Diffusion Tensor Imaging in Amyotrophic Lateral Sclerosis: Machine Learning for Biomarker Development
Abstract
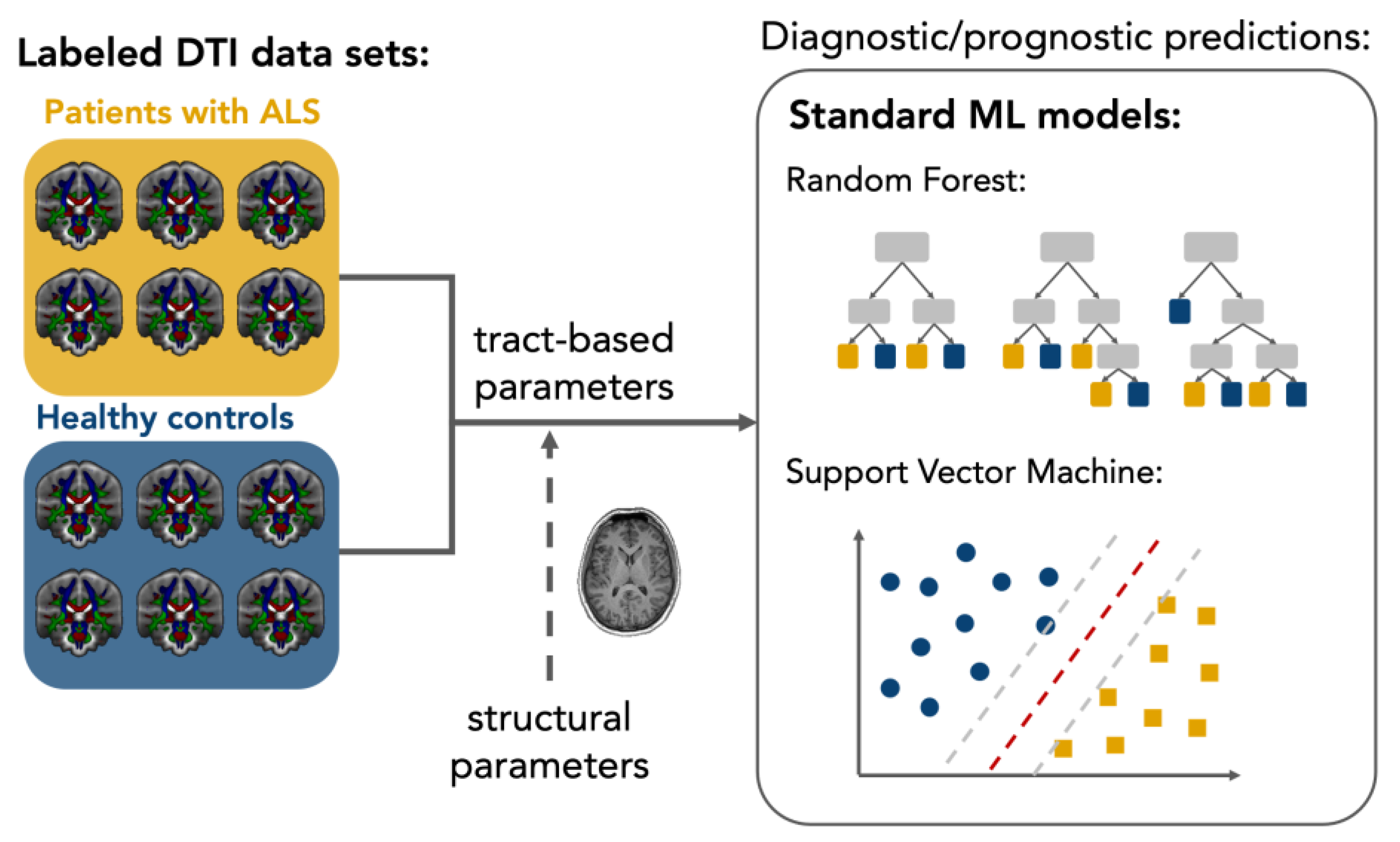
:1. Introduction
2. Diagnostic Models
3. Phenotypic Differentiation
Combinations with Non-Imaging Parameters
4. Longitudinal Monitoring
4.1. Study Conceptualization
4.2. Influences of Aging
5. Limitations
6. Further Perspectives
7. Summary
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Myszczynska, M.A.; Ojamies, P.N.; Lacoste, A.M.B.; Neil, D.; Saffari, A.; Mead, R.; Hautbergue, G.M.; Holbrook, J.D.; Ferraiuolo, L. Applications of Machine Learning to Diagnosis and Treatment of Neurodegenerative Diseases. Nat. Rev. Neurol. 2020, 16, 440–456. [Google Scholar] [CrossRef] [PubMed]
- Van Es, M.A.; Hardiman, O.; Chio, A.; Al-Chalabi, A.; Pasterkamp, R.J.; Veldink, J.H.; van den Berg, L.H. Amyotrophic Lateral Sclerosis. Lancet 2017, 390, 2084–2098. [Google Scholar] [CrossRef] [PubMed]
- Brettschneider, J.; Del Tredici, K.; Toledo, J.B.; Robinson, J.L.; Irwin, D.J.; Grossman, M.; Suh, E.; Van Deerlin, V.M.; Wood, E.M.; Baek, Y.; et al. Stages of PTDP-43 Pathology in Amyotrophic Lateral Sclerosis. Ann. Neurol. 2013, 74, 20–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braak, H.; Brettschneider, J.; Ludolph, A.C.; Lee, V.M.; Trojanowski, J.Q.; Tredici, K.D. Amyotrophic Lateral Sclerosis—A Model of Corticofugal Axonal Spread. Nat. Rev. Neurol. 2013, 9, 708–714. [Google Scholar] [CrossRef] [Green Version]
- Grossman, M. Amyotrophic Lateral Sclerosis—A Multisystem Neurodegenerative Disorder. Nat. Rev. Neurol. 2019, 15, 5–6. [Google Scholar] [CrossRef]
- Bendotti, C.; Bonetto, V.; Pupillo, E.; Logroscino, G.; Al-Chalabi, A.; Lunetta, C.; Riva, N.; Mora, G.; Lauria, G.; Weishaupt, J.H.; et al. Focus on the Heterogeneity of Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Front. Degener. 2020, 21, 485–495. [Google Scholar] [CrossRef]
- Goyal, N.A.; Berry, J.D.; Windebank, A.; Staff, N.P.; Maragakis, N.J.; Berg, L.H.; Genge, A.; Miller, R.; Baloh, R.H.; Kern, R.; et al. Addressing Heterogeneity in Amyotrophic Lateral Sclerosis CLINICAL TRIALS. Muscle Nerve 2020, 62, 156–166. [Google Scholar] [CrossRef] [Green Version]
- Basser, P.J. Inferring Microstructural Features and the Physiological State of Tissues from Diffusion-Weighted Images. NMR Biomed. 1995, 8, 333–344. [Google Scholar] [CrossRef]
- Le Bihan, D.; Mangin, J.F.; Poupon, C.; Clark, C.A.; Pappata, S.; Molko, N.; Chabriat, H. Diffusion Tensor Imaging: Concepts and Applications. J. Magn. Reason. Imaging 2001, 13, 534–546. [Google Scholar] [CrossRef]
- Song, S.-K.; Sun, S.-W.; Ramsbottom, M.J.; Chang, C.; Russell, J.; Cross, A.H. Dysmyelination Revealed through MRI as Increased Radial (but Unchanged Axial) Diffusion of Water. Neuroimage 2002, 17, 1429–1436. [Google Scholar] [CrossRef]
- Kassubek, J.; Müller, H.-P.; Del Tredici, K.; Lulé, D.; Gorges, M.; Braak, H.; Ludolph, A.C. Imaging the Pathoanatomy of Amyotrophic Lateral Sclerosis in Vivo: Targeting a Propagation-Based Biological Marker. J. Neurol. Neurosurg. Psychiatry 2018, 89, 374–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menke, R.A.L.; Agosta, F.; Grosskreutz, J.; Filippi, M.; Turner, M.R. Neuroimaging Endpoints in Amyotrophic Lateral Sclerosis. Neurotherapeutics 2017, 14, 11–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocar, T.D.; Müller, H.-P.; Ludolph, A.C.; Kassubek, J. Feature Selection from Magnetic Resonance Imaging Data in ALS: A Systematic Review. Ther. Adv. Chronic Dis. 2021, 12, 204062232110510. [Google Scholar] [CrossRef] [PubMed]
- Cardenas-Blanco, A.; Machts, J.; Acosta-Cabronero, J.; Kaufmann, J.; Abdulla, S.; Kollewe, K.; Petri, S.; Heinze, H.-J.; Dengler, R.; Vielhaber, S.; et al. Central White Matter Degeneration in Bulbar- and Limb-Onset Amyotrophic Lateral Sclerosis. J. Neurol. 2014, 261, 1961–1967. [Google Scholar] [CrossRef]
- Müller, H.-P.; Turner, M.R.; Grosskreutz, J.; Abrahams, S.; Bede, P.; Govind, V.; Prudlo, J.; Ludolph, A.C.; Filippi, M.; Kassubek, J.; et al. A Large-Scale Multicentre Cerebral Diffusion Tensor Imaging Study in Amyotrophic Lateral Sclerosis. J. Neurol. Neurosurg. Psychiatry 2016, 87, 570–579. [Google Scholar] [CrossRef] [Green Version]
- Filippini, N.; Douaud, G.; Mackay, C.E.; Knight, S.; Talbot, K.; Turner, M.R. Corpus Callosum Involvement Is a Consistent Feature of Amyotrophic Lateral Sclerosis. Neurology 2010, 75, 1645–1652. [Google Scholar] [CrossRef] [Green Version]
- Cardenas-Blanco, A.; Machts, J.; Acosta-Cabronero, J.; Kaufmann, J.; Abdulla, S.; Kollewe, K.; Petri, S.; Schreiber, S.; Heinze, H.-J.; Dengler, R.; et al. Structural and Diffusion Imaging versus Clinical Assessment to Monitor Amyotrophic Lateral Sclerosis. Neuroimage Clin. 2016, 11, 408–414. [Google Scholar] [CrossRef] [Green Version]
- Van Eijk, R.P.A.; Kliest, T.; van den Berg, L.H. Current Trends in the Clinical Trial Landscape for Amyotrophic Lateral Sclerosis. Curr. Opin. Neurol. 2020, 33, 655–661. [Google Scholar] [CrossRef]
- Steinacker, P.; Huss, A.; Mayer, B.; Grehl, T.; Grosskreutz, J.; Borck, G.; Kuhle, J.; Lulé, D.; Meyer, T.; Oeckl, P.; et al. Diagnostic and Prognostic Significance of Neurofilament Light Chain NF-L, but Not Progranulin and S100B, in the Course of Amyotrophic Lateral Sclerosis: Data from the German MND-Net. Amyotroph. Lateral Scler. Front. Degener. 2017, 18, 112–119. [Google Scholar] [CrossRef]
- Foerster, B.R.; Dwamena, B.A.; Petrou, M.; Carlos, R.C.; Callaghan, B.C.; Pomper, M.G. Diagnostic Accuracy Using Diffusion Tensor Imaging in the Diagnosis of ALS: A Meta-Analysis. Acad. Radiol. 2012, 19, 1075–1086. [Google Scholar] [CrossRef]
- De Marchi, F.; Stecco, A.; Falaschi, Z.; Filippone, F.; Pasché, A.; Bebeti, A.; Leigheb, M.; Cantello, R.; Mazzini, L. Detection of White Matter Ultrastructural Changes for Amyotrophic Lateral Sclerosis Characterization: A Diagnostic Study from Dti-Derived Data. Brain Sci. 2020, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Tahedl, M.; Murad, A.; Lope, J.; Hardiman, O.; Bede, P. Evaluation and Categorisation of Individual Patients Based on White Matter Profiles: Single-Patient Diffusion Data Interpretation in Neurodegeneration. J. Neurol. Sci. 2021, 428, 117584. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.-H.; Beam, A.L.; Kohane, I.S. Artificial Intelligence in Healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Grollemund, V.; Pradat, P.-F.; Querin, G.; Delbot, F.; Le Chat, G.; Pradat-Peyre, J.-F.; Bede, P. Machine Learning in Amyotrophic Lateral Sclerosis: Achievements, Pitfalls, and Future Directions. Front. Neurosci. 2019, 13, 135. [Google Scholar] [CrossRef] [Green Version]
- Davatzikos, C. Machine Learning in Neuroimaging: Progress and Challenges. Neuroimage 2019, 197, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Orrù, G.; Pettersson-Yeo, W.; Marquand, A.F.; Sartori, G.; Mechelli, A. Using Support Vector Machine to Identify Imaging Biomarkers of Neurological and Psychiatric Disease: A Critical Review. Neurosci. Biobehav. Rev. 2012, 36, 1140–1152. [Google Scholar] [CrossRef] [PubMed]
- Sarica, A.; Cerasa, A.; Quattrone, A. Random Forest Algorithm for the Classification of Neuroimaging Data in Alzheimer’s Disease: A Systematic Review. Front. Aging Neurosci. 2017, 9, 329. [Google Scholar] [CrossRef] [Green Version]
- Raudys, Š. Statistical and Neural Classifiers; Advances in Pattern Recognition; Springer London: London, UK, 2001; ISBN 978-1-85233-297-6. [Google Scholar]
- Chen, Q.-F.; Zhang, X.-H.; Huang, N.-X.; Chen, H.-J. Identification of Amyotrophic Lateral Sclerosis Based on Diffusion Tensor Imaging and Support Vector Machine. Front. Neurol. 2020, 11, 275. [Google Scholar] [CrossRef]
- Sarica, A.; Cerasa, A.; Valentino, P.; Yeatman, J.; Trotta, M.; Barone, S.; Granata, A.; Nisticò, R.; Perrotta, P.; Pucci, F.; et al. The Corticospinal Tract Profile in Amyotrophic Lateral Sclerosis. Hum. Brain Mapp. 2017, 38, 727–739. [Google Scholar] [CrossRef]
- Ferraro, P.M.; Agosta, F.; Riva, N.; Copetti, M.; Spinelli, E.G.; Falzone, Y.; Sorarù, G.; Comi, G.; Chiò, A.; Filippi, M. Multimodal Structural MRI in the Diagnosis of Motor Neuron Diseases. Neuroimage Clin. 2017, 16, 240–247. [Google Scholar] [CrossRef]
- Kocar, T.D.; Behler, A.; Ludolph, A.C.; Müller, H.-P.; Kassubek, J. Multiparametric Microstructural MRI and Machine Learning Classification Yields High Diagnostic Accuracy in Amyotrophic Lateral Sclerosis: Proof of Concept. Front. Neurol. 2021, 12, 745475. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.H.G.; Westeneng, H.; Nitert, A.D.; van Veenhuijzen, K.; Meier, J.M.; van der Burgh, H.K.; van Zandvoort, M.J.E.; van Es, M.A.; Veldink, J.H.; van den Berg, L.H. MRI Clustering Reveals Three ALS Subtypes With Unique Neurodegeneration Patterns. Ann. Neurol. 2022, 92, ana.26488. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wei, Q.; Hou, Y.; Lei, D.; Yuan, A.; Yuan, A.; Ai, Y.; Qin, K.; Yang, J.; Yang, J.; et al. Disruption of the White Matter Structural Network and Its Correlation with Baseline Progression Rate in Patients with Sporadic Amyotrophic Lateral Sclerosis. Transl. Neurodegener. 2021, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Fratello, M.; Caiazzo, G.; Trojsi, F.; Russo, A.; Tedeschi, G.; Tagliaferri, R.; Esposito, F. Multi-View Ensemble Classification of Brain Connectivity Images for Neurodegeneration Type Discrimination. Neuroinform 2017, 15, 199–213. [Google Scholar] [CrossRef] [Green Version]
- Gabel, M.C.; Broad, R.J.; Young, A.L.; Abrahams, S.; Bastin, M.E.; Menke, R.A.L.; Al-Chalabi, A.; Goldstein, L.H.; Tsermentseli, S.; Alexander, D.C.; et al. Evolution of White Matter Damage in Amyotrophic Lateral Sclerosis. Ann. Clin. Transl. Neurol. 2020, 7, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Behler, A.; Müller, H.-P.; Ludolph, A.C.; Lulé, D.; Kassubek, J. A Multivariate Bayesian Classification Algorithm for Cerebral Stage Prediction by Diffusion Tensor Imaging in Amyotrophic Lateral Sclerosis. Neuroimage Clin. 2022, 35, 103094. [Google Scholar] [CrossRef]
- Bede, P.; Murad, A.; Hardiman, O. Pathological Neural Networks and Artificial Neural Networks in ALS: Diagnostic Classification Based on Pathognomonic Neuroimaging Features. J. Neurol. 2022, 269, 2440–2452. [Google Scholar] [CrossRef]
- Del Tredici, K.; Braak, H. Neuropathology and Neuroanatomy of TDP-43 Amyotrophic Lateral Sclerosis. Curr. Opin. Neurol. 2022, 35, 660–671. [Google Scholar] [CrossRef]
- Welsh, R.C.; Jelsone-Swain, L.; Foerster, B.R. The Utility of Independent Component Analysis and Machine Learning in the Identification of the Amyotrophic Lateral Sclerosis Diseased Brain. Front. Hum. Neurosci. 2013, 7, 251. [Google Scholar] [CrossRef] [Green Version]
- Bede, P.; Iyer, P.M.; Finegan, E.; Omer, T.; Hardiman, O. Virtual Brain Biopsies in Amyotrophic Lateral Sclerosis: Diagnostic Classification Based on In Vivo Pathological Patterns. Neuroimage Clin. 2017, 15, 653–658. [Google Scholar] [CrossRef]
- Schuster, C.; Hardiman, O.; Bede, P. Development of an Automated MRI-Based Diagnostic Protocol for Amyotrophic Lateral Sclerosis Using Disease-Specific Pathognomonic Features: A Quantitative Disease-State Classification Study. PLoS ONE 2016, 11, e0167331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Burgh, H.K.; Schmidt, R.; Westeneng, H.-J.; de Reus, M.A.; van den Berg, L.H.; van den Heuvel, M.P. Deep Learning Predictions of Survival Based on MRI in Amyotrophic Lateral Sclerosis. NeuroImage Clin. 2017, 13, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Wirth, A.M.; Khomenko, A.; Baldaranov, D.; Kobor, I.; Hsam, O.; Grimm, T.; Johannesen, S.; Bruun, T.-H.; Schulte-Mattler, W.; Greenlee, M.W.; et al. Combinatory Biomarker Use of Cortical Thickness, MUNIX, and ALSFRS-R at Baseline and in Longitudinal Courses of Individual Patients With Amyotrophic Lateral Sclerosis. Front. Neurol. 2018, 9, 614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behler, A.; Müller, H.; Del Tredici, K.; Braak, H.; Ludolph, A.C.; Lulé, D.; Kassubek, J. Multimodal in Vivo Staging in Amyotrophic Lateral Sclerosis Using Artificial Intelligence. Ann. Clin. Transl. Neurol. 2022, 9, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.; Hardiman, O.; Bede, P. Survival Prediction in Amyotrophic Lateral Sclerosis Based on MRI Measures and Clinical Characteristics. BMC Neurol. 2017, 17, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bede, P.; Elamin, M.; Byrne, S.; Hardiman, O. Sexual Dimorphism in ALS: Exploring Gender-Specific Neuroimaging Signatures. Amyotroph. Lateral Scler. Front. Degener. 2014, 15, 235–243. [Google Scholar] [CrossRef]
- Arbabshirani, M.R.; Plis, S.; Sui, J.; Calhoun, V.D. Single Subject Prediction of Brain Disorders in Neuroimaging: Promises and Pitfalls. NeuroImage 2017, 145, 137–165. [Google Scholar] [CrossRef] [Green Version]
- Brooks, B.R.; Miller, R.G.; Swash, M.; Munsat, T.L. El Escorial Revisited: Revised Criteria for the Diagnosis of Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2000, 1, 293–299. [Google Scholar] [CrossRef]
- Hannaford, A.; Pavey, N.; Bos, M.; Geevasinga, N.; Menon, P.; Shefner, J.M.; Kiernan, M.C.; Vucic, S. Diagnostic Utility of Gold Coast Criteria in Amyotrophic Lateral Sclerosis. Ann. Neurol. 2021, 89, 979–986. [Google Scholar] [CrossRef]
- De Jongh, A.D.; Braun, N.; Weber, M.; van Es, M.A.; Masrori, P.; Veldink, J.H.; van Damme, P.; van den Berg, L.H.; van Eijk, R.P.A. Characterising ALS Disease Progression According to El Escorial and Gold Coast Criteria. J Neurol Neurosurg Psychiatry 2022, 93, 865–870. [Google Scholar] [CrossRef]
- Traynor, B.J.; Codd, M.B.; Corr, B.; Forde, C.; Frost, E.; Hardiman, O. Amyotrophic Lateral Sclerosis Mimic Syndromes: A Population-Based Study. Arch. Neurol. 2000, 57, 109. [Google Scholar] [CrossRef] [Green Version]
- Ludolph, A.; Drory, V.; Hardiman, O.; Nakano, I.; Ravits, J.; Robberecht, W.; Shefner, J. For The WFN Research Group On ALS/MND A Revision of the El Escorial Criteria—2015. Amyotroph. Lateral Scler. Front. Degener. 2015, 16, 291–292. [Google Scholar] [CrossRef] [PubMed]
- De Vries, B.S.; Rustemeijer, L.M.M.; Bakker, L.A.; Schröder, C.D.; Veldink, J.H.; van den Berg, L.H.; Nijboer, T.C.W.; van Es, M.A. Cognitive and Behavioural Changes in PLS and PMA:Challenging the Concept of Restricted Phenotypes. J. Neurol. Neurosurg. Psychiatry 2019, 90, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Finegan, E.; Chipika, R.H.; Shing, S.L.H.; Hardiman, O.; Bede, P. Primary Lateral Sclerosis: A Distinct Entity or Part of the ALS Spectrum? Amyotroph. Lateral Scler. Front. Degener. 2019, 20, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Kassubek, J.; Müller, H.-P. Advanced Neuroimaging Approaches in Amyotrophic Lateral Sclerosis: Refining the Clinical Diagnosis. Expert Rev. Neurother. 2020, 20, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Rosenbohm, A.; Del Tredici, K.; Braak, H.; Huppertz, H.-J.; Ludolph, A.C.; Müller, H.-P.; Kassubek, J. Involvement of Cortico-Efferent Tracts in Flail Arm Syndrome: A Tract-of-Interest-Based DTI Study. J. Neurol. 2022, 269, 2619–2626. [Google Scholar] [CrossRef] [PubMed]
- Sarica, A.; Valentino, P.; Nisticò, R.; Barone, S.; Pucci, F.; Quattrone, A.; Cerasa, A.; Quattrone, A. Assessment of the Corticospinal Tract Profile in Pure Lower Motor Neuron Disease: A Diffusion Tensor Imaging Study. Neurodegener Dis. 2019, 19, 128–138. [Google Scholar] [CrossRef]
- Münch, M.; Müller, H.-P.; Behler, A.; Ludolph, A.C.; Kassubek, J. Segmental Alterations of the Corpus Callosum in Motor Neuron Disease: A DTI and Texture Analysis in 575 Patients. Neuroimage Clin. 2022, 35, 103061. [Google Scholar] [CrossRef]
- Temp, A.G.M.; Naumann, M.; Hermann, A.; Glaß, H. Applied Bayesian Approaches for Research in Motor Neuron Disease. Front. Neurol. 2022, 13, 796777. [Google Scholar] [CrossRef]
- Gromicho, M.; Leão, T.; Oliveira Santos, M.; Pinto, S.; Carvalho, A.M.; Madeira, S.C.; De Carvalho, M. Dynamic Bayesian Networks for Stratification of Disease Progression in Amyotrophic Lateral Sclerosis. Eur. J. Neurol. 2022, 29, 2201–2210. [Google Scholar] [CrossRef]
- Baek, S.-H.; Park, J.; Kim, Y.H.; Seok, H.Y.; Oh, K.-W.; Kim, H.-J.; Kwon, Y.-J.; Sim, Y.; Tae, W.-S.; Kim, S.H.; et al. Usefulness of Diffusion Tensor Imaging Findings as Biomarkers for Amyotrophic Lateral Sclerosis. Sci. Rep. 2020, 10, 5199. [Google Scholar] [CrossRef] [Green Version]
- Alruwaili, A.R.; Pannek, K.; Henderson, R.D.; Gray, M.; Kurniawan, N.D.; McCombe, P.A. Tract Integrity in Amyotrophic Lateral Sclerosis: 6–Month Evaluation Using MR Diffusion Tensor Imaging. BMC Med. Imaging 2019, 19, 19. [Google Scholar] [CrossRef] [Green Version]
- Blain, C.R.V.; Williams, V.C.; Johnston, C.; Stanton, B.R.; Ganesalingam, J.; Jarosz, J.M.; Jones, D.K.; Barker, G.J.; Williams, S.C.R.; Leigh, N.P.; et al. A Longitudinal Study of Diffusion Tensor MRI in ALS. Amyotroph. Lateral. Scler. 2007, 8, 348–355. [Google Scholar] [CrossRef]
- Distaso, E.; Milella, G.; Mezzapesa, D.M.; Introna, A.; D’Errico, E.; Fraddosio, A.; Zoccolella, S.; Dicuonzo, F.; Simone, I.L. Magnetic Resonance Metrics to Evaluate the Effect of Therapy in Amyotrophic Lateral Sclerosis: The Experience with Edaravone. J. Neurol. 2021, 268, 3307–3315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schuff, N.; Woolley, S.C.; Chiang, G.C.; Boreta, L.; Laxamana, J.; Katz, J.S.; Weiner, M.W. Progression of White Matter Degeneration in Amyotrophic Lateral Sclerosis: A Diffusion Tensor Imaging Study. Amyotroph. Lateral Scler. 2011, 12, 421–429. [Google Scholar] [CrossRef]
- Zhang, Y.; Hedo, R.; Rivera, A.; Rull, R.; Richardson, S.; Tu, X.M. Post Hoc Power Analysis: Is It an Informative and Meaningful Analysis? Gen. Psychiatry 2019, 32, e100069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behler, A.; Lulé, D.; Ludolph, A.C.; Kassubek, J.; Müller, H.-P. Longitudinal Monitoring of Amyotrophic Lateral Sclerosis by Diffusion Tensor Imaging: Power Calculations for Group Studies. Front. Neurosci. 2022, 16, 929151. [Google Scholar] [CrossRef]
- Müller, H.-P.; Behler, A.; Landwehrmeyer, G.B.; Huppertz, H.-J.; Kassubek, J. How to Arrange Follow-Up Time-Intervals for Longitudinal Brain MRI Studies in Neurodegenerative Diseases. Front. Neurosci. 2021, 15, 682812. [Google Scholar] [CrossRef] [PubMed]
- Westlye, L.T.; Walhovd, K.B.; Dale, A.M.; Bjørnerud, A.; Due-Tønnessen, P.; Engvig, A.; Grydeland, H.; Tamnes, C.K.; Ostby, Y.; Fjell, A.M. Life-Span Changes of the Human Brain White Matter: Diffusion Tensor Imaging (DTI) and Volumetry. Cereb. Cortex 2010, 20, 2055–2068. [Google Scholar] [CrossRef] [Green Version]
- Cox, S.R.; Ritchie, S.J.; Tucker-Drob, E.M.; Liewald, D.C.; Hagenaars, S.P.; Davies, G.; Wardlaw, J.M.; Gale, C.R.; Bastin, M.E.; Deary, I.J. Ageing and Brain White Matter Structure in 3,513 UK Biobank Participants. Nat. Commun. 2016, 7, 13629. [Google Scholar] [CrossRef]
- Kalra, S.; Müller, H.-P.; Ishaque, A.; Zinman, L.; Korngut, L.; Genge, A.; Beaulieu, C.; Frayne, R.; Graham, S.J.; Kassubek, J. A Prospective Harmonized Multicenter DTI Study of Cerebral White Matter Degeneration in ALS. Neurology 2020, 95, e943–e952. [Google Scholar] [CrossRef]
- Behler, A.; Kassubek, J.; Müller, H.-P. Age-Related Alterations in DTI Metrics in the Human Brain—Consequences for Age Correction. Front. Aging Neurosci. 2021, 13, 682109. [Google Scholar] [CrossRef]
- Franke, K.; Gaser, C. Ten Years of BrainAGE as a Neuroimaging Biomarker of Brain Aging: What Insights Have We Gained? Front. Neurol. 2019, 10, 789. [Google Scholar] [CrossRef] [Green Version]
- Cole, J.H.; Franke, K. Predicting Age Using Neuroimaging: Innovative Brain Ageing Biomarkers. Trends Neurosci. 2017, 40, 681–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, J.H. Multimodality Neuroimaging Brain-Age in UK Biobank: Relationship to Biomedical, Lifestyle, and Cognitive Factors. Neurobiol. Aging 2020, 92, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Kocar, T.D.; Behler, A.; Leinert, C.; Denkinger, M.; Ludolph, A.C.; Müller, H.-P.; Kassubek, J. Artificial Neural Networks for Non-Linear Age Correction of Diffusion Metrics in the Brain. Front. Aging Neurosci. 2022, 14, 999787. [Google Scholar] [CrossRef]
- Fang, F.; Ingre, C.; Roos, P.; Kamel, F.; Piehl, F. Risk Factors for Amyotrophic Lateral Sclerosis. CLEP 2015, 7, 181. [Google Scholar] [CrossRef] [Green Version]
- Hermann, A.; Tarakdjian, G.N.; Temp, A.G.M.; Kasper, E.; Machts, J.; Kaufmann, J.; Vielhaber, S.; Prudlo, J.; Cole, J.H.; Teipel, S.; et al. Cognitive and Behavioural but Not Motor Impairment Increases Brain Age in Amyotrophic Lateral Sclerosis. Brain Commun. 2022, 4, fcac239. [Google Scholar] [CrossRef] [PubMed]
- Franke, K.; Ristow, M.; Gaser, C. Gender-Specific Impact of Personal Health Parameters on Individual Brain Aging in Cognitively Unimpaired Elderly Subjects. Front. Aging Neurosci. 2014, 6, 94. [Google Scholar] [CrossRef] [Green Version]
- Beck, D.; de Lange, A.-M.G.; Alnæs, D.; Maximov, I.I.; Pedersen, M.L.; Leinhard, O.D.; Linge, J.; Simon, R.; Richard, G.; Ulrichsen, K.M.; et al. Adipose Tissue Distribution from Body MRI Is Associated with Cross-Sectional and Longitudinal Brain Age in Adults. NeuroImage Clin. 2022, 33, 102949. [Google Scholar] [CrossRef]
- McEvoy, L.K.; Fennema-Notestine, C.; Elman, J.A.; Eyler, L.T.; Franz, C.E.; Hagler, D.J.; Hatton, S.N.; Lyons, M.J.; Panizzon, M.S.; Dale, A.M.; et al. Alcohol Intake and Brain White Matter in Middle Aged Men: Microscopic and Macroscopic Differences. NeuroImage Clin. 2018, 18, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, X.; Qian, W.; Shen, Z.; Zhang, M. Altered Human Brain Anatomy in Chronic Smokers: A Review of Magnetic Resonance Imaging Studies. Neurol. Sci. 2015, 36, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Topiwala, A.; Allan, C.L.; Valkanova, V.; Zsoldos, E.; Filippini, N.; Sexton, C.; Mahmood, A.; Fooks, P.; Singh-Manoux, A.; Mackay, C.E.; et al. Moderate Alcohol Consumption as Risk Factor for Adverse Brain Outcomes and Cognitive Decline: Longitudinal Cohort Study. BMJ 2017, 357, j2353. [Google Scholar] [CrossRef] [Green Version]
- Grumbach, P.; Opel, N.; Martin, S.; Meinert, S.; Leehr, E.J.; Redlich, R.; Enneking, V.; Goltermann, J.; Baune, B.T.; Dannlowski, U.; et al. Sleep Duration Is Associated with White Matter Microstructure and Cognitive Performance in Healthy Adults. Hum. Brain Mapp. 2020, 41, 4397–4405. [Google Scholar] [CrossRef] [PubMed]
- Dalmaijer, E.S.; Nord, C.L.; Astle, D.E. Statistical Power for Cluster Analysis. BMC Bioinform. 2022, 23, 205. [Google Scholar] [CrossRef]
- Pinaya, W.H.L.; Scarpazza, C.; Garcia-Dias, R.; Vieira, S.; Baecker, L.; F da Costa, P.; Redolfi, A.; Frisoni, G.B.; Pievani, M.; Calhoun, V.D.; et al. Using Normative Modelling to Detect Disease Progression in Mild Cognitive Impairment and Alzheimer’s Disease in a Cross-Sectional Multi-Cohort Study. Sci. Rep. 2021, 11, 15746. [Google Scholar] [CrossRef]
- Filippi, M.; Agosta, F.; Grosskreutz, J.; Benatar, M.; Kassubek, J.; Verstraete, E.; Turner, M.R. Progress towards a Neuroimaging Biomarker for Amyotrophic Lateral Sclerosis. Lancet Neurol. 2015, 14, 786–788. [Google Scholar] [CrossRef]
- Steinbach, R.; Gaur, N.; Stubendorff, B.; Witte, O.W.; Grosskreutz, J. Developing a Neuroimaging Biomarker for Amyotrophic Lateral Sclerosis: Multi-Center Data Sharing and the Road to a “Global Cohort”. Front. Neurol. 2018, 9, 1055. [Google Scholar] [CrossRef] [Green Version]
- Pinto, M.S.; Paolella, R.; Billiet, T.; Van Dyck, P.; Guns, P.-J.; Jeurissen, B.; Ribbens, A.; den Dekker, A.J.; Sijbers, J. Harmonization of Brain Diffusion MRI: Concepts and Methods. Front. Neurosci. 2020, 14, 396. [Google Scholar] [CrossRef]
- Zaharchuk, G.; Gong, E.; Wintermark, M.; Rubin, D.; Langlotz, C.P. Deep Learning in Neuroradiology. Am. J. Neuroradiol. 2018, 39, 1776–1784. [Google Scholar] [CrossRef]
- Wang, S.; Tang, L.; Majety, A.; Rousseau, J.F.; Shih, G.; Ding, Y.; Peng, Y. Trustworthy Assertion Classification through Prompting. J. Biomed. Inform. 2022, 132, 104139. [Google Scholar] [CrossRef]
- Kamagata, K.; Andica, C.; Kato, A.; Saito, Y.; Uchida, W.; Hatano, T.; Lukies, M.; Ogawa, T.; Takeshige-Amano, H.; Akashi, T.; et al. Diffusion Magnetic Resonance Imaging-Based Biomarkers for Neurodegenerative Diseases. IJMS 2021, 22, 5216. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-J.; Zhan, C.; Cai, L.-M.; Lin, J.-H.; Zhou, M.-X.; Zou, Z.-Y.; Yao, X.-F.; Lin, Y.-J. White Matter Microstructural Impairments in Amyotrophic Lateral Sclerosis: A Mean Apparent Propagator MRI Study. NeuroImage Clin. 2021, 32, 102863. [Google Scholar] [CrossRef]
- Welton, T.; Maller, J.J.; Lebel, R.M.; Tan, E.T.; Rowe, D.B.; Grieve, S.M. Diffusion Kurtosis and Quantitative Susceptibility Mapping MRI Are Sensitive to Structural Abnormalities in Amyotrophic Lateral Sclerosis. NeuroImage Clin. 2019, 24, 101953. [Google Scholar] [CrossRef]
- Huang, N.; Zou, Z.; Xue, Y.; Chen, H. Abnormal Cerebral Microstructures Revealed by Diffusion Kurtosis Imaging in Amyotrophic Lateral Sclerosis. J. Magn. Reson. Imaging 2020, 51, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Juengling, F.D.; Wuest, F.; Kalra, S.; Agosta, F.; Schirrmacher, R.; Thiel, A.; Thaiss, W.; Müller, H.-P.; Kassubek, J. Simultaneous PET/MRI: The Future Gold Standard for Characterizing Motor Neuron Disease—A Clinico-Radiological and Neuroscientific Perspective. Front. Neurol. 2022, 13, 890425. [Google Scholar] [CrossRef]
- Hübers, A.; Kassubek, J.; Müller, H.-P.; Broc, N.; Dreyhaupt, J.; Ludolph, A.C. The Ipsilateral Silent Period: An Early Diagnostic Marker of Callosal Disconnection in ALS. Ther. Adv. Chronic Dis. 2021, 12, 20406223211044070. [Google Scholar] [CrossRef] [PubMed]
- Kiernan, M.C.; Vucic, S.; Talbot, K.; McDermott, C.J.; Hardiman, O.; Shefner, J.M.; Al-Chalabi, A.; Huynh, W.; Cudkowicz, M.; Talman, P.; et al. Improving Clinical Trial Outcomes in Amyotrophic Lateral Sclerosis. Nat. Rev. Neurol. 2021, 17, 104–118. [Google Scholar] [CrossRef]


| Study | Algorithm | Task | Features | Sample Size | Model Validation | Accuracy |
|---|---|---|---|---|---|---|
| Chen et al. [29] | SVM | Diagnostic prediction | FA values of all white matter voxels | 22 patients with ALS; 26 HC | Leave-one-out cross-validation | 83% |
| Ferraro et al. [31] | SVM | Diagnostic prediction | FA, MD, AD, and RD of CST and motor callosal tracts | 123 patients with ALS; 44 patients with PUMN; 20 ALS-mimics; 78 HC | Independent test sample | 78% |
| Kocar et al. [32] | SVM | Diagnostic prediction | FA of CST, corticopontine tract, corticorubral tract, corticostriatal pathway, proximal perforant path, CC area II, and CC area III | 98 patients with ALS; 98 HC | Leave-one-out cross-validation | 66% |
| Münch et al. [33] | SVM | Diagnostic prediction | FA of CC area I–III | 432 patients with ALS; 55 patients with PLS; 45 patients with FAS; 22 patients with PBP; 21 patients with LMND; 112 HC | Independent test sample | 65% |
| Li et al. [34] | SVM | Prediction of progression | White matter network matrices | 73 patients with ALS | Nested cross-validation | 85% |
| Sarica et al. [30] | RF | Diagnostic prediction | FA, MD, AD, and RD of CST voxels | 24 patients with ALS; 24 HC | 5-fold cross-validation | 80% |
| Fratello et al. [35] | Multi-view models with RF | Diagnostic prediction | whole-brain FA maps | 41 patients with ALS; 37 patients with Parkinson’s disease; 43 HC | n/a | 58% |
| Gabel et al. [36] | Event-based modeling | Ordering of events, i.e., regional involvement | FA of CST (inferior/middle/superior), CC (genu/body/splenium), cingulum (dorsal section), superior longitudinal fasciculus, inferior longitudinal fasciculus, inferior fronto-occipital fasciculus, and uncinate fasciculus | 154 patients with ALS; 128 HC | Cross-validation | n/a |
| Behler et al. [37] | Multivariate Bayesian classification | Cerebral stage prediction | FA of CST, corticopontine tract, corticorubral tract, corticostriatal pathway, and proximal perforant path | 325 patients with ALS; 130 HC | Comparison to threshold-based DTI staging | n/a |
| Bede et al. [38] | Multilayer perceptron | Diagnostic prediction | FA, MD, AD, and RD of 30 white matter regions | 214 patients with ALS; 37 patients with a non-ALS neurodegenerative diagnosis; 127 HC | Independent test sample | 79% |
| Study | Algorithm | Task | Modalities | Sample Size | Model Validation | Accuracy |
|---|---|---|---|---|---|---|
| Ferraro et al. [31] | SVM | Diagnostic prediction | DTI (tract-based diffusion metrics) + T1w MRI (cortical thickness) | 123 patients with ALS, 44 patients with PUMN, 20 ALS mimics; 78 HC | Independent test sample | 91% |
| Kocar et al. [32] | SVM | Diagnostic prediction | DTI (tract-based diffusion metrics) + T1w MRI (texture parameters) | 98 patients with ALS; 98 HC | Leave-one-out cross-validation | 80% |
| Münch et al. [33] | SVM | Diagnostic prediction | DTI (tract-based diffusion metrics) + T1w MRI (texture parameters) | 432 patients with ALS, 55 patients with PLS, 45 patients with FAS, 22 patients with PBP, 21 patients with LMND; 112 HC | Independent test sample | 84% |
| Bede et al. [41] | Canonical discriminant function | Diagnostic prediction | DTI (ROI-based diffusion metrics) + T1w MRI (ROI-based signal intensity + basal ganglia volumetrics) | 75 patients with ALS; 75 HC | Independent test sample | 90% |
| Fratello et al. [35] | Multi-view models with RF | Diagnostic prediction | DTI (whole-brain FA maps) + fMRI (whole-brain default-mode networks) | 41 patients with ALS, 37 patients with Parkinson’s disease; 43 HC | 5-fold cross-validation | 67% |
| Schuster et al. [42] | Binary logistic regression | Diagnostic prediction | DTI (ROI-based diffusion metrics) + T1w MRI (regional grey matter densities) | 81 patients with ALS; 66 HC | Independent test sample | 78% |
| Schuster et al. [43] | Binary logistic ridge regression | Survival prediction | DTI (regional diffusion metrics) + T1w MRI (regional cortical thickness) | 60 patients with ALS; 69 HC | Independent test sample | 58% |
| Kocar et al. [32] | Multilayer perceptron | Diagnostic prediction | DTI (tract-based diffusion metrics) + T1w MRI (texture parameters) | 98 patients with ALS; 98 HC | Independent test sample | 72% |
| Bede et al. [38] | Multilayer perceptron model | Diagnostic prediction | DTI (ROI-based diffusion metrics) + T1w MRI (cerebral volumes + cortical thicknesses) | 214 patients with ALS, 37 patients with a non-ALS neurodegenerative diagnosis; 127 HC | Independent test sample | 75% |
| Van der Burgh et al. [44] | Deep learning networks | Survival prediction | DTI (tract-based FA) + T1w MRI (cortical thicknesses + subcortical volumes) + clinical parameters | 135 patients with ALS | Independent test sample | 84% |
| Tan et al. [45] | Probabilistic network-based clustering | Divide patients into subgroups of similar neurodegeneration patterns | DTI (white matter connectome FA) + T1w MRI (whole-brain cortical thickness) | 488 patients with ALS; 338 HC | Longitudinal subsample | 90% in the validation sample |
| Behler et al. [46] | Hierarchical clustering | Divide patients into subgroups of similar neurodegeneration patterns | DTI (tract-based FA) + video-oculography + cognitive scores | 245 patients with ALS | No | n/a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Behler, A.; Müller, H.-P.; Ludolph, A.C.; Kassubek, J. Diffusion Tensor Imaging in Amyotrophic Lateral Sclerosis: Machine Learning for Biomarker Development. Int. J. Mol. Sci. 2023, 24, 1911. https://doi.org/10.3390/ijms24031911
Behler A, Müller H-P, Ludolph AC, Kassubek J. Diffusion Tensor Imaging in Amyotrophic Lateral Sclerosis: Machine Learning for Biomarker Development. International Journal of Molecular Sciences. 2023; 24(3):1911. https://doi.org/10.3390/ijms24031911
Chicago/Turabian StyleBehler, Anna, Hans-Peter Müller, Albert C. Ludolph, and Jan Kassubek. 2023. "Diffusion Tensor Imaging in Amyotrophic Lateral Sclerosis: Machine Learning for Biomarker Development" International Journal of Molecular Sciences 24, no. 3: 1911. https://doi.org/10.3390/ijms24031911
APA StyleBehler, A., Müller, H.-P., Ludolph, A. C., & Kassubek, J. (2023). Diffusion Tensor Imaging in Amyotrophic Lateral Sclerosis: Machine Learning for Biomarker Development. International Journal of Molecular Sciences, 24(3), 1911. https://doi.org/10.3390/ijms24031911






