibitor, Abt-737, Efficiently Induces Apoptosis and Suppresses Growth of Hepatoma Cells in Combination with Sorafenib. Hepatology 2010, 52, 1310–1321. [Google Scholar] [CrossRef]
- Yu, Y.; Xu, L.; Qi, L.; Wang, C.; Xu, N.; Liu, S.; Li, S.; Tian, H.; Liu, W.; Xu, Y.; et al. Abt737 Induces Mitochondrial Pathway Apoptosis and Mitophagy by Regulating Drp1-Dependent Mitochondrial Fission in Human Ovarian Cancer Cells. Biomed. Pharmacother. 2017, 96, 22–29. [Google Scholar] [CrossRef]
- Xu, Y.; Gao, W.; Zhang, Y.; Wu, S.; Liu, Y.; Deng, X.; Xie, L.; Yang, J.; Yu, H.; Su, J.; et al. Abt737 Reverses Cisplatin Resistance by Targeting Glucose Metabolism of Human Ovarian Cancer Cells. Int. J. Oncol. 2018, 53, 1055–1068. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zhang, X.; Wang, M.; Xu, G.; Zhao, S.; Feng, Y.; Pan, C.; Yang, W.; Zhou, J.; Shang, L.; et al. Afatinib Combined with Anti-Pd1 Enhances Immunotherapy of Hepatocellular Carcinoma Via Erbb2/Stat3/Pd-L1 Signaling. Front. Oncol. 2023, 13, 1198118. [Google Scholar] [CrossRef]
- Xu, H.; Chen, K.; Shang, R.; Chen, X.; Zhang, Y.; Song, X.; Evert, M.; Zhong, S.; Li, B.; Calvisi, D.F.; et al. Alpelisib Combination Treatment as Novel Targeted Therapy against Hepatocellular Carcinoma. Cell Death Dis. 2021, 12, 920. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.L.; Zhu, Z.M. Comprehensive Analysis of N6-Methyladenosine -Related Long Non-Coding Rnas and Immune Cell Infiltration in Hepatocellular Carcinoma. Bioengineered 2021, 12, 1708–1724. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhang, Q.; Liu, F.; Quan, J. A Novel Cuproptosis-Related Lncrna Signature Predicts the Prognosis and Immune Landscape in Bladder Cancer. Front. Immunol. 2022, 13, 1027449. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A Survey of Best Practices for Rna-Seq Data Analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Xin, S.; Li, R.; Su, J.; Cao, Q.; Wang, H.; Wei, Z.; Li, G.; Qin, W.; Zhang, Z.; Wang, C.; et al. Novel Model Based on Disulfidptosis-Related Genes to Predict Prognosis and Therapy of Bladder Urothelial Carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 13925–13942. [Google Scholar] [CrossRef]
- Xue, W.; Qiu, K.; Dong, B.; Guo, D.; Fu, J.; Zhu, C.; Niu, Z. Disulfidptosis-Associated Long Non-Coding Rna Signature Predicts the Prognosis, Tumor Microenvironment, and Immunotherapy and Chemotherapy Options in Colon Adenocarcinoma. Cancer Cell. Int. 2023, 23, 218. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, W.; Yan, Y. Identification and Characterization of a Novel Molecular Classification Based on Disulfidptosis-Related Genes to Predict Prognosis and Immunotherapy Efficacy in Hepatocellular Carcinoma. Aging 2023, 15, 6135–6151. [Google Scholar] [CrossRef] [PubMed]
- Grammas, P.; Moore, P.; Weigel, P.H. Microvessels from Alzheimer’s Disease Brains Kill Neurons in Vitro. Am. J. Pathol. 1999, 154, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. The Lasso Method for Variable Selection in the Cox Model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Xu, F.; Huang, X.; Li, Y.; Chen, Y.; Lin, L. M(6)a-Related Lncrnas Are Potential Biomarkers for Predicting Prognoses and Immune Responses in Patients with Luad. Mol. Ther. Nucleic Acids 2021, 24, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Ringner, M. What Is Principal Component Analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Rich, J.T.; Neely, J.G.; Paniello, R.C.; Voelker, C.C.J.; Nussenbaum, B.; Wang, E.W. A Practical Guide to Understanding Kaplan-Meier Curves. Otolaryngol. Head Neck. Surg. 2010, 143, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, P.C.; Jager, K.J.; Zwinderman, A.H.; Zoccali, C.; Dekker, F.W. The Analysis of Survival Data in Nephrology: Basic Concepts and Methods of Cox Regression. Kidney Int. 2008, 74, 705–709. [Google Scholar] [CrossRef]
- Yadav, V.K.; De, S. An Assessment of Computational Methods for Estimating Purity and Clonality Using Genomic Data Derived from Heterogeneous Tumor Tissue Samples. Brief Bioinform. 2015, 16, 232–241. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring Tumour Purity and Stromal and Immune Cell Admixture from Expression Data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. Xcell: Digitally Portraying the Tissue Cellular Heterogeneity Landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the Population Abundance of Tissue-Infiltrating Immune and Stromal Cell Populations Using Gene Expression. Genome Biol. 2016, 17, 218. [Google Scholar]
- Li, B.; Severson, E.; Pignon, J.-C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive Analyses of Tumor Immunity: Implications for Cancer Immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Allgäuer, M.; Budczies, J.; Christopoulos, P.; Endris, V.; Lier, A.; Rempel, E.; Volckmar, A.-L.; Kirchner, M.; von Winterfeld, M.; Leichsenring, J.; et al. Implementing Tumor Mutational Burden (Tmb) Analysis in Routine Diagnostics-a Primer for Molecular Pathologists and Clinicians. Transl. Lung Cancer Res. 2018, 7, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Geeleher, P.; Cox, N.; Huang, R.S. Prrophetic: An R Package for Prediction of Clinical Chemotherapeutic Response from Tumor Gene Expression Levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef]
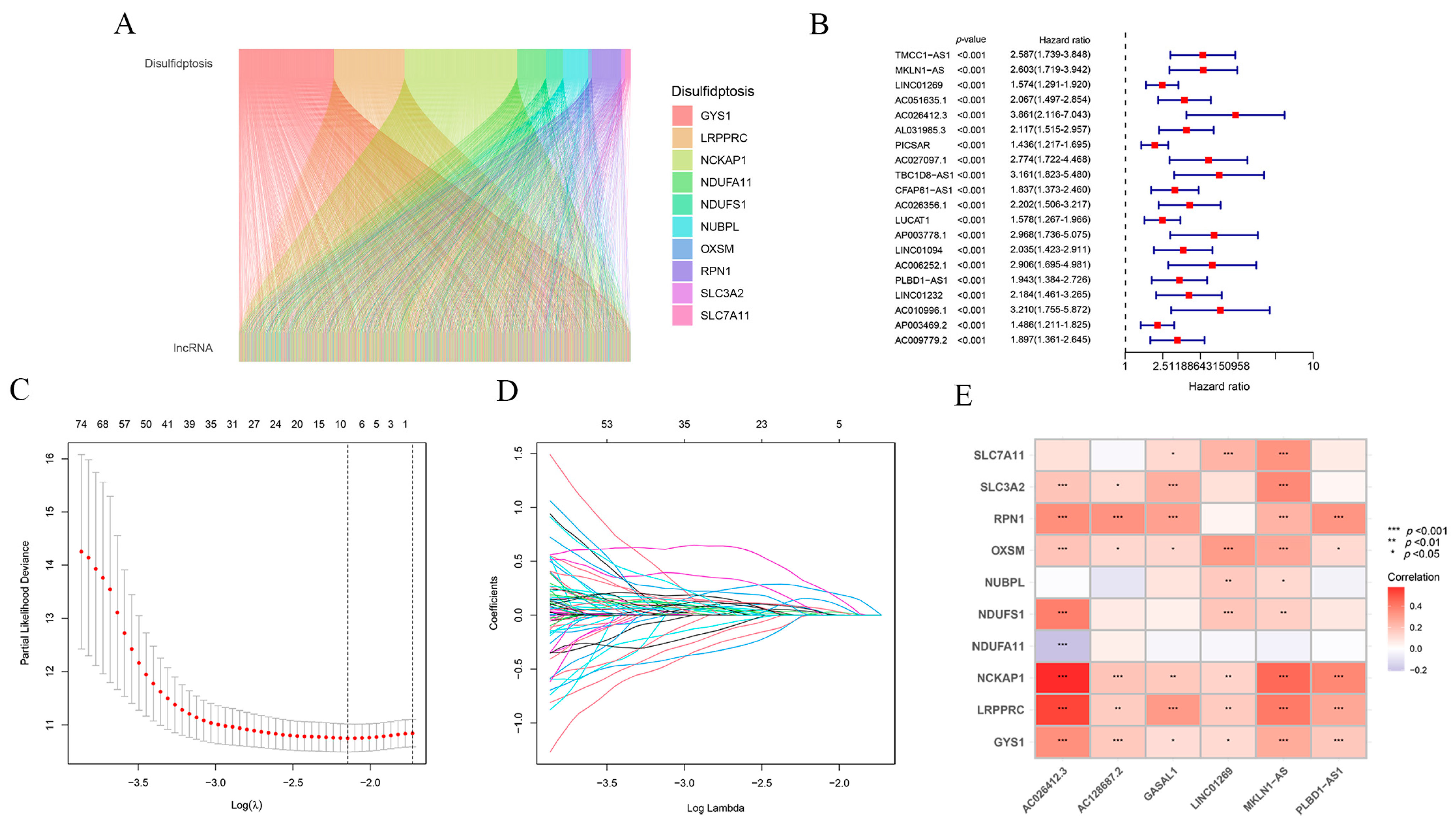


| Disulfidptosis | lncRNA | Cor | p-Value | Regulation |
|---|---|---|---|---|
| NCKAP1 | FGD5-AS1 | 0.755540944 | 2.54 × 10−70 | positive |
| NCKAP1 | AC092614.1 | 0.700809965 | 1.58 × 10−56 | positive |
| NCKAP1 | Z68871.1 | 0.697550131 | 8.32 × 10−56 | positive |
| NCKAP1 | NORAD | 0.697398002 | 8.99 × 10−56 | positive |
| NCKAP1 | AC005670.3 | 0.695765397 | 2.05 × 10−55 | positive |
| NUBPL | AC008549.1 | 0.688460193 | 7.66 × 10−54 | positive |
| NDUFA11 | SNHG25 | 0.68845343 | 7.68 × 10−54 | positive |
| NCKAP1 | EBLN3P | 0.674755577 | 5.15 × 10−51 | positive |
| NCKAP1 | AC004596.1 | 0.670828403 | 3.12 × 10−50 | positive |
| NCKAP1 | FAM111A-DT | 0.663170449 | 9.68 × 10−49 | positive |
| NCKAP1 | AC073046.1 | 0.658844394 | 6.44 × 10−48 | positive |
| NCKAP1 | NRAV | 0.658106969 | 8.87 × 10−48 | positive |
| NCKAP1 | FBXL19-AS1 | 0.656078366 | 2.13 × 10−47 | positive |
| NCKAP1 | AC112220.2 | 0.642313785 | 6.81 × 10−45 | positive |
| NCKAP1 | NNT-AS1 | 0.641857367 | 8.21 × 10−45 | positive |
| NCKAP1 | LINC01560 | 0.641104761 | 1.11 × 10−44 | positive |
| NCKAP1 | AC007406.4 | 0.638614637 | 3.05 × 10−44 | positive |
| NUBPL | AC010501.2 | 0.638601193 | 3.07 × 10−44 | positive |
| LRPPRC | ZNF337-AS1 | 0.637634279 | 4.53 × 10−44 | positive |
| NCKAP1 | LINC02035 | 0.636883618 | 6.12 × 10−44 | positive |
| LRPPRC | MCM3AP-AS1 | 0.635449648 | 1.08 × 10−43 | positive |
| NUBPL | TPRG1-AS1 | 0.632600955 | 3.35 × 10−43 | positive |
| NCKAP1 | AC108463.2 | 0.629554128 | 1.10 × 10−42 | positive |
| NCKAP1 | AC010834.3 | 0.62874749 | 1.51 × 10−42 | positive |
| LRPPRC | FGD5-AS1 | 0.628682804 | 1.55 × 10−42 | positive |
| NCKAP1 | NRSN2-AS1 | 0.627176632 | 2.78 × 10−42 | positive |
| NCKAP1 | LINC01278 | 0.626164716 | 4.10 × 10−42 | positive |
| NCKAP1 | AL157392.3 | 0.624970416 | 6.49 × 10−42 | positive |
| NCKAP1 | STARD7-AS1 | 0.623197317 | 1.28 × 10−41 | positive |
| NCKAP1 | OIP5-AS1 | 0.623017826 | 1.37 × 10−41 | positive |
| LRPPRC | AC092614.1 | 0.621999965 | 2.01 × 10−41 | positive |
| GYS1 | NRSN2-AS1 | 0.621082258 | 2.85 × 10−41 | positive |
| NCKAP1 | CTBP1-DT | 0.619754599 | 4.70 × 10−41 | positive |
| NUBPL | LINC01124 | 0.619531729 | 5.11 × 10−41 | positive |
| NCKAP1 | AC011462.5 | 0.616702842 | 1.47 × 10−40 | positive |
| NCKAP1 | AC107027.3 | 0.613268075 | 5.23 × 10−40 | positive |
| NCKAP1 | RNF213-AS1 | 0.612672379 | 6.51 × 10−40 | positive |
| NCKAP1 | SMARCA5-AS1 | 0.61225062 | 7.59 × 10−40 | positive |
| NCKAP1 | LINC00205 | 0.609943603 | 1.76 × 10−39 | positive |
| NUBPL | GCC2-AS1 | 0.609278159 | 2.24 × 10−39 | positive |
| NCKAP1 | ZEB1-AS1 | 0.60887102 | 2.60 × 10−39 | positive |
| LRPPRC | AC005670.3 | 0.608849702 | 2.62 × 10−39 | positive |
| NCKAP1 | MCM3AP-AS1 | 0.608419664 | 3.06 × 10−39 | positive |
| NDUFS1 | OIP5-AS1 | 0.604600905 | 1.20 × 10−38 | positive |
| NCKAP1 | HCG18 | 0.603891569 | 1.55 × 10−38 | positive |
| NCKAP1 | ZNF32-AS2 | 0.600052829 | 5.99 × 10−38 | positive |
| LRPPRC | AC016747.1 | 0.599723623 | 6.73 × 10−38 | positive |
| NCKAP1 | LINC01521 | 0.599402932 | 7.52 × 10−38 | positive |
| NCKAP1 | AC097448.1 | 0.599076405 | 8.43 × 10−38 | positive |
| NCKAP1 | AC006008.1 | 0.598792563 | 9.31 × 10−38 | positive |
| NCKAP1 | AC107959.3 | 0.598162326 | 1.16 × 10−37 | positive |
| NDUFA11 | TP53TG1 | 0.596806014 | 1.86 × 10−37 | positive |
| NCKAP1 | SBF2-AS1 | 0.596758995 | 1.89 × 10−37 | positive |
| NCKAP1 | WAC-AS1 | 0.59524344 | 3.19 × 10−37 | positive |
| NCKAP1 | ZNF337-AS1 | 0.594297989 | 4.41 × 10−37 | positive |
| NDUFA11 | AC108673.3 | 0.594176279 | 4.60 × 10−37 | positive |
| NCKAP1 | AP001469.2 | 0.593039476 | 6.79 × 10−37 | positive |
| GYS1 | STARD7-AS1 | 0.590300693 | 1.73 × 10−36 | positive |
| LRPPRC | HCG18 | 0.588923912 | 2.75 × 10−36 | positive |
| LRPPRC | AL133243.2 | 0.588483387 | 3.19 × 10−36 | positive |
| NDUFS1 | PAXIP1-AS2 | 0.587674426 | 4.18 × 10−36 | positive |
| LRPPRC | AC114763.1 | 0.585416633 | 8.90 × 10−36 | positive |
| NCKAP1 | AC073254.1 | 0.58450427 | 1.20 × 10−35 | positive |
| NUBPL | AL132800.1 | 0.584445842 | 1.23 × 10−35 | positive |
| GYS1 | AC090409.1 | 0.584364815 | 1.26 × 10−35 | positive |
| NCKAP1 | AP003392.1 | 0.584330402 | 1.28 × 10−35 | positive |
| GYS1 | AC005261.1 | 0.583205211 | 1.85 × 10−35 | positive |
| NCKAP1 | WARS2-AS1 | 0.582826349 | 2.10 × 10−35 | positive |
| LRPPRC | EBLN3P | 0.580224203 | 4.94 × 10−35 | positive |
| GYS1 | AL353748.3 | 0.579802946 | 5.67 × 10−35 | positive |
| LRPPRC | AC073046.1 | 0.578759756 | 7.96 × 10−35 | positive |
| LRPPRC | SNHG16 | 0.577307032 | 1.28 × 10−34 | positive |
| GYS1 | WDFY3-AS2 | 0.577261915 | 1.29 × 10−34 | positive |
| NCKAP1 | AC026412.3 | 0.577055681 | 1.38 × 10−34 | positive |
| NCKAP1 | EIF3J-DT | 0.575128924 | 2.58 × 10−34 | positive |
| SLC7A11 | AC016717.2 | 0.574785212 | 2.88 × 10−34 | positive |
| NCKAP1 | NIPBL-DT | 0.573977047 | 3.73 × 10−34 | positive |
| NCKAP1 | PAXIP1-AS2 | 0.572860551 | 5.33 × 10−34 | positive |
| LRPPRC | AC007406.4 | 0.57199109 | 7.02 × 10−34 | positive |
| NDUFA11 | SNHG9 | 0.570739778 | 1.04 × 10−33 | positive |
| NCKAP1 | AC026979.4 | 0.569894433 | 1.36 × 10−33 | positive |
| NCKAP1 | AC016705.2 | 0.56982153 | 1.40 × 10−33 | positive |
| NDUFS1 | AC135050.5 | 0.569725317 | 1.44 × 10−33 | positive |
| NCKAP1 | AC005034.5 | 0.569677444 | 1.46 × 10−33 | positive |
| GYS1 | LINC01772 | 0.569075281 | 1.77 × 10−33 | positive |
| NCKAP1 | WDFY3-AS2 | 0.56880377 | 1.92 × 10−33 | positive |
| LRPPRC | NORAD | 0.568364614 | 2.21 × 10−33 | positive |
| NCKAP1 | AC025171.2 | 0.567681518 | 2.74 × 10−33 | positive |
| NCKAP1 | AC114763.1 | 0.566813614 | 3.59 × 10−33 | positive |
| LRPPRC | CTBP1-DT | 0.566805009 | 3.60 × 10−33 | positive |
| NCKAP1 | AL499602.1 | 0.566769896 | 3.64 × 10−33 | positive |
| NCKAP1 | LINC00342 | 0.566560491 | 3.89 × 10−33 | positive |
| NCKAP1 | AL133243.2 | 0.565705344 | 5.07 × 10−33 | positive |
| NCKAP1 | AC120114.1 | 0.565556097 | 5.31 × 10−33 | positive |
| LRPPRC | AC073254.1 | 0.562259089 | 1.47 × 10−32 | positive |
| GYS1 | WARS2-AS1 | 0.561505107 | 1.86 × 10−32 | positive |
| NCKAP1 | LINC00265 | 0.560909226 | 2.23 × 10−32 | positive |
| GYS1 | AP002761.3 | 0.560771952 | 2.33 × 10−32 | positive |
| NCKAP1 | NPTN-IT1 | 0.560266675 | 2.71 × 10−32 | positive |
| NCKAP1 | TRAPPC12-AS1 | 0.559882531 | 3.05 × 10−32 | positive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, X.; Wang, Y.; Yang, Y.; Fu, Y.; Liu, Y. Constructed Risk Prognosis Model Associated with Disulfidptosis lncRNAs in HCC. Int. J. Mol. Sci. 2023, 24, 17626. https://doi.org/10.3390/ijms242417626
Jia X, Wang Y, Yang Y, Fu Y, Liu Y. Constructed Risk Prognosis Model Associated with Disulfidptosis lncRNAs in HCC. International Journal of Molecular Sciences. 2023; 24(24):17626. https://doi.org/10.3390/ijms242417626
Chicago/Turabian StyleJia, Xiao, Yiqi Wang, Yang Yang, Yueyue Fu, and Yijin Liu. 2023. "Constructed Risk Prognosis Model Associated with Disulfidptosis lncRNAs in HCC" International Journal of Molecular Sciences 24, no. 24: 17626. https://doi.org/10.3390/ijms242417626






