Role of Melatonin in Cancer: Effect on Clock Genes
Abstract
1. Introduction
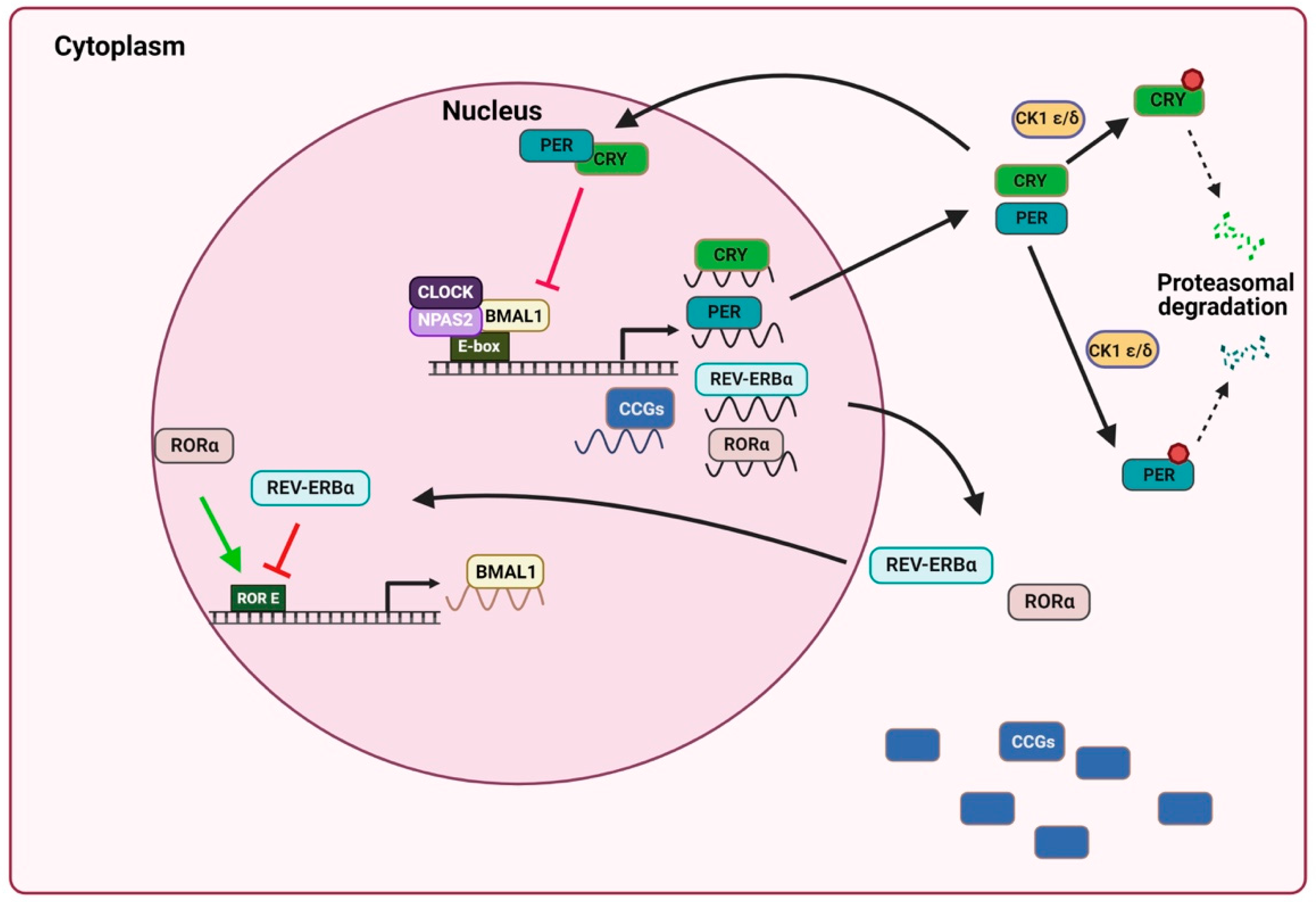
2. General Mechanism of the Circadian Clock
3. Clock Genes and Cancer
3.1. Clock Genes, Proliferation and Apoptosis
3.2. Clock Genes and Metastasis
3.3. Clock Genes and Tumor Immunity
3.4. Clock Genes and Chronotherapy
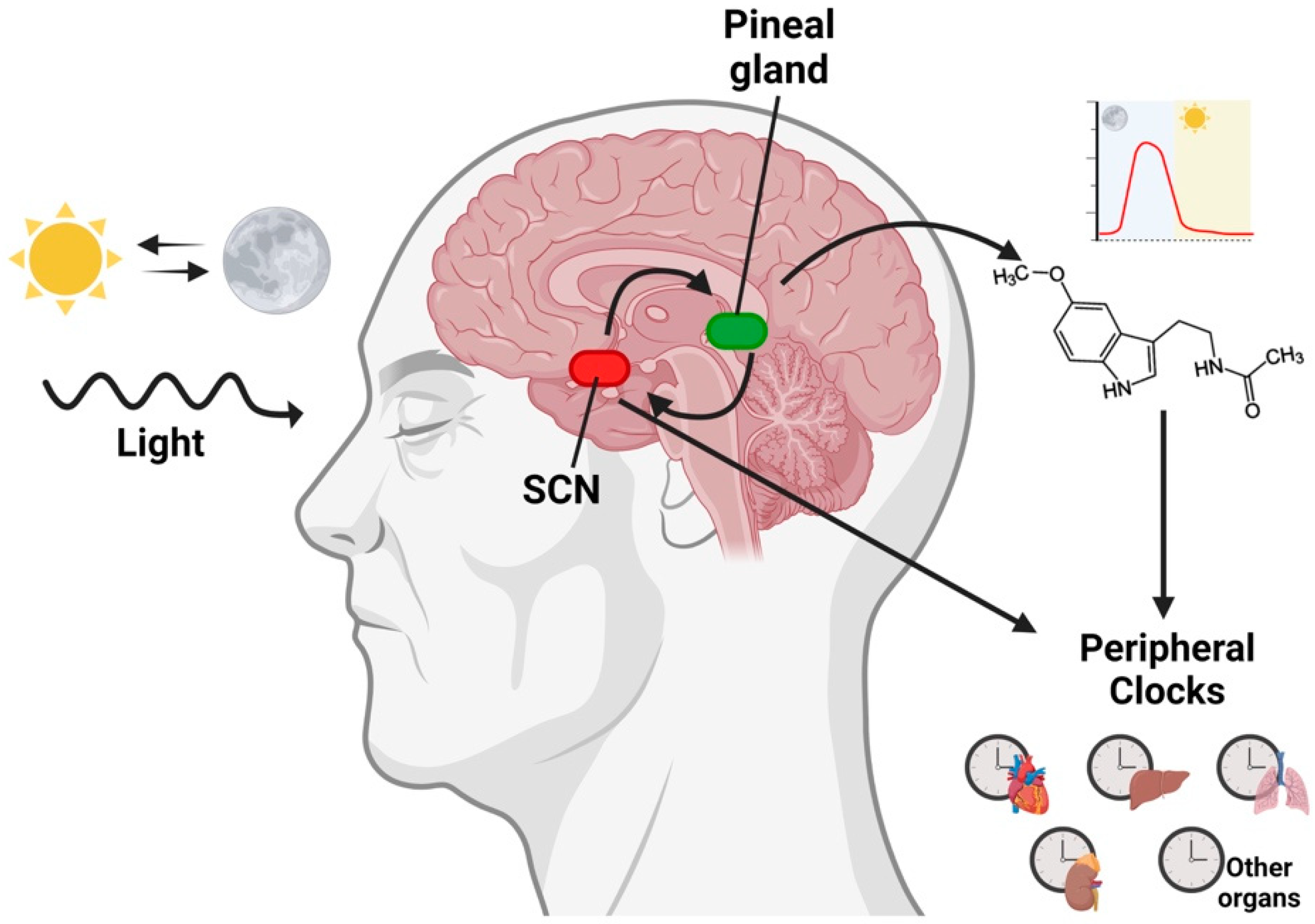
4. Melatonin and Circadian Rhythms
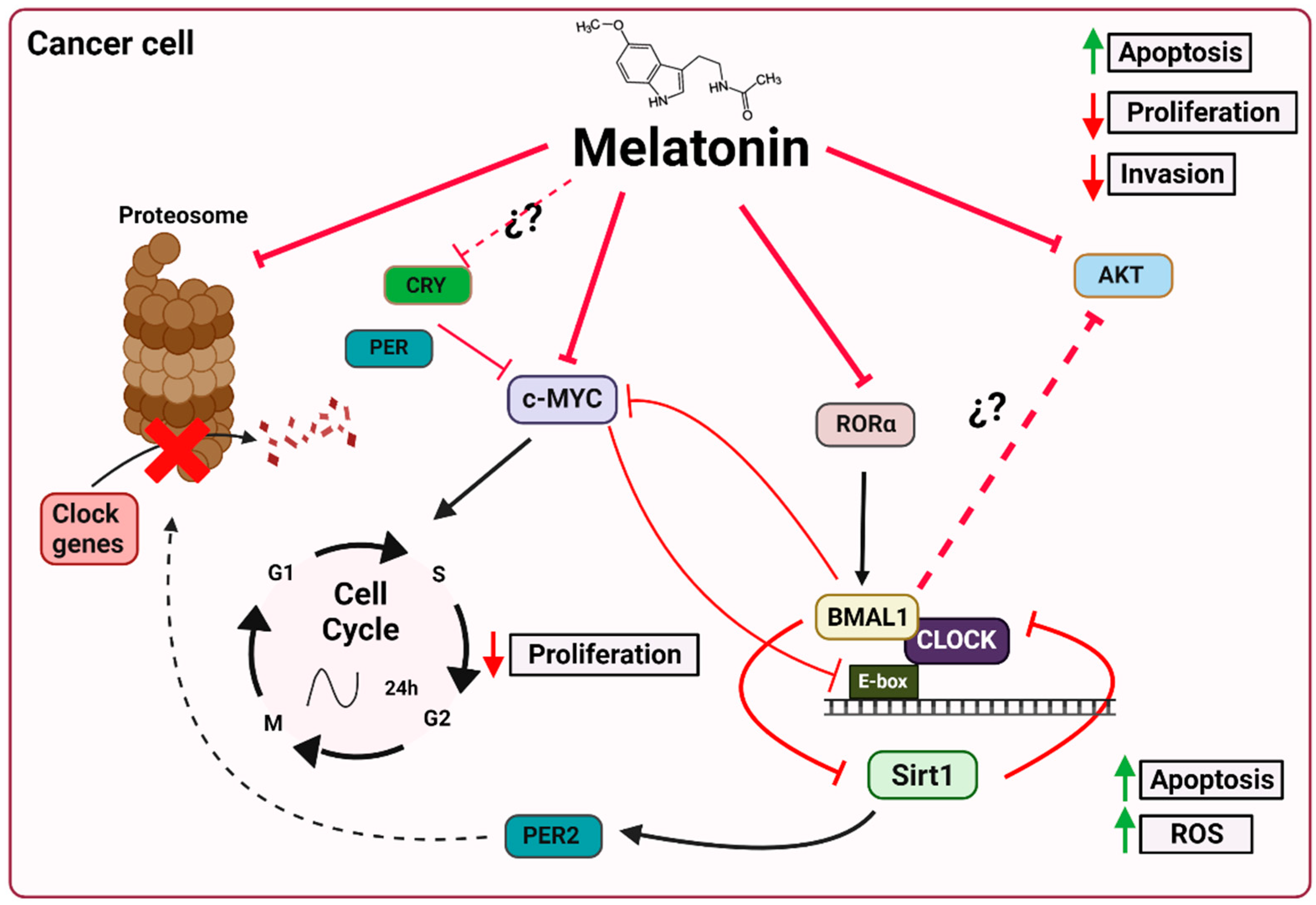
5. Melatonin, Clock Genes and Cancer
5.1. Melatonin and the Ubiquitin–Proteasome System
5.2. Melatonin and c-Myc
5.3. Melatonin and Sirt1
5.4. Melatonin and AKT
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AANAT | arylalkylamine N-acetyltransferase |
| aMT | melatonin |
| BIM | Bcl-2-interacting mediator of cell death |
| Bmal1 brain and muscle aryl hydrocarbon receptor nuclear translocator-like protein 1 | |
| CCGs | clock-controlled genes |
| CK1 ε/δ | cysteine kinase 1 ε/δ |
| Clock | circadian locomotor output cycles kaput |
| Cry | Chryptochrome |
| DC | dendritic cell |
| IARC | International Agency for Research on Cancer |
| MMP9 | matrix metalloproteinase 9 |
| NAD+ | nicotinamide adenine dinucleotide |
| NAMPT | nicotinamide phosphoribosyl transferase |
| NPAS2 | neuronal PAS domain protein 2 |
| Per | Period |
| Rev-erbα | reverse strand of protein ERB alpha |
| RORα | orphan retinoic acid receptor-related alpha |
| ROS | reactive oxygen species |
| SCN | suprachiasmatic nucleus |
| SIRT1 | sirtuin 1 |
References
- Cadenas, C.; Van De Sandt, L.; Edlund, K.; Lohr, M.; Hellwig, B.; Marchan, R.; Schmidt, M.; Rahnenführer, J.; Oster, H.; Hengstler, J.G. Loss of Circadian Clock Gene Expression Is Associated with Tumor Progression in Breast Cancer. Cell Cycle 2014, 13, 3282–3291. [Google Scholar] [CrossRef] [PubMed]
- Acuña-Castroviejo, D.; Escames, G.; Venegas, C.; Díaz-Casado, M.E.; Lima-Cabello, E.; López, L.C.; Rosales-Corral, S.; Tan, D.X.; Reiter, R.J. Extrapineal Melatonin: Sources, Regulation, and Potential Functions. Cell. Mol. Life Sci. 2014, 71, 2997–3025. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J. Pineal Melatonin: Cell Biology of Its Synthesis and of Its Physiological Interactions. Endocr. Rev. 1991, 12, 151–180. [Google Scholar] [CrossRef] [PubMed]
- Cardinali, D.P.; Hardeland, R. Inflammaging, Metabolic Syndrome and Melatonin: A Call for Treatment Studies. Neuroendocrinology 2017, 104, 382–397. [Google Scholar] [CrossRef]
- Rahbarghazi, A.; Siahkouhian, M.; Rahbarghazi, R.; Ahmadi, M.; Bolboli, L.; Keyhanmanesh, R.; Mahdipour, M.; Rajabi, H. Role of Melatonin in the Angiogenesis Potential; Highlights on the Cardiovascular Disease. J. Inflamm. 2021, 18, 4. [Google Scholar] [CrossRef]
- Moslehi, M.; Moazamiyanfar, R.; Dakkali, M.S.; Rezaei, S.; Rastegar-Pouyani, N.; Jafarzadeh, E.; Mouludi, K.; Khodamoradi, E.; Taeb, S.; Najafi, M. Modulation of the Immune System by Melatonin; Implications for Cancer Therapy. Int. Immunopharmacol. 2022, 108, 108890. [Google Scholar] [CrossRef]
- Florido, J.; Rodriguez-Santana, C.; Martinez-Ruiz, L.; López-Rodríguez, A.; Acuña-Castroviejo, D.; Rusanova, I.; Escames, G. Understanding the Mechanism of Action of Melatonin, Which Induces ROS Production in Cancer Cells. Antioxidants 2022, 11, 1621. [Google Scholar] [CrossRef]
- Schernhammer, E.S.; Kroenke, C.H.; Laden, F.; Hankinson, S.E. Night Work and Risk of Breast Cancer. Epidemiology 2006, 17, 108–111. [Google Scholar] [CrossRef]
- LeMasters, G.K.; Genaidy, A.M.; Succop, P.; Deddens, J.; Sobeih, T.; Barriera-Viruet, H.; Dunning, K.; Lockey, J. Cancer Risk among Firefighters: A Review and Meta-Analysis of 32 Studies. J. Occup. Environ. Med. 2006, 48, 1189–1202. [Google Scholar] [CrossRef]
- Conlon, M.; Lightfoot, N.; Kreiger, N. Rotating Shift Work and Risk of Prostate Cancer [1]. Epidemiology 2007, 18, 182–183. [Google Scholar] [CrossRef]
- Viswanathan, A.N.; Schernhammer, E.S. Circulating Melatonin and the Risk of Breast and Endometrial Cancer in Women. Cancer Lett. 2009, 281, 1–7. [Google Scholar] [CrossRef]
- Touitou, Y.; Reinberg, A.; Touitou, D. Association between Light at Night, Melatonin Secretion, Sleep Deprivation, and the Internal Clock: Health Impacts and Mechanisms of Circadian Disruption. Life Sci. 2017, 173, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Lowrey, P.L.; Takahashi, J.S. Mammalian Circadian Biology: Elucidating Genome-Wide Levels of Temporal Organization. Annu. Rev. Genom. Hum. Genet. 2004, 5, 407–441. [Google Scholar] [CrossRef] [PubMed]
- Stratmann, M.; Schibler, U. Properties, Entrainment, and Physiological Functions of Mammalian Peripheral Oscillators. J. Biol. Rhythms 2006, 21, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Fishbein, A.B.; Knutson, K.L.; Zee, P.C. Circadian Disruption and Human Health. J. Clin. Investig. 2021, 131, 1–12. [Google Scholar] [CrossRef]
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular Architecture of the Mammalian Circadian Clock. Trends Cell Biol 2014, 24, 90–99. [Google Scholar] [CrossRef]
- Patke, A.; Young, M.W.; Axelrod, S. Molecular Mechanisms and Physiological Importance of Circadian Rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef]
- Cho, H.; Zhao, X.; Hatori, M.; Yu, R.T.; Barish, G.D.; Lam, M.T.; Chong, L.; Ditacchio, L.; Atkins, A.R.; Glass, C.K.; et al. Regulation of Circadian Behavior and Metabolism by Rev-Erbα and Rev-Erbβ. Nature 2012, 485, 123–127. [Google Scholar] [CrossRef]
- Koike, N.; Yoo, S.-H.; Huang, H.-C.; Kumar, V.; Lee, C.; Kim, T.-K.; Takahashi, J.S. Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef]
- Tokunaga, H.; Takebayashi, Y.; Utsunomiya, H.; Akahira, J.I.; Higashimoto, M.; Mashiko, M.; Ito, K.; Niikura, H.; Takenoshita, S.I.; Yaegashi, N. Clinicopathological Significance of Circadian Rhythm-Related Gene Expression Levels in Patients with Epithelial Ovarian Cancer. Acta Obstet. Gynecol. Scand. 2008, 87, 1060–1070. [Google Scholar] [CrossRef]
- Relles, D.; Sendecki, J.; Chipitsyna, G.; Hyslop, T.; Yeo, C.J.; Arafat, H.A. Circadian Gene Expression and Clinicopathologic Correlates in Pancreatic Cancer. J. Gastrointest. Surg. 2013, 17, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Zhang, H.; Hyland, P.L.; Berndt, S.; Gapstur, S.M.; Wheeler, W.; The ELLIPSE Consortium; Amos, C.I.; Bezieau, S.; Bickeböller, H.; et al. Inherited Variation in Circadian Rhythm Genes and Risks of Prostate Cancer and Three Other Cancer Sites in Combined Cancer Consortia. Int. J. Cancer 2017, 141, 1794–1802. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Kraljevi, S. Circadian (De) Regulation in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci 2019, 20, 2662. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Volume 98: Painting, Firefighting and Shiftwork; International Agency for Research on Cancer: Lyon, France, 2010; Volume 98.
- Fagundo-Rivera, J.; Gómez-Salgado, J.; García-Iglesias, J.J.; Gómez-Salgado, C.; Camacho-Martín, S.; Ruiz-Frutos, C. Relationship between Night Shifts and Risk of Breast Cancer among Nurses: A Systematic Review. Medicina 2020, 56, 680. [Google Scholar] [CrossRef]
- Mocellin, S.; Tropea, S.; Benna, C.; Rossi, C.R. Circadian Pathway Genetic Variation and Cancer Risk: Evidence from Genome-Wide Association Studies. BMC Med. 2018, 16, 1–8. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the Path to Cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef]
- Gotoh, T.; Vila-Caballer, M.; Liu, J.; Schiffhauer, S.; Finkielstein, C.V. Association of the Circadian Factor Period 2 to P53 Influences P53’s Function in DNA-Damage Signaling. Mol. Biol. Cell 2015, 26, 359–372. [Google Scholar] [CrossRef]
- Li, H.X. The Role of Circadian Clock Genes in Tumors. Onco. Targets. Ther. 2019, 12, 3645–3660. [Google Scholar] [CrossRef]
- Kiessling, S.; Beaulieu-Laroche, L.; Blum, I.D.; Landgraf, D.; Welsh, D.K.; Storch, K.F.; Labrecque, N.; Cermakian, N. Enhancing Circadian Clock Function in Cancer Cells Inhibits Tumor Growth. BMC Biol. 2017, 15, 1–18. [Google Scholar] [CrossRef]
- Mazzoccoli, G.; Panza, A.; Valvano, M.R.; Palumbo, O.; Carella, M.; Pazienza, V.; Biscaglia, G.; Tavano, F.; Di Sebastiano, P.; Andriulli, A.; et al. Clock Gene Expression Levels and Relationship with Clinical and Pathological Features in Colorectal Cancer Patients. Chronobiol. Int. 2011, 28, 841–851. [Google Scholar] [CrossRef]
- Yi, C.; Mu, L.; de la Longrais, I.A.R.; Sochirca, O.; Arisio, R.; Yu, H.; Hoffman, A.E.; Zhu, Y.; Katsaro, D. The Circadian Gene NPAS2 Is a Novel Prognostic Biomarker for Breast Cancer. Breast Cancer Res. Treat. 2010, 120, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Barul, C.; Richard, H.; Parent, M.E. Night-Shift Work and Risk of Prostate Cancer: Results From a Canadian Case- Control Study, the Prostate Cancer and Environment Study. Am. J. Epidemiol. 2019, 188, 1801–1811. [Google Scholar] [CrossRef] [PubMed]
- Dun, A.; Zhao, X.; Jin, X.; Wei, T.; Gao, X.; Wang, Y.; Hou, H. Association between Night-Shift Work and Cancer Risk: Updated Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Erren, T.C.; Lewis, P. Can Yesterday’s Smoking Research Inform Today’s Shiftwork Research? Epistemological Consequences for Exposures and Doses Due to Circadian Disruption at and off Work. J. Occup. Med. Toxicol. 2017, 12, 1–8. [Google Scholar] [CrossRef]
- Soták, M.; Sumová, A.; Pácha, J. Cross-Talk between the Circadian Clock and the Cell Cycle in Cancer. Ann. Med. 2014, 46, 221–232. [Google Scholar] [CrossRef]
- Gréchez-Cassiau, A.; Rayet, B.; Guillaumond, F.; Teboul, M.; Delaunay, F. The Circadian Clock Component BMAL1 Is a Critical Regulator of P21 WAF1/CIP1 Expression and Hepatocyte Proliferation. J. Biol. Chem. 2008, 283, 4535–4542. [Google Scholar] [CrossRef]
- Bretones, G.; Delgado, M.D.; León, J. Myc and Cell Cycle Control. Biochim. Biophys. Acta-Gene Regul. Mech. 2015, 1849, 506–516. [Google Scholar] [CrossRef]
- Gery, S.; Komatsu, N.; Baldjyan, L.; Yu, A.; Koo, D.; Koeffler, H.P. The Circadian Gene Per1 Plays an Important Role in Cell Growth and DNA Damage Control in Human Cancer Cells. Mol. Cell 2006, 22, 375–382. [Google Scholar] [CrossRef]
- Sato, F.; Wu, Y.; Kumar, U.; Liu, Y.; Morohashi, S.; Kato, Y.; Kijima, H. Has Pro-Apoptotic Effects during Cisplatin (CDDP) Treatment in Human Gingival Cancer CA9-22 Cells. Eur. J. Cancer 2011, 47, 1747–1758. [Google Scholar] [CrossRef]
- Hua, H.; Wang, Y.; Wan, C.; Liu, Y.; Zhu, B.; Yang, C.; Wang, X.; Wang, Z.; Cornelissen-Guillaume, G.; Halberg, F. Circadian Gene MPer2 Overexpression Induces Cancer Cell Apoptosis. Cancer Sci. 2006, 97, 589–596. [Google Scholar] [CrossRef]
- Sato, F.; Nagata, C.; Liu, Y.; Suzuki, T.; Kondo, J.; Morohashi, S.; Imaizumi, T.; Kato, Y.; Kijima, H. PERIOD1 Is an Anti-Apoptotic Factor in Human Pancreatic and Hepatic Cancer Cells. J. Biochem. 2009, 146, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Li, H.X.; Fu, X.J.; Yang, K.; Chen, D.; Tang, H.; Zhao, Q. The Clock Gene PER1 Suppresses Expression of Tumor-Related Genes in Human Oral Squamous Cell Carcinoma. Oncotarget 2016, 7, 20574–20583. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, C.S.; Almeida, L.O.; Guimarães, D.M.; Martins, M.D.; Papagerakis, P.; Papagerakis, S.; Leopoldino, A.M.; Castilho, R.M.; Squarize, C.H. PI3K-PTEN Dysregulation Leads to MTOR-Driven Upregulation of the Core Clock Gene BMAL1 in Normal and Malignant Epithelial Cells. Oncotarget 2016, 7, 42393–42407. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, F.; Wei, M.; Zhang, S.; Wang, T. Circadian Clock Protein Period2 Suppresses the Pi3k/Akt Pathway and Promotes Cisplatin Sensitivity in Ovarian Cancer. Cancer Manag. Res. 2020, 12, 11897–11908. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gong, X.; Yang, K. Overexpression of the Clock Gene Per2 Suppresses Oral Squamous Cell Carcinoma Progression by Activating Autophagy via the PI3K/AKT/MTOR Pathway. J. Cancer 2020, 11, 3655–3666. [Google Scholar] [CrossRef]
- Wang, J.; Li, S.; Li, X.; Li, B.; Li, Y.; Xia, K.; Yang, Y.; Aman, S.; Wang, M.; Wu, H. Circadian Protein BMAL1 Promotes Breast Cancer Cell Invasion and Metastasis by Up-Regulating Matrix Metalloproteinase9 Expression. Cancer Cell Int. 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Gwon, D.H.; Lee, W.Y.; Shin, N.; Kim, S.I.; Jeong, K.; Lee, W.H.; Kim, D.W.; Hong, J.; Lee, S.Y. BMAL1 Suppresses Proliferation, Migration, and Invasion of U87MG Cells by Downregulating Cyclin B1, Phospho-AKT, and Metalloproteinase-9. Int. J. Mol. Sci. 2020, 21, 2352. [Google Scholar] [CrossRef]
- Diamantopoulou, Z.; Castro-Giner, F.; Schwab, F.D.; Foerster, C.; Saini, M.; Budinjas, S.; Strittmatter, K.; Krol, I.; Seifert, B.; Heinzelmann-Schwarz, V.; et al. The Metastatic Spread of Breast Cancer Accelerates during Sleep. Nature 2022, 607, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Broadberry, E.; Mcconnell, J.; Williams, J.; Yang, N.; Zindy, E.; Leek, A.; Waddington, R.; Joseph, L.; Howe, M.; Meng, Q.; et al. Disrupted Circadian Clocks and Altered Tissue Mechanics in Primary Human Breast Tumours. Breast Cancer Res. 2018, 20, 125. [Google Scholar] [CrossRef]
- Xia, H.; Niu, Z.; Ma, H.; Cao, S.; Hao, S.; Liu, Z.; Wang, F. Deregulated Expression of the Per1 and Per2 in Human Gliomas. Can J. Neurol. Sci. 2010, 37, 365–370. [Google Scholar] [CrossRef]
- Zhao, H.; Zeng, Z.; Yang, J.; Jin, Y.; Qiu, M.; Hu, X.; Han, J.; Liu, K. Prognostic Relevance of Period1 (Per1) and Period2 (Per2) Expression in Human Gastric Cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 619–630. [Google Scholar]
- Xiang, R.U.N.; Cui, Y.U.E.; Wang, Y.; Xie, T.; Yang, X.; Wang, Z.H.U.; Li, J.; Li, Q. Circadian Clock Gene Per2 Downregulation in Non-Small Cell Lung Cancer Is Associated with Tumour Progression and Metastasis. Oncol. Rep. 2018, 40, 3040–3048. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Wang, Y.; Liu, Y.; Zhu, C.; Lin, J.; Qian, R.; Hua, L.; Lu, C. BMAL1 Induces Colorectal Cancer Metastasis by Stimulating Exosome Secretion. Mol. Biol. Rep. 2022, 49, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Scheiermann, C.; Gibbs, J.; Ince, L.; Loudon, A. Clocking in to Immunity. Nat. Rev. Immunol. 2018, 18, 423–437. [Google Scholar] [CrossRef]
- Wang, C.; Barnoud, C.; Cenerenti, M.; Sun, M.; Caffa, I.; Kizil, B.; Bill, R.; Liu, Y.; Pick, R.; Garnier, L.; et al. Dendritic Cells Direct Circadian Anti-Tumor Immune Response. Nature 2022. [Google Scholar] [CrossRef]
- Taniguchi, H.; Fernández, A.F.; Setién, F.; Ropero, S.; Ballestar, E.; Villanueva, A.; Yamamoto, H.; Imai, K.; Shinomura, Y.; Esteller, M. Epigenetic Inactivation of the Circadian Clock Gene BMAL1 in Hematologic Malignancies. Cancer Res. 2009, 69, 8447–8454. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yang, Z.; Niu, Z.; Peng, J.; Li, Q.; Xiong, W.; Langnas, A.N.; Ma, M.Y.; Zhao, Y. MOP3, a Component of the Molecular Clock, Regulates the Development of B Cells. Immunology 2006, 119, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Cermakian, N.; Lange, T.; Golombek, D.; Sarkar, D.; Nakao, A.; Shibata, S.; Mazzoccoli, G.; Cronobiologia, D.; Aires, B.; Brunswick, N.; et al. Crosstalk between the circadian clock circuitry and the immune system. Chronobiol. Int. J. Biol. Med. Rhythm. Res. 2020, 30, 870–888. [Google Scholar] [CrossRef]
- Cardinali, D.P.; Brown, G.M.; Pandi-Perumal, S.R. Chronotherapy. Handb. Clin. Neurol. 2021, 179, 357–370. [Google Scholar] [CrossRef]
- Redondo, J.A.; Bibes, R.; Drubbel, A.V.; Dassy, B.; Bisteau, X.; Maury, E.; Beck, B. Per2 Circadian Oscillation Sensitizes Esophageal Cancer Cells to Chemotherapy. Biology 2021, 10, 266. [Google Scholar] [CrossRef]
- Dakup, P.P.; Porter, K.I.; Little, A.A.; Gajula, R.P.; Zhang, H.; Skornyakov, E.; Kemp, M.G.; Van Dongen, H.P.A.; Gaddameedhi, S. The Circadian Clock Regulates Cisplatin-Induced Toxicity and Tumor Regression in Melanoma Mouse and Human Models. Oncotarget 2018, 9, 14524–14538. [Google Scholar] [CrossRef]
- Thakur, S.; Singh, B.; Mishra, V.; Yadav, N.; Giri, N.; Sharma, P.; Saini, A.; Garg, L.K. Bilayer Tablet Based Chronotherapeutics in the Management of Nocturnal Asthma: An Overview. Recent Pat. Drug Deliv. Formul. 2019, 13, 74–82. [Google Scholar] [CrossRef]
- Whibley, D.; Braley, T.J.; Kratz, A.L.; Murphy, S.L. Transient Effects of Sleep on Next-Day Pain and Fatigue in Older Adults with Symptomatic Osteoarthritis. J. Pain 2019, 20, 1373–1382. [Google Scholar] [CrossRef]
- Levi, F.; Benavides, M.; Chevelle, C.; Le Saunier, F.; Bailleul, F.; Missetc Regensberg, J.L.; Regensberg, C.; Vannetzel, J.M.; Reinberg, A.; Mathe, G. Chemotherapy of Advanced Ovarian Cancer with 4’-0-Tetrahydropyranyl Doxorubicin and Cisplatin: A Randomized Phase II Trial with an Evaluation of Circadian Timing and Dose-Intensity. J. Clin. Oncol. 1990, 8, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Giacchetti, S.; Dugué, P.A.; Innominato, P.F.; Bjarnason, G.A.; Focan, C.; Garufi, C.; Tumolo, S.; Coudert, B.; Iacobelli, S.; Smaaland, R.; et al. Sex Moderates Circadian Chemotherapy Effects on Survival of Patients with Metastatic Colorectal Cancer: A Meta-Analysis. Ann. Oncol. 2012, 23, 3110–3116. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A.; Van Gelder, R.N. Clocks, Cancer, and Chronochemotherapy. Science 2021, 371, 42. [Google Scholar] [CrossRef] [PubMed]
- Venegas, C.; García, J.A.; Escames, G.; Ortiz, F.; López, A.; Doerrier, C.; Garcıía-Corzo, L.; López, L.C.; Reiter, R.J.; Acuna-Castroviejo, D. Extrapineal Melatonin: Analysis of Its Subcellular Distribution and Daily Fluctuations. J. Pineal Res. 2012, 52, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Acuña-Castroviejo, D.; Rahim, I.; Acuña-Fernández, C.; Fernández-Ortiz, M.; Solera-Marín, J.; Sayed, R.K.A.; Díaz-Casado, M.E.; Rusanova, I.; López, L.C.; Escames, G. Melatonin, Clock Genes and Mitochondria in Sepsis. Cell. Mol. Life Sci. 2017, 74, 3965–3987. [Google Scholar] [CrossRef]
- Klein, D.C. Arylalkylamine N-Acetyltransferase: “The Timezyme”. J. Biol. Chem. 2007, 282, 4233–4237. [Google Scholar] [CrossRef]
- Tan, D.X.; Xu, B.; Zhou, X.; Reiter, R.J. Associated Health Consequences and Rejuvenation of the Pineal Gland. Molecules 2018, 23, 301. [Google Scholar] [CrossRef]
- Kandalepas, P.C.; Mitchell, J.W.; Gillette, M.U.G. Melatonin Signal Transduction Pathways Require E-Box-Mediated Transcription of Per1 and Per2 to Reset the SCN Clock at Dusk. PLoS ONE 2016, 11, e0157824. [Google Scholar] [CrossRef]
- Escames, G.; León, J.; López, L.C.; Acuña-Castroviejo, D. Mechanisms of N-Methyl-D-Aspartate Receptor Inhibition by Melatonin in the Rat Striatum. J. Neuroendocrinol. 2004, 16, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Targhazeh, N.; Reiter, R.J.; Rahimi, M.; Qujeq, D.; Yousefi, T.; Shahavi, M.H.; Mir, S.M. Oncostatic Activities of Melatonin: Roles in Cell Cycle, Apoptosis, and Autophagy. Biochimie 2022, 202, 34–48. [Google Scholar] [CrossRef]
- Liu, C.; Weaver, D.R.; Jin, X.; Shearman, L.P.; Pieschl, R.L.; Gribkoff, V.K.; Reppert, S.M. Molecular Dissection of Two Distinct Actions of Melatonin on the Suprachiasmatic Circadian Clock. Neuron 1997, 19, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Vriend, J.; Reiter, R.J. Melatonin Feedback on Clock Genes: A Theory Involving the Proteasome. J. Pineal Res. 2015, 58, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Xian, D.; Seok, T.; Kim, J.; Cruz, M.H.C. Delivery of Pineal Melatonin to the Brain and SCN: Role of Canaliculi, Cerebrospinal Fluid, Tanycytes and Virchow—Robin Perivascular Spaces. Brain Struct. Funct. 2014, 219, 1873–1887. [Google Scholar] [CrossRef]
- Hardeland, R.; Madrid, J.A.; Tan, D.-X.; Reiter, R.J. Melatonin, the Circadian Multioscillator System and Health: The Need for Detailed Analyses of Peripheral Melatonin Signaling. J. Pineal Res. 2012, 52, 139–166. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Weaver, D.R. Coordination of Circadian Clocks in Mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef]
- Schernhammer, E.; Chen, Æ.H.; Ritz, Æ.B. Circulating Melatonin Levels: Possible Link between Parkinson’ s Disease and Cancer Risk? Cancer Causes Control 2006, 17, 577–582. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Zhou, Y.; Meng, X.; Zhang, J.J.; Xu, D.P.; Li, H. Bin Melatonin for the Prevention and Treatment of Cancer. Oncotarget 2017, 8, 39896–39921. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Patel, K.K.; Dehari, D.; Agrawal, A.K.; Singh, S. Melatonin and Its Ubiquitous Anticancer Effects. Mol. Cell. Biochem. 2019, 462, 133–155. [Google Scholar] [CrossRef]
- Xiang, S.; Mao, L.; Duplessis, T.; Yuan, L.; Dauchy, R.; Dauchy, E.; Blask, D.; Frasch, T.; Hhill, S.M. Oscillation of Clock and Clock Controlled Genes Induced by Serum Shock in Human Breast Epithelial and Breast Cancer Cells: Regulation by Melatonin. Breast Cancer Basic Clin. Res. 2012, 6, 137–150. [Google Scholar] [CrossRef]
- Jung-Hynes, B.; Huang, W.; Reiter, R.J.; Ahmad, N. Melatonin Resynchronizes Dysregulated Circadian Rhythm Circuitry in Human Prostate Cancer Cells. J. Pineal Res. 2010, 49, 60–68. [Google Scholar] [CrossRef]
- Hill, S.M.; Frasch, T.; Xiang, S.; Yuan, L.; Duplessis, T.; Mao, L. Molecular Mechanisms of Melatonin Anticancer Effects. Integr. Cancer Ther. 2009, 8, 337–346. [Google Scholar] [CrossRef]
- Sánchez, D.I.; González-Fernández, B.; Crespo, I.; San-Miguel, B.; Álvarez, M.; González-Gallego, J.; Tuñón, M.J. Melatonin Modulates Dysregulated Circadian Clocks in Mice with Diethylnitrosamine-Induced Hepatocellular Carcinoma. J. Pineal Res. 2018, 65, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.J.; Seo, E.B.; Kwon, S.H.; Lee, S.H.; Kim, S.K.; Park, S.K.; Kim, K.; Park, S.G.; Park, I.C.; Park, J.W.; et al. Extracellular Acidosis Promotes Metastatic Potency via Decrease of the BMAL1 Circadian Clock Gene in Breast Cancer. Cells 2020, 9, 989. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Woo, S.M.; Min, K.J.; Kwon, T.K. Transcriptional and Post-Translational Regulation of Bim Controls Apoptosis in Melatonin-Treated Human Renal Cancer Caki Cells. J. Pineal Res. 2014, 56, 97–106. [Google Scholar] [CrossRef]
- Vriend, J.; Reiter, R.J. Melatonin as a Proteasome Inhibitor. Is There Any Clinical Evidence? Life Sci. 2014, 115, 8–14. [Google Scholar] [CrossRef]
- Smith, A.; Simanski, S.; Fallahi, M.; Ayad, N.G.; Smith, A.; Simanski, S.; Fallahi, M.; Ayad, N.G. Wee1 Degradation and Mitotic Entry Redundant Ubiquitin Ligase Activities Regulate Wee1 Degradation and Mitotic Entry. Cell Cycle 2007, 6, 2795–2799. [Google Scholar] [CrossRef]
- Lee, H.; Lee, H.J.; Jung, J.H.; Shin, E.A.; Kim, S.H. Melatonin Disturbs SUMOylation-Mediated Crosstalk between c-Myc and Nestin via MT1 Activation and Promotes the Sensitivity of Paclitaxel in Brain Cancer Stem Cells. J. Pineal Res. 2018, 65, 1–20. [Google Scholar] [CrossRef]
- Cini, G.; Neri, B.; Pacini, A.; Cesati, V.; Sassoli, C.; Quattrone, S.; D’Apolito, M.; Fazio, A.; Scapagnini, G.; Provenzani, A.; et al. Antiproliferative Activity of Melatonin by Transcriptional Inhibition of Cyclin D1 Expression: A Molecular Basis for Melatonin-Induced Oncostatic Effects. J. Pineal Res. 2005, 39, 12–20. [Google Scholar] [CrossRef]
- Rögelsperger, O.; Wlcek, K.; Ekmekcioglu, C.; Humpeler, S.; Svoboda, M.; Königsberg, R.; Klimpfinger, M.; Jäger, W.; Thalhammer, T. Melatonin Receptors, Melatonin Metabolizing Enzymes and Cyclin D1 in Human Breast Cancer. J. Recept. Signal Transduct. 2011, 31, 180–187. [Google Scholar] [CrossRef]
- Ferreira, L.C.; Orso, F.; Dettori, D.; Lacerda, J.Z.; Borin, T.F.; Taverna, D.; Zuccari, D.A.P.C. The Role of Melatonin on MiRNAs Modulation in Triple-Negative Breast Cancer Cells. PLoS ONE 2020, 15, e0228062. [Google Scholar] [CrossRef]
- Liu, Z.; Selby, C.P.; Yang, Y.; Lindsey-Boltz, L.A.; Cao, X.; Eynullazada, K.; Sancar, A. Circadian Regulation of C-MYC in Mice. Proc. Natl. Acad. Sci. USA 2020, 117, 21609–21617. [Google Scholar] [CrossRef]
- Wolf, E.; Lin, C.Y.; Eilers, M.; Levens, D.L. Taming of the Beast: Shaping Myc-Dependent Amplification. Trends Cell Biol. 2015, 25, 241–248. [Google Scholar] [CrossRef]
- Fu, L.; Pelicano, H.; Liu, J.; Huang, P.; Lee, C.C. The Circadian Gene Period2 Plays an Important Role in Tumor Suppression and DNA Damage Response In Vivo Lates Bmal1 Transcription through PAS-Mediated Reac-Tions with Other Transcription Factors. In the Best-Known Negative Feedback Loop, Cytoplasmic CRY1. Cell 2002, 111, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Altman, B.J.; Hsieh, A.L.; Sengupta, A.; Krishnanaiah, S.Y.; Stine, Z.E.; Walton, Z.E.; Gouw, A.M.; Venkataraman, A.; Li, B.; Goraksha-Hicks; et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab. 2015, 22, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, Y.; Sahar, S.; Astarita, G.; Kaluzova, M.; Sassone-Corsi, P. Circadian Control of the Nad+ Salvage Pathway by Clock-Sirt1. Science 2009, 324, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, K.M.; Yoshino, J.; Brace, C.S.; Abrassart, D.; Kobayashi, Y.; Marcheva, B.; Hong, H.-K.; Chong, J.L.; Buhr, E.D.; Lee, C.; et al. Circadian Clock Feedback Cycle through NAMPT-Mediated NAD + Biosynthesis. Science 2009, 324, 651–654. [Google Scholar] [CrossRef]
- Jung-Hynes, B.; Ahmad, N. SIRT1 Controls Circadian Clock Circuitry and Promotes Cell Survival: A Connection with Age-Related Neoplasms. FASEB J. 2009, 23, 2803–2809. [Google Scholar] [CrossRef]
- Imai, S.-i.; Guarente, L. NAD+ and Sirtuins in Aging and Disease. Trends Cell Biol. 2014, 24, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Zhang, L.; Gao, W.; Huang, C.; Huber, P.E.; Zhou, X.; Li, C.; Shen, G.; Zou, B. NAD+ Metabolism: Pathophysiologic Mechanisms and Therapeutic Potential. Signal Transduct. Target. Ther. 2020, 5, 227. [Google Scholar] [CrossRef]
- Cheng, Y.; Cai, L.; Jiang, P.; Wang, J.; Gao, C.; Feng, H.; Wang, C.; Pan, H.; Yang, Y. SIRT1 Inhibition by Melatonin Exerts Antitumor Activity in Human Osteosarcoma Cells. Eur. J. Pharmacol. 2013, 715, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Proietti, S.; Cucina, A.; Dobrowolny, G.; D’Anselmi, F.; Dinicola, S.; Masiello, M.G.; Pasqualato, A.; Palombo, A.; Morini, V.; Reiter, R.J.; et al. Melatonin Down-Regulates MDM2 Gene Expression and Enhances P53 Acetylation in MCF-7 Cells. J. Pineal Res. 2014, 57, 120–129. [Google Scholar] [CrossRef]
- Dauchy, R.T.; Xiang, S.; Mao, L.; Brimer, S.; Wren, M.A.; Yuan, L.; Anbalagan, M.; Hauch, A.; Frasch, T.; Rowan, B.G.; et al. Circadian and Melatonin Disruption by Exposure to Light at Night Drives Intrinsic Resistance to Tamoxifen Therapy in Breast Cancer. Cancer Res. 2014, 74, 4099–4110. [Google Scholar] [CrossRef]
- Shen, Y.-Q.; Guerra-Librero, A.; Fernandez-Gil, B.I.; Florido, J.; García-López, S.; Martinez-Ruiz, L.; Mendivil-Perez, M.; Soto-Mercado, V.; Acuña-Castroviejo, D.; Ortega-Arellano, H.; et al. Combination of Melatonin and Rapamycin for Head and Neck Cancer Therapy: Suppression of AKT/MTOR Pathway Activation, and Activation of Mitophagy and Apoptosis via Mitochondrial Function Regulation. J. Pineal Res. 2018, 64, e12461. [Google Scholar] [CrossRef]
- Chen, K.; Zhu, P.; Chen, W.; Luo, K.; Shi, X.-J.; Zhai, W. Melatonin Inhibits Proliferation, Migration, and Invasion by Inducing ROS-Mediated Apoptosis via Suppression of the PI3K/Akt/MTOR Signaling Pathway in Gallbladder Cancer Cells. Aging 2021, 13, 22502–22515. [Google Scholar] [CrossRef]
- Phiboonchaiyanan, P.P.; Puthongking, P.; Chawjarean, V.; Harikarnpakdee, S.; Sukprasansap, M.; Chanvorachote, P.; Priprem, A.; Govitrapong, P. Melatonin and Its Derivative Disrupt Cancer Stem-like Phenotypes of Lung Cancer Cells via AKT Downregulation. Clin. Exp. Pharmacol. Physiol. 2021, 48, 1712–1723. [Google Scholar] [CrossRef]
- Jung, C.H.; Kim, E.M.; Park, J.K.; Hwang, S.G.; Moon, S.K.; Kim, W.J.; Um, H.D. Bmal1 Suppresses Cancer Cell Invasion by Blocking the Phosphoinositide 3-Kinase-Akt-MMP-2 Signaling Pathway. Oncol. Rep. 2013, 29, 2109–2113. [Google Scholar] [CrossRef]
| Tumor Type | Study Design | Dose | Results | Author |
|---|---|---|---|---|
| Breast cancer | In vitro | 10 nM | -↓ SIRT1 protein levels | [85] |
| Prostate cancer | In vitro | 100 μM, 1 mM or 2 mM | -↑ Per2 and Clock mRNA and protein levels -↓ Bmal1 mRNA levels -Rhythmic oscillation of Per2 and Dbp | [84] |
| Breast cancer | In vitro | 1 nM | -↓ Bmal1 and RORα mRNA levels -↑ Rev-erbα and Per2 mRNA levels at 20 h after treatment | [83] |
| Hepatocellular carcinoma | In vivo Mice | 5, 10 mg/kg | -↓ Bmal1, Clock and RORα mRNA and protein levels -↑ Per1, Per2, Per3, Cry1, CK1ε mRNA levels -↓ Sirt1 mRNA levels -↑ Rev-Erbα and Rev-Erbβ mRNA and protein levels | [86] |
| Breast cancer | In vitro | 1, 2 mM | Inhibition of LDH-A prevents ↓ Bmal1 under hypoxia conditions | [87] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Santana, C.; Florido, J.; Martínez-Ruiz, L.; López-Rodríguez, A.; Acuña-Castroviejo, D.; Escames, G. Role of Melatonin in Cancer: Effect on Clock Genes. Int. J. Mol. Sci. 2023, 24, 1919. https://doi.org/10.3390/ijms24031919
Rodríguez-Santana C, Florido J, Martínez-Ruiz L, López-Rodríguez A, Acuña-Castroviejo D, Escames G. Role of Melatonin in Cancer: Effect on Clock Genes. International Journal of Molecular Sciences. 2023; 24(3):1919. https://doi.org/10.3390/ijms24031919
Chicago/Turabian StyleRodríguez-Santana, César, Javier Florido, Laura Martínez-Ruiz, Alba López-Rodríguez, Darío Acuña-Castroviejo, and Germaine Escames. 2023. "Role of Melatonin in Cancer: Effect on Clock Genes" International Journal of Molecular Sciences 24, no. 3: 1919. https://doi.org/10.3390/ijms24031919
APA StyleRodríguez-Santana, C., Florido, J., Martínez-Ruiz, L., López-Rodríguez, A., Acuña-Castroviejo, D., & Escames, G. (2023). Role of Melatonin in Cancer: Effect on Clock Genes. International Journal of Molecular Sciences, 24(3), 1919. https://doi.org/10.3390/ijms24031919







