The Sertoli Cell Complement Signature: A Suspected Mechanism in Xenograft Survival
Abstract
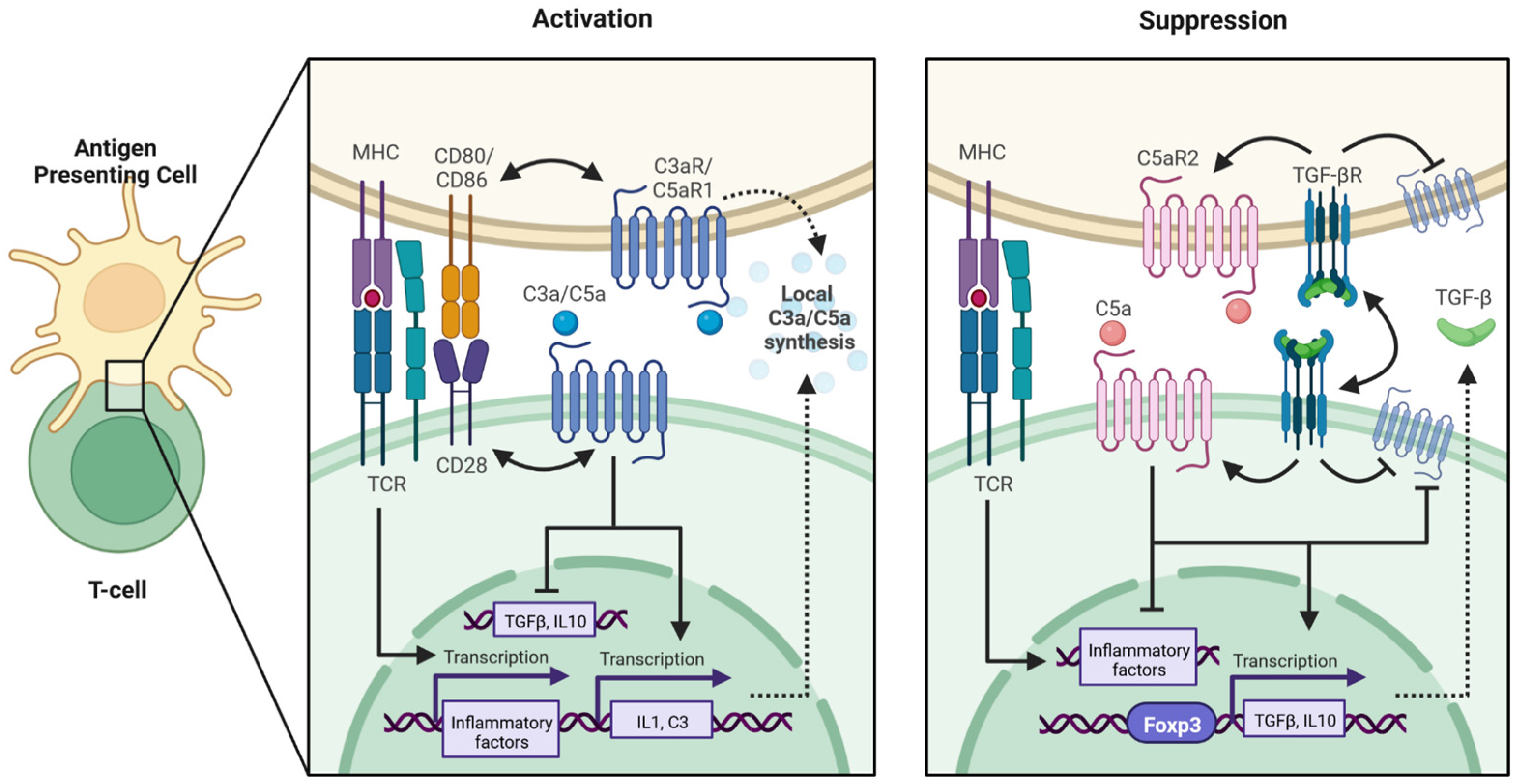
:1. Introduction
2. Results
2.1. Pig Sertoli Cells Survive Human Complement
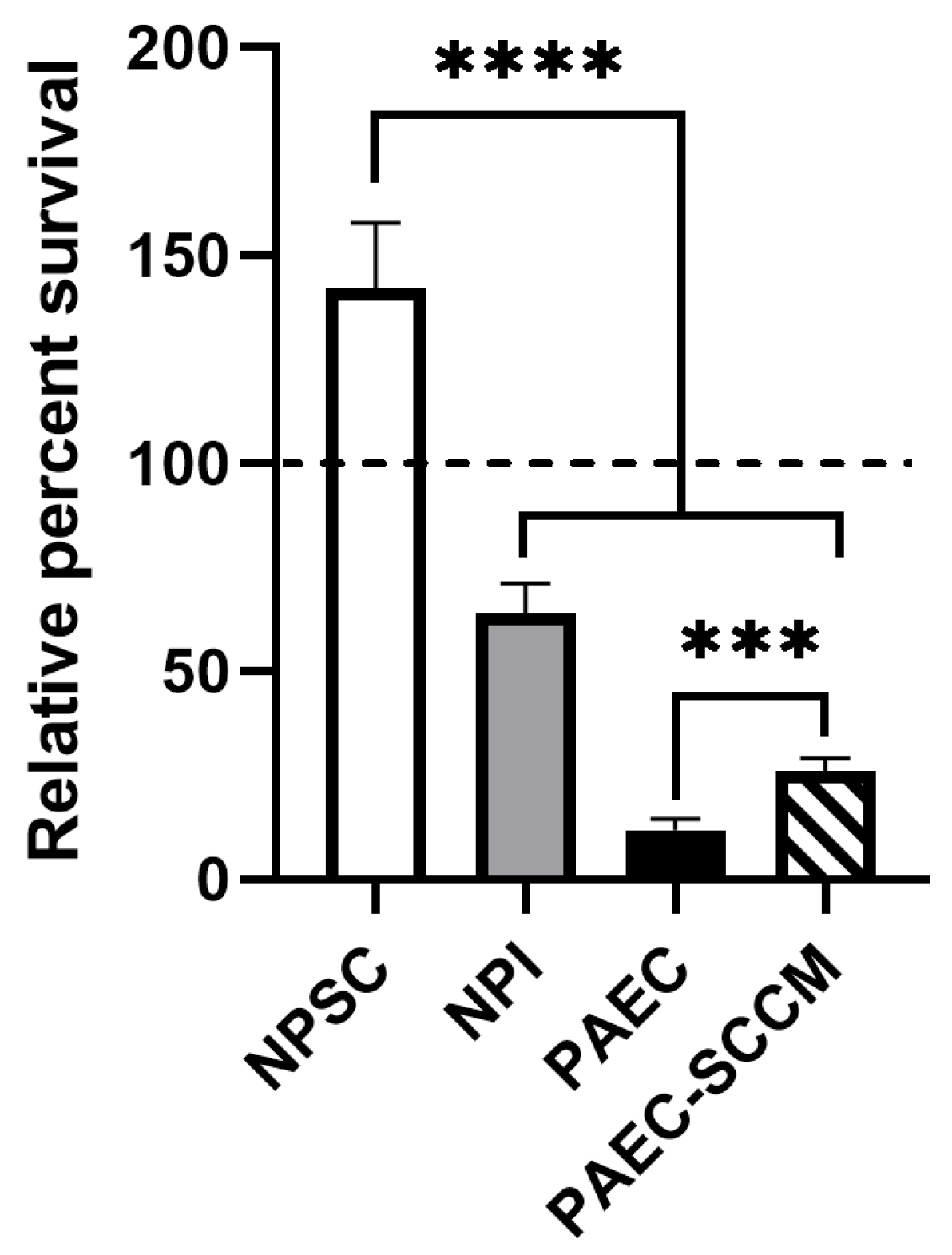
2.2. Pig SCCM Significantly Increases PAEC Survival of Human Complement
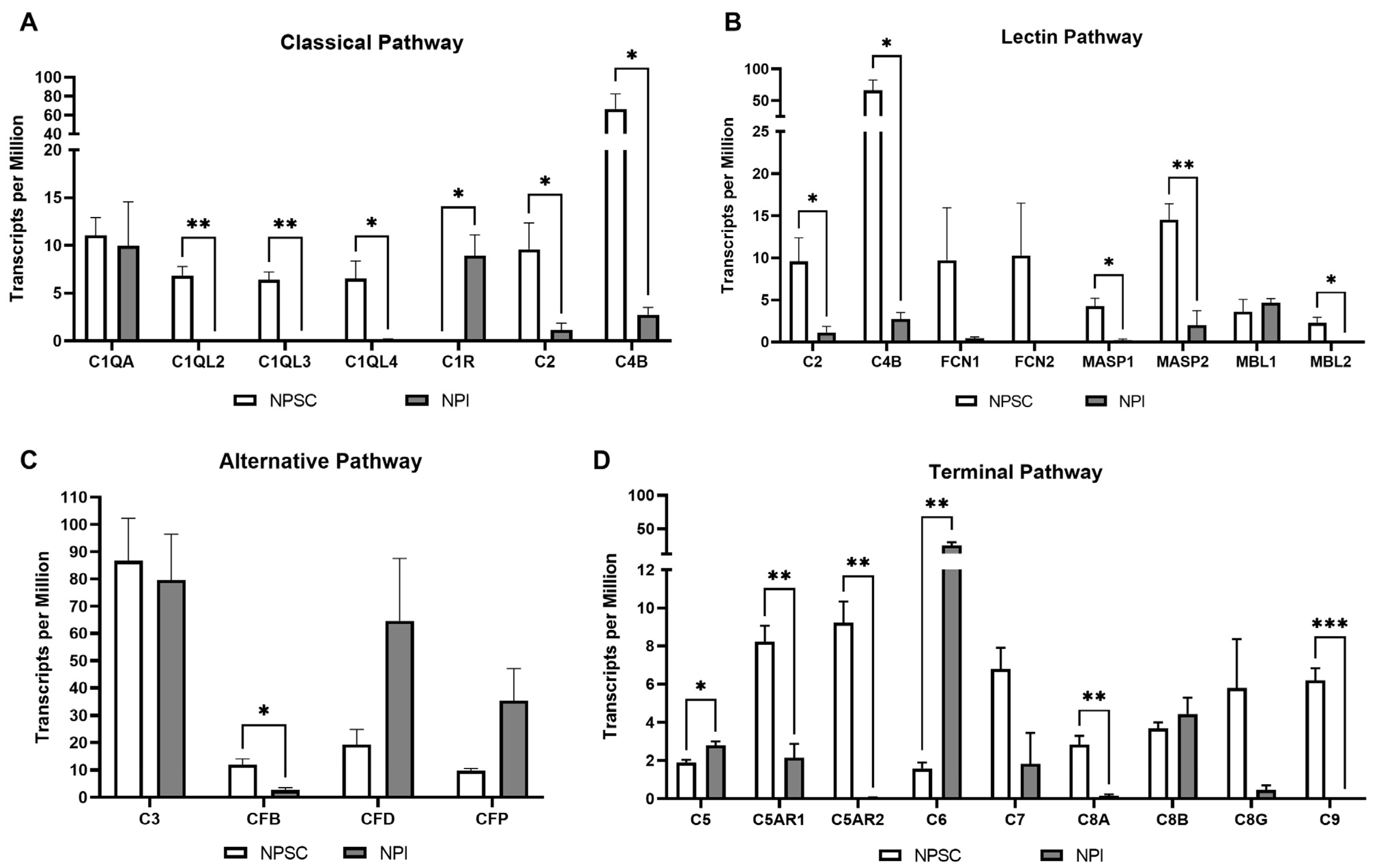
2.3. RNA Sequencing Analysis Identified That Pig Sertoli Cells Express Genes for at Least 22 Complement Cascade Components and 3 Complement Receptors
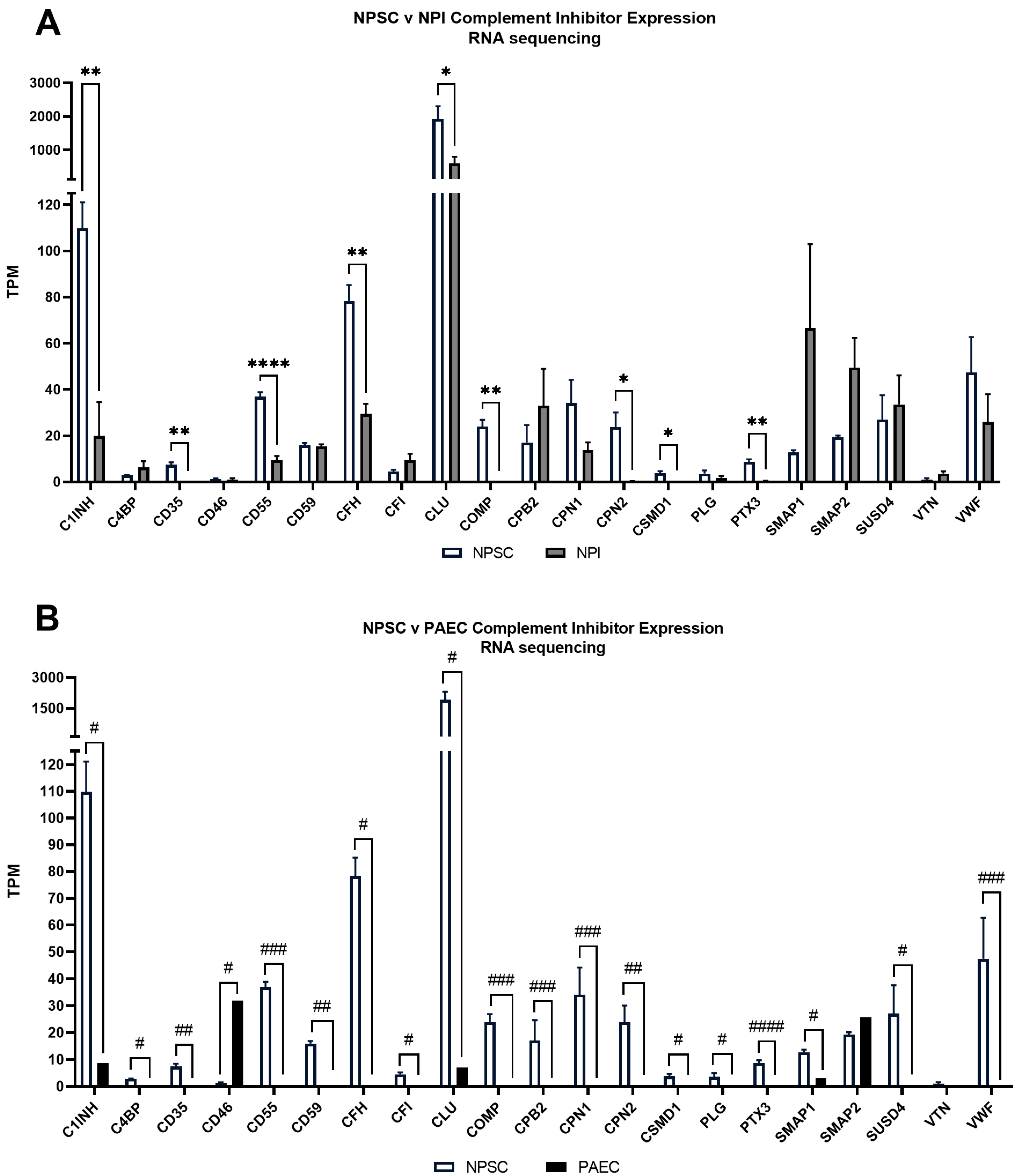
2.4. RNA Sequencing Identified Pig Sertoli Cells Express mRNA for 21 Complement Inhibitors
2.5. PCR Confirms That Pig Sertoli Cells Have Elevated Gene Expression of Several Complement Inhibitors
2.6. Pig Sertoli Cells Secrete Complement Inhibtory Proteins
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Pig Sertoli Cell, Islet, and Aortic Endothelial Cell Isolation
4.3. Human Complement Serum Cytotoxicity Assay
4.4. NPSC-Conditioned Media in HCS Cytotoxicity Assay
4.5. RNA Sequencing
4.6. RNA Isolation and PCR of Complement Inhibitors
4.7. Protein Isolation and Quantification with ELISA
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaur, G.; Wright, K.; Verma, S.; Haynes, A.; Dufour, J.M. The Good, the Bad and the Ugly of Testicular Immune Regulation: A Delicate Balance Between Immune Function and Immune Privilege. Adv. Exp. Med. Biol. 2021, 1288, 21–47. [Google Scholar] [CrossRef]
- Dufour, J.M.; Rajotte, R.V.; Seeberger, K.; Kin, T.; Korbutt, G.S. Long-term survival of neonatal porcine Sertoli cells in non-immunosuppressed rats. Xenotransplantation 2003, 10, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Tönshoff, B. Immunosuppressants in Organ Transplantation. Handb. Exp. Pharmacol. 2020, 261, 441–469. [Google Scholar] [CrossRef] [PubMed]
- Grafals, M.; Thurman, J.M. The Role of Complement in Organ Transplantation. Front. Immunol. 2019, 10, 2380. [Google Scholar] [CrossRef] [PubMed]
- Dufour, J.M.; Hamilton, M.; Rajotte, R.V.; Korbutt, G.S. Neonatal porcine Sertoli cells inhibit human natural antibody-mediated lysis. Biol. Reprod. 2005, 72, 1224–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, K.; Dziuk, R.; Mital, P.; Kaur, G.; Dufour, J.M. Xenotransplanted Pig Sertoli Cells Inhibit Both the Alternative and Classical Pathways of Complement-Mediated Cell Lysis While Pig Islets Are Killed. Cell Transplant. 2016, 25, 2027–2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnum, S.; Schein, T. The Complement FactsBook, 2nd ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 1–480. [Google Scholar]
- Morgan, B.P.; Harris, C.L. Chapter 1-The complement system: A brief overview. In Complement Regulatory Proteins; Morgan, B.P., Harris, C.L., Eds.; Academic Press: London, UK, 1999; pp. 1–31. [Google Scholar]
- Merle, N.S.; Noe, R.; Halbwachs-Mecarelli, L.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement System Part II: Role in Immunity. Front. Immunol. 2015, 6, 257. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Hara, H.; Cooper, D.K.C. The complex functioning of the complement system in xenotransplantation. Xenotransplantation 2019, 26, e12517. [Google Scholar] [CrossRef]
- Washburn, R.L.; Kaur, G.; Dufour, J.M. Mouse Sertoli Cells Inhibit Humoral-Based Immunity. Int. J. Mol. Sci. 2022, 23, 12760. [Google Scholar] [CrossRef]
- Yancey, K.B.; Lawley, T.J.; Dersookian, M.; Harvath, L. Analysis of the interaction of human C5a and C5a des Arg with human monocytes and neutrophils: Flow cytometric and chemotaxis studies. J. Investig. Dermatol. 1989, 92, 184–189. [Google Scholar] [CrossRef]
- Gao, S.; Cui, Z.; Zhao, M.H. The Complement C3a and C3a Receptor Pathway in Kidney Diseases. Front. Immunol. 2020, 11, 1875. [Google Scholar] [CrossRef]
- Inforzato, A.; Doni, A.; Barajon, I.; Leone, R.; Garlanda, C.; Bottazzi, B.; Mantovani, A. PTX3 as a paradigm for the interaction of pentraxins with the complement system. Semin. Immunol. 2013, 25, 79–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inforzato, A.; Peri, G.; Doni, A.; Garlanda, C.; Mantovani, A.; Bastone, A.; Carpentieri, A.; Amoresano, A.; Pucci, P.; Roos, A.; et al. Structure and function of the long pentraxin PTX3 glycosidic moiety: Fine-tuning of the interaction with C1q and complement activation. Biochemistry 2006, 45, 11540–11551. [Google Scholar] [CrossRef] [PubMed]
- Doni, A.; Stravalaci, M.; Inforzato, A.; Magrini, E.; Mantovani, A.; Garlanda, C.; Bottazzi, B. The Long Pentraxin PTX3 as a Link Between Innate Immunity, Tissue Remodeling, and Cancer. Front. Immunol. 2019, 10, 712. [Google Scholar] [CrossRef] [Green Version]
- Dufour, J.M.; Lord, S.J.; Kin, T.; Rayat, G.R.; Dixon, D.E.; Bleackley, R.C.; Korbutt, G.S.; Rajotte, R.V. Comparison of Successful and Unsuccessful Islet/Sertoli Cell Cotransplant Grafts in Streptozotocin-Induced Diabetic Mice. Cell Transplant. 2007, 16, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Korbutt, G.S.; Elliott, J.F.; Rajotte, R.V. Cotransplantation of allogeneic islets with allogeneic testicular cell aggregates allows long-term graft survival without systemic immunosuppression. Diabetes 1997, 46, 317–322. [Google Scholar] [CrossRef] [Green Version]
- West, E.E.; Kolev, M.; Kemper, C. Complement and the Regulation of T Cell Responses. Annu. Rev. Immunol. 2018, 36, 309–338. [Google Scholar] [CrossRef] [PubMed]
- Arbore, G.; Kemper, C.; Kolev, M. Intracellular complement-the complosome-in immune cell regulation. Mol. Immunol. 2017, 89, 2–9. [Google Scholar] [CrossRef]
- Lu, J.; Wu, X.; Teh, B.K. The regulatory roles of C1q. Immunobiology 2007, 212, 245–252. [Google Scholar] [CrossRef]
- Kouser, L.; Madhukaran, S.P.; Shastri, A.; Saraon, A.; Ferluga, J.; Al-Mozaini, M.; Kishore, U. Emerging and Novel Functions of Complement Protein C1q. Front. Immunol. 2015, 6, 317. [Google Scholar] [CrossRef]
- Zhou, Z.; Xu, M.J.; Gao, B. Hepatocytes: A key cell type for innate immunity. Cell. Mol. Immunol. 2016, 13, 301–315. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Marsh, J.E.; Sacks, S.H. Intrarenal synthesis of complement. Kidney Int. 2001, 59, 1227–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lubbers, R.; van Essen, M.F.; van Kooten, C.; Trouw, L.A. Production of complement components by cells of the immune system. Clin. Exp. Immunol. 2017, 188, 183–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbore, G.; West, E.E.; Rahman, J.; Le Friec, G.; Niyonzima, N.; Pirooznia, M.; Tunc, I.; Pavlidis, P.; Powell, N.; Li, Y.; et al. Complement receptor CD46 co-stimulates optimal human CD8(+) T cell effector function via fatty acid metabolism. Nat. Commun. 2018, 9, 4186. [Google Scholar] [CrossRef] [Green Version]
- Ghannam, A.; Fauquert, J.L.; Thomas, C.; Kemper, C.; Drouet, C. Human complement C3 deficiency: Th1 induction requires T cell-derived complement C3a and CD46 activation. Mol. Immunol. 2014, 58, 98–107. [Google Scholar] [CrossRef]
- Kemper, C.; Atkinson, J.P. T-cell regulation: With complements from innate immunity. Nat. Rev. Immunol. 2007, 7, 9–18. [Google Scholar] [CrossRef]
- Kwan, W.H.; van der Touw, W.; Paz-Artal, E.; Li, M.O.; Heeger, P.S. Signaling through C5a receptor and C3a receptor diminishes function of murine natural regulatory T cells. J. Exp. Med. 2013, 210, 257–268. [Google Scholar] [CrossRef]
- Strainic, M.G.; Shevach, E.M.; An, F.; Lin, F.; Medof, M.E. Absence of signaling into CD4(+) cells via C3aR and C5aR enables autoinductive TGF-beta1 signaling and induction of Foxp3(+) regulatory T cells. Nat. Immunol. 2013, 14, 162–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, G.; Wright, K.; Mital, P.; Hibler, T.; Miranda, J.M.; Thompson, L.A.; Halley, K.; Dufour, J.M. Neonatal Pig Sertoli Cells Survive Xenotransplantation by Creating an Immune Modulatory Environment Involving CD4 and CD8 Regulatory T Cells. Cell Transplant. 2020, 29, 963689720947102. [Google Scholar] [CrossRef]
- Guo, J.; Grow, E.J.; Mlcochova, H.; Maher, G.J.; Lindskog, C.; Nie, X.; Guo, Y.; Takei, Y.; Yun, J.; Cai, L.; et al. The adult human testis transcriptional cell atlas. Cell Res. 2018, 28, 1141–1157. [Google Scholar] [CrossRef]
- Li, R.; Coulthard, L.G.; Wu, M.C.; Taylor, S.M.; Woodruff, T.M. C5L2: A controversial receptor of complement anaphylatoxin, C5a. FASEB J. 2013, 27, 855–864. [Google Scholar] [CrossRef]
- Lesher, A.M.; Song, W.C. Review: Complement and its regulatory proteins in kidney diseases. Nephrology 2010, 15, 663–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorgersen, E.B.; Barratt-Due, A.; Haugaa, H.; Harboe, M.; Pischke, S.E.; Nilsson, P.H.; Mollnes, T.E. The Role of Complement in Liver Injury, Regeneration, and Transplantation. Hepatology 2019, 70, 725–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Dufour, J.M.; Dass, B.; Halley, K.R.; Korbutt, G.S.; Dixon, D.E.; Rajotte, R.V. Sertoli cell line lacks the immunoprotective properties associated with primary Sertoli cells. Cell Transplant. 2008, 17, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Korbutt, G.S.; Elliott, J.F.; Ao, Z.; Smith, D.K.; Warnock, G.L.; Rajotte, R.V. Large scale isolation, growth, and function of porcine neonatal islet cells. J. Clin. Investig. 1996, 97, 2119–2129. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Whitener, R.L.; Peiris, H.; Gu, X.; Chang, C.A.; Lam, J.Y.; Camunas-Soler, J.; Park, I.; Bevacqua, R.J.; Tellez, K.; et al. Molecular and genetic regulation of pig pancreatic islet cell development. Development 2020, 147, dev186213. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Ding, P.; Liang, X.; Ding, X.; Brandt, C.B.; Sjöstedt, E.; Zhu, J.; Bolund, S.; Zhang, L.; de Rooij, L.; et al. Endothelial cell heterogeneity and microglia regulons revealed by a pig cell landscape at single-cell level. Nat. Commun. 2022, 13, 3620. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
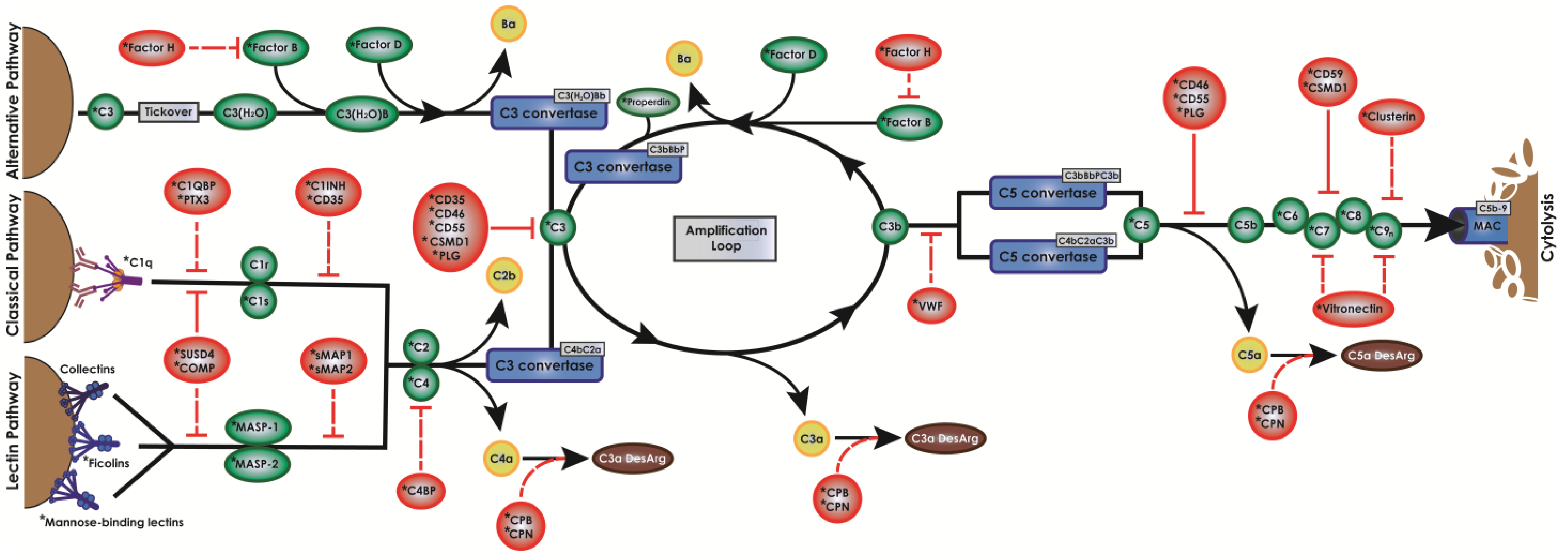


| Inhibitor | Mass (kD) | Distribution | Target | Function in Complement |
|---|---|---|---|---|
| C1INH | 85–105 | Plasma | C1r, C1s | Dissociates C1r/s from C1q |
| C4BP | 540–590 | Plasma | C4b | Prevents assembly of CP C3 convertase |
| CD35 | 190–250 | Membrane | C1q, MBL, C4b, C3b | Competitively binds C1q/MBL/C4b/C3b |
| CD46 | 51–68 | Membrane | C4b, C3b | Cleaves C3b/C4b |
| CD55 | 60–75 | Membrane | C3b | Decays convertases, inhibits C3 cleavage |
| CD59 | 18–23 | Membrane | C8 | Suicide inhibition binding of C8 |
| CFH | 115 | Plasma | C3b, Bb | Cleaves C3b, dissociates Bb from C3b |
| CFI * | 88 | Plasma | C4b, C3b | Inactivates C4b/C3b by cleaving α chains |
| CLU | 34–80 | Both | C6, C7, C8, C9 | Inhibits MAC assembly, prevents C9 insertion |
| COMP | 524 | Plasma | C1q, MBL, C3 | Prevents C1q/MBL binding, stabilizes C3 |
| CPB2/N1/N2 | 47–60 | Plasma | C3a, C5a | Removes carboxy terminus to form desArg |
| CSMD1 | 389 | Membrane | C3b, C7 | Cleaves C3b, prevents C7 binding to C6 |
| PLG | 92 | Both | C3, C3b, C3d, C5 | Decays convertases, inhibits C3 |
| PTX3 | 175 | Plasma | C1q | Prevents C1q activation/binding |
| SMAP1/2 | 94 | Plasma | MASP1-3 | Binds MASP1-3 and prevents their action |
| SUSD4 | 49 | Membrane | C1q, MBL | Prevents C1q/MBL binding and C2 cleavage |
| VTN | 75 | Plasma | C7 | Binds C7 to prevent C8 recruitment |
| VWF | 500–20,000 | Plasma | C3b | Cleaves C3b |
| Cell Type | Complement Inhibitors | Complement Factors | Complement Receptors |
|---|---|---|---|
| Sertoli cells (pig) | C1INH, C4BP, CD35, CD46, CD55, CD59, CFH, CFI, CLU, COMP, CPB2, CPN1, CPN2, CSMD1, PLG, PTX3, SMAP1, SMAP2, SUSD4, VTN, VWF | C1q, C1s, C2, C3, C4, C5, C6, C7, C8, C9, CFB, CFD, CFP, FCN1, FCN2, MASP1, MASP2, MBL1, MBL2 | CD35, C5aR1, C5aR2 |
| Testicular tissue cells (human) | C1INH, CD35, CD46, CD55, CD59, CRIg, CFH, CFHR1, CFI, CLU, COMP, CPB1, CPN1, CSMD1, PTX3, SMAP1, SMAP2, SUSD4, VTN, VWF | C1q, C1s, C2, C3, C4, C5, C6, C7, C8, C9, CFD, CFP, FCN1, MASP1 | CD21, CD35, C3aR, C5aR1, C5aR2, CRIg |
| Complement-producing cells | |||
| Hepatocytes | C1INH, C4BP, CD46, CD55, CD59, CFH, CFHR1-5, CFI, CLU, PLG, PTX3, SMAP1, SMAP2, VTN | C1r, C1s, C2, C3, C4, C5, C6, C8, C9, CFB, FCN1, FCN2, FCN3, MBL, MASP1, MASP2, MASP3 | C5aR2 |
| Renal cells | CD35, CD46, CD55, CD59, CLU, PLG, PTX3, SMAP1, SMAP2, VTN, VWF | C1q, C2, C3, C4, CFB, CFD, C9 | CD35 |
| Immune cells | |||
| Neutrophils | CD35, C4BP, CD46, CD55, CD59, CLU, PTX3, SMAP1, SMAP2 | C3, C6, C7, CFB, CFP, FCN1 | CD35, CR3, CR4, C3aR, C5aR1 |
| Monocytes | CD35, C1INH, C4BP, CFH, CFI, CLU, PTX3, SMAP1, SMAP2 | C1q, C1r, C1s, C2, C3, C4, C5, C6, C7, C8, C9, CFB, CFD, CFP | CD35, CR3, CR4, C3aR, C5aR1 |
| Macrophages | C1INH, CD35, CFH, CFI, CLU, PTX3, SMAP1, SMAP2 | C1q, C1r, C1s, C2, C3, C4, C5, CFB, CFD, CFP | CD35, CR3, CR4, CRIg, C3aR, C5aR1, C5aR2 |
| Dendritic cells | C4BP, CD35, CFH, CFI, CLU, PTX3, SMAP1, SMAP2 | C1q, C1r, C1s, C2, C3, C4, C5, C7, C8, C9, CFB, CFD, CFP | CD35, CR3, CR4, CRIg, C3aR, C5aR1 |
| B cells | CD35, CD46, CD55, CD59, CLU, PTX3, SMAP1, SMAP2 | C5 | CD35, CD21, CR4, C5aR1 |
| T cells | C1QBP, CD35, CD46, CD55, CD59, CFH, CLU, PTX3, SMAP1, SMAP2, SUSD4 | C3, C5, CFB, CFD, CFP | C1qBP, CD35, C3aR, C5aR1, C5aR2 |
| Gene | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) |
|---|---|---|
| C1INH 1 | GAC CAA GTT CTA TCC CAC TCA C | CAG GTT GAG GTC GTA GGT AAA G |
| CD35 | GAG TTT GGA GCA GCC TTC CT | GGC ACC CAT GTG TTG TTG AC |
| CD46 | ATC GCT GCA ATT GTT GTG GG | GAT TCC ACG TCC TCT CAG CA |
| CD55 | GAC TTG TTA TTT GGC GCA TCC | ATC GCC AGT CGC AGG TAA AT |
| CFH | GAC ACG ACC TCA TTC CCA TTA C | GTT CTG ACC ACY GTC CAT TCT C |
| CLU | GCA GAA TGA CGA CCG CTA CT | CGT GAG ATT CAC CTC TCA CTC C |
| COMP | GGA CAG TGA TGG TGA TGG TAT AG | TCA CAT GCA TCT CCC ACA AA |
| CPN2 | TGC TTC ATC CAC GAG GTG TT | ACC ACT TTG GTC AGG TTG GG |
| CSMD1 | GGC TTT GTG GAA AAT GCC GT | TGC ACG AGT AGT GAA CCA CC |
| GAPDH 2 | CCT CCC CGT TCG ACA GAC A | GAT GCG GCC AAA TCC GTT |
| PTX3 | AGC AGG TTG TGA AAC AGC GA | GTT TCA TTG GTG TTG CCG GG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Washburn, R.L.; Martinez-Marin, D.; Korać, K.; Sniegowski, T.; Rodriguez, A.R.; Chilton, B.S.; Hibler, T.; Pruitt, K.; Bhutia, Y.D.; Dufour, J.M. The Sertoli Cell Complement Signature: A Suspected Mechanism in Xenograft Survival. Int. J. Mol. Sci. 2023, 24, 1890. https://doi.org/10.3390/ijms24031890
Washburn RL, Martinez-Marin D, Korać K, Sniegowski T, Rodriguez AR, Chilton BS, Hibler T, Pruitt K, Bhutia YD, Dufour JM. The Sertoli Cell Complement Signature: A Suspected Mechanism in Xenograft Survival. International Journal of Molecular Sciences. 2023; 24(3):1890. https://doi.org/10.3390/ijms24031890
Chicago/Turabian StyleWashburn, Rachel L., Dalia Martinez-Marin, Ksenija Korać, Tyler Sniegowski, Alexis R. Rodriguez, Beverly S. Chilton, Taylor Hibler, Kevin Pruitt, Yangzom D. Bhutia, and Jannette M. Dufour. 2023. "The Sertoli Cell Complement Signature: A Suspected Mechanism in Xenograft Survival" International Journal of Molecular Sciences 24, no. 3: 1890. https://doi.org/10.3390/ijms24031890
APA StyleWashburn, R. L., Martinez-Marin, D., Korać, K., Sniegowski, T., Rodriguez, A. R., Chilton, B. S., Hibler, T., Pruitt, K., Bhutia, Y. D., & Dufour, J. M. (2023). The Sertoli Cell Complement Signature: A Suspected Mechanism in Xenograft Survival. International Journal of Molecular Sciences, 24(3), 1890. https://doi.org/10.3390/ijms24031890








