Protective Effects of Lactobacillus gasseri against High-Cholesterol Diet-Induced Fatty Liver and Regulation of Host Gene Expression Profiles
Abstract
:1. Introduction
2. Results
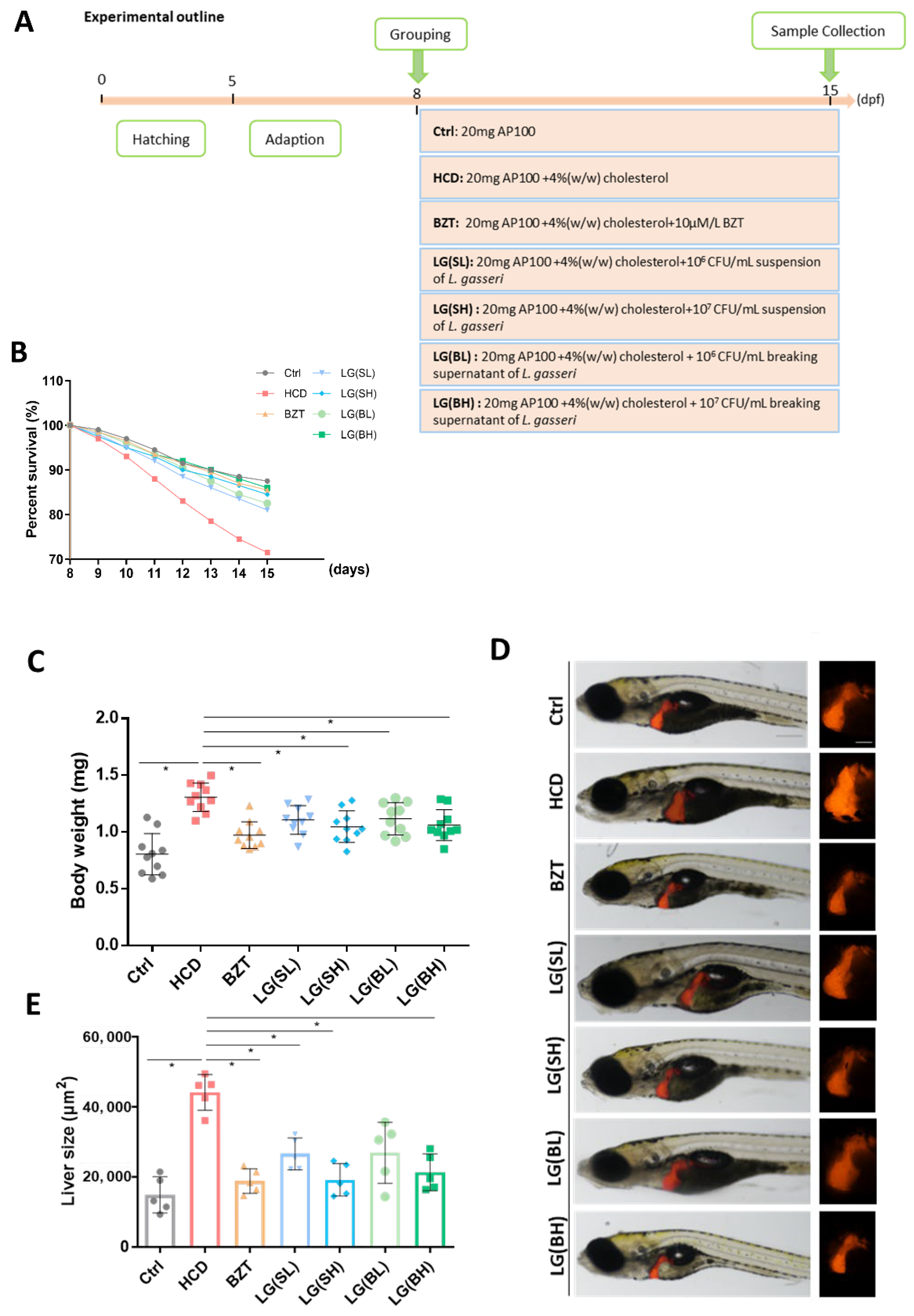
2.1. Treatment with L. gasseri Ameliorated HCD-Induced Mortality and Morphological Changes of Zebrafish Larvae
2.2. L. gasseri Administration Attenuated HCD-Induced Fatty Liver on Lipid Accumulation and Histological Changes in Zebrafish Larvae
2.3. L. gasseri Supplementation Alleviated HCD-Induced Fatty Liver on Oxidative Stress and Macrophage Aggregation in the Liver of Zebrafish Larvae
2.4. Transcriptome Profiling of HCD Revealed Hepatic Steatosis Injuries in Zebrafish Liver
2.5. Potential Functional Mechanisms of L. gasseri on Gene Expression Profiles of HCD-Induced Hepatic Steatosis Injury
3. Discussion
4. Materials and Methods
4.1. Bacteria Cultivation
4.2. Zebrafish Husbandry
4.3. Preparation of High Cholesterol Diet and Bacteria
4.4. Grouping of Experimental Animals
4.5. Biochemical Analysis
4.6. Histopathological Analysis
4.7. Nile Red Staining
4.8. Quantification of Reactive Oxygen Species (ROS)
4.9. Real-Time Quantitative PCR (qRT-PCR) Analysis
4.10. RNA Transcriptomic Analysis
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goh, B.B.; Mccullough, A.J. Natural History of Nonalcoholic Fatty Liver Disease. Dig. Dis. Sci. 2016, 61, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Longo, D.L.; Diehl, A.M.; Day, C. Cause, Pathogenesis, and Treatment of Nonalcoholic Steatohepatitis. N. Engl. J. Med. 2017, 377, 2063–2072. [Google Scholar]
- Ying, L.; Yan, F.; Zhao, Y.; Gao, H.; Williams, B.R.; Hu, Y.; Li, X.; Tian, R.; Xu, P.; Wang, Y. (-)-Epigallocatechin-3-gallate and atorvastatin treatment down-regulates liver fibrosis-related genes in non-alcoholic fatty liver disease. Clin. Exp. Pharmacol. Physiol. 2017, 44, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.J.; Aguilar, M.; Cheung, R.; Perumpail, R.B.; Harrison, S.A.; Younossi, Z.M.; Ahmed, A. Onalcoholic Steatohepatitis Is the Second Leading Etiology of Liver Disease Among Adults Awaiting Liver Transplantation in the United States. Gastroenterology 2015, 148, 547–555. [Google Scholar] [CrossRef]
- Haddad, T.M.; Hamdeh, S.; Kanmanthareddy, A.; Alla, V.M. Nonalcoholic fatty liver disease and the risk of clinical cardiovascular events: A systematic review and meta-analysis. Diabetes Metab. Syndr. 2017, 11, S209–S216. [Google Scholar] [CrossRef]
- Miyoshi, M.; Ogawa, A.; Higurashi, S.; Kadooka, Y. Anti-obesity effect of Lactobacillus gasseri SBT2055 accompanied by inhibition of pro-inflammatory gene expression in the visceral adipose tissue in diet-induced obese mice. Eur. J. Nutr. 2014, 53, 599–606. [Google Scholar] [CrossRef]
- Malaguarnera, M.; Rosa, M.D.; Nicoletti, F.; Malaguarnera, L. Molecular mechanisms involved in NAFLD progression. J. Mol. Med. 2009, 87, 679. [Google Scholar] [CrossRef]
- Qin, G.; Ma, J.; Huang, Q.; Yin, H.; Han, J.; Li, M.; Deng, Y.; Wang, B.; Hassan, W.; Shang, J. Isoquercetin Improves Hepatic Lipid Accumulation by Activating AMPK Pathway and Suppressing TGF-β Signaling on an HFD-Induced Nonalcoholic Fatty Liver Disease Rat Model. Int. J. Mol. Sci. 2018, 19, 4126. [Google Scholar] [CrossRef] [Green Version]
- Park, S.S.; Lee, Y.J.; Song, S.; Kim, B.; Kang, H.; Oh, S.; Eungseok, K. Lactobacillus acidophilus ns1 attenuates diet-induced obesity and fatty liver. J. Endocrinol. 2018, 237, 87–100. [Google Scholar] [CrossRef]
- Yu, Y.; Dunaway, S.; Champer, J.; Kim, J.; Alikhan, A. Changing our microbiome: Probiotics in dermatology. Br. J. Dermatol. 2020, 182, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Tilg, H.; Cani, P.D.; Mayer, E.A. Gut microbiome and liver diseases. Gut 2016, 65, 2035. [Google Scholar] [CrossRef] [PubMed]
- Harakeh, S.M.; Imran, K.; Taha, K.; Elie, B.; Almasaudi, S.B.; Bahijri, S.M.; Alfadul, S.M.; Ajabnoor, G.M.A.; Azhar, E.I. Gut Microbiota: A Contributing Factor to Obesity. Front. Cell. Infect. Microbiol. 2016, 6, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, C.; Rivera, L.; Furness, J.B.; Angus, P.W. The role of the gut microbiota in NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Lin, A.; Kong, M.; Yao, X.; Liu, H. Intestinal microbiome and NAFLD: Molecular insights and therapeutic perspectives. J. Gastroenterol. 2020, 55, 142–158. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, C.F.; Shahani, K.M.; Amer, M.A. Therapeutic role of dietary lactobacilli and lactobacillic fermented dairy products. Fems Microbiol. Lett. 1987, 46, 343–356. [Google Scholar] [CrossRef]
- Kato, I.; Kobayashi, S.; Yokokura, T.; Mutai, M. Antitumor activity of Lactobacillus casei in mice. Gan. Gann Jpn. J. Cancer Res. 1981, 72, 517–523. [Google Scholar]
- Perdigon, G.; De Macias, M.E.; Alvarez, S.; Oliver, G.; de Ruiz Holgado, A.A. Effect of perorally administered lactobacilli on macrophage activation in mice. Infect. Immun. 1986, 53, 404–410. [Google Scholar] [CrossRef] [Green Version]
- Perdigon, G.; Alvarez, S.; Nader, D.; Roux, M.E.; Pesce, D. The Oral Administration of Lactic Acid Bacteria Increase the Mucosal Intestinal Immunity in Response to Enteropathogens. J. Food Prot. 1990, 53, 404–410. [Google Scholar] [CrossRef]
- Sato, K. Enhancement of host resistance against Listeria infection by Lactobacillus casei: Role of macrophages. Infect. Immun. 1984, 44, 445–451. [Google Scholar] [CrossRef] [Green Version]
- Wong, W.S.; Wong, L.H.; Chim, M.L.; Chu, C.W.; Yeung, K.W.; Li, C.T.; Chan, L.Y. Treatment of nonalcoholic steatohepatitis with probiotics. A proof-of-concept study. Ann. Hepatol. 2013, 12, 256–262. [Google Scholar] [CrossRef]
- Miyazawa, K.; Kawase, M.; Kubota, A.; Yoda, K.; He, F. Heat-killed Lactobacillus gasseri can enhance immunity in the elderly in a doubleblind, placebo-controlled clinical study. Benef. Microbes 2012, 6, 1–9. [Google Scholar]
- Motevaseli, E.; Shirzad, M.; Akrami, S.M.; Mousavi, A.S.; Mirsalehian, A.; Modarressi, M.H. Normal and tumour cervical cells respond differently to vaginal lactobacilli, independent of pH and lactate. J. Med. Microbiol. 2013, 62, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, G.; Tanaka, M.; Harada, Y.; Nagashima, H.; Masuda, T. Homologous Expression and Characterization of Gassericin T and Gassericin S, a Novel Class IIb Bacteriocin Produced by Lactobacillus gasseri LA327. Appl. Environ. Microbiol. 2019, 85, e02815-18. [Google Scholar] [CrossRef] [PubMed]
- Reuter, G. The Lactobacillus and Bifidobacterium microflora of the human intestine: Composition and succession. Curr. Issues Intest. Microbiol. 2001, 2, 43–53. [Google Scholar]
- Marín, M.; Arroyo, R.; Jiménez, E.; Gómez, A.; Fernández, L.; Rodríguez, J. Cold storage of human milk: Effect on its bacterial composition. J. Pediatr. Gastroenterol. Nutr. 2009, 49, 343–348. [Google Scholar] [CrossRef]
- Sato, M.; Uzu, K.; Yoshida, T.; Hamad, E.M.; Kawakami, H.; Matsuyama, H.; Abd El-Gawad, I.A.; Imaizumi, K. Effects of milk fermented by Lactobacillus gasseri SBT2055 on adipocyte size in rats. Br. J. Nutr. 2008, 99, 1013–1017. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, A.; Kadooka, Y.; Kato, K.; Shirouchi, B.; Sato, M. Lactobacillus gasseri SBT2055 reduces postprandial and fasting serum non-esterified fatty acid levels in Japanese hypertriacylglycerolemic subjects. Lipids Heal. Dis. 2014, 13, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.Y.; Shin, M.J.; Youn, G.S.; Yoon, S.J.; Choi, Y.R.; Kim, H.S.; Gupta, H.; Han, S.H.; Kim, B.K.; Lee, D.Y.; et al. Lactobacillus attenuates progression of nonalcoholic fatty liver disease by lowering cholesterol and steatosis. Clin. Mol. Hepatol. 2021, 27, 110–124. [Google Scholar] [CrossRef]
- Arguello, G.; Balboa, E.; Arrese, M.; Zanlungo, S. Recent insights on the role of cholesterol in non-alcoholic fatty liver disease. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2015, 1852, 1765–1778. [Google Scholar] [CrossRef] [Green Version]
- Tomita, K.; Teratani, T.; Suzuki, T.; Shimizu, M.; Sato, H.; Narimatsu, K.; Okada, Y.; Kurihara, C.; Irie, R.; Yokoyama, H. Free cholesterol accumulation in hepatic stellate cells: Mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology 2014, 59, 154–169. [Google Scholar] [CrossRef]
- Liong, M.-T.; Dunshea, F.R.; Shah, N.P. Effects of a synbiotic containing Lactobacillus acidophilus ATCC 4962 on plasma lipid profiles and morphology of erythrocytes in hypercholesterolaemic pigs on high-and low-fat diets. Br. J. Nutr. 2007, 98, 736–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simons, L.A.; Amansec, S.G.; Conway, P. Effect of Lactobacillus fermentum on serum lipids in subjects with elevated serum cholesterol. Nutr. Metab. Cardiovasc. Dis. 2006, 16, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Mahboubi, M. Lactobacillus gasseri as a Functional Food and Its Role in Obesity. Int. J. Med. Rev. 2019, 6, 59–64. [Google Scholar] [CrossRef]
- Park, Y.-H.; Kim, J.-G.; Shin, Y.-W.; Kim, S.-H.; Whang, K.-Y. Effect of dietary inclusion of Lactobacillus acidophilus ATCC 43121 on cholesterol metabolism in rats. J. Microbiol. Biotechnol. 2007, 17, 655–662. [Google Scholar]
- Palaniyandi, S.A.; Damodharan, K.; Suh, J.-W.; Yang, S.H. Probiotic characterization of cholesterol-lowering Lactobacillus fermentum MJM60397. Probiotics Antimicrob. Proteins 2020, 12, 1161–1172. [Google Scholar] [CrossRef]
- Fuentes, M.C.; Lajo, T.; Carrión, J.M.; Cuné, J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Br. J. Nutr. 2013, 109, 1866–1872. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, A.; Kobayashi, T.; Sakai, F.; Kadooka, Y.; Kawasaki, Y. Lactobacillus gasseri SBT2055 suppresses fatty acid release through enlargement of fat emulsion size in vitro and promotes fecal fat excretion in healthy Japanese subjects. Lipids Health Dis. 2015, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- MacArthur, J.M.; Bishop, J.R.; Stanford, K.I.; Wang, L.; Bensadoun, A.; Witztum, J.L.; Esko, J.D. Liver heparan sulfate proteoglycans mediate clearance of triglyceride-rich lipoproteins independently of LDL receptor family members. J. Clin. Investig. 2007, 117, 153–164. [Google Scholar] [CrossRef]
- Sunny, N.E.; Parks, E.J.; Browning, J.D.; Burgess, S.C. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metab. 2011, 14, 804–810. [Google Scholar] [CrossRef] [Green Version]
- Saponaro, C.; Sabatini, S.; Gaggini, M.; Carli, F.; Rosso, C.; Positano, V.; Armandi, A.; Caviglia, G.P.; Faletti, R.; Bugianesi, E. Adipose tissue dysfunction and visceral fat are associated to hepatic insulin resistance and severity of NASH even in lean individuals. Liver Int. 2022, 42, 2418–2427. [Google Scholar] [CrossRef]
- Paul, B.; Henne, W.M. How cells flex their PEX to fine-tune lipolysis in NAFLD. Nat. Metab. 2021, 3, 1591–1593. [Google Scholar] [CrossRef] [PubMed]
- Aregger, M.; Lawson, K.A.; Billmann, M.; Costanzo, M.; Tong, A.H.; Chan, K.; Rahman, M.; Brown, K.R.; Ross, C.; Usaj, M. Systematic mapping of genetic interactions for de novo fatty acid synthesis identifies C12orf49 as a regulator of lipid metabolism. Nat. Metab. 2020, 2, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Campagnoli, L.I.M.; Marchesi, N.; Vairetti, M.; Pascale, A.; Ferrigno, A.; Barbieri, A. Age-Related NAFLD: The Use of Probiotics as a Supportive Therapeutic Intervention. Cells 2022, 11, 2827. [Google Scholar] [CrossRef]
- Azad, M.A.K.; Sarker, M.; Wan, D. Immunomodulatory Effects of Probiotics on Cytokine Profiles. BioMed. Res. Int. 2018, 2018, 8063647. [Google Scholar] [CrossRef] [Green Version]
- María Remes-Troche, J.; Coss-Adame, E.; Ángel Valdovinos-Díaz, M.; Gómez-Escudero, O.; Eugenia Icaza-Chávez, M.; Antonio Chávez-Barrera, J.; Zárate-Mondragón, F.; Antonio Velarde-Ruíz Velasco, J.; Rafael Aceves-Tavares, G.; Antonio Lira-Pedrín, M.; et al. Lactobacillus acidophilus LB: A useful pharmabiotic for the treatment of digestive disorders. Ther. Adv. Gastroenterol. 2020, 13, 1756284820971201. [Google Scholar] [CrossRef]
- Sánchez, B.; Delgado, S.; Blanco-Míguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2017, 61, 1600240. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Li, H.; Li, J.; Chen, Y.; Yang, Y. Lactobacillus rhamnosus GG treatment improves intestinal permeability and modulates microbiota dysbiosis in an experimental model of sepsis. Int. J. Mol. Med. 2019, 43, 1139–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsyth, C.B.; Farhadi, A.; Jakate, S.M.; Tang, Y.; Shaikh, M.; Keshavarzian, A. Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol 2009, 43, 163–172. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Drabik, K.A.; Waypa, T.S.; Musch, M.W.; Alverdy, J.C.; Schneewind, O.; Chang, E.B.; Petrof, E.O. Soluble factors from Lactobacillus GG activate MAPKs and induce cytoprotective heat shock proteins in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2006, 290, C1018–C1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.S.; Choi, J.W.; Jhun, J.; Kwon, J.Y.; Lee, B.I.; Yang, C.W.; Park, S.H.; Cho, M.L. Lactobacillus acidophilus Improves Intestinal Inflammation in an Acute Colitis Mouse Model by Regulation of Th17 and Treg Cell Balance and Fibrosis Development. J. Med. Food 2018, 21, 215–224. [Google Scholar] [CrossRef] [Green Version]
- Vemuri, R.; Shinde, T.; Gundamaraju, R.; Gondalia, S.V.; Karpe, A.V.; Beale, D.J.; Martoni, C.J.; Eri, R. Lactobacillus acidophilus DDS-1 Modulates the Gut Microbiota and Improves Metabolic Profiles in Aging Mice. Nutrients 2018, 10, 1255. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, A.; Huerta-Salgado, C.; Orozco-Aguilar, J.; Aguirre, F.; Tacchi, F.; Simon, F.; Cabello-Verrugio, C. Role of Oxidative Stress in Hepatic and Extrahepatic Dysfunctions during Nonalcoholic Fatty Liver Disease (NAFLD). Oxidative Med. Cell. Longev. 2020, 2020, 1617805. [Google Scholar] [CrossRef]
- Conti, V.; Izzo, V.; Corbi, G.; Russomanno, G.; Manzo, V.; De Lise, F.; Di Donato, A.; Filippelli, A. Antioxidant Supplementation in the Treatment of Aging-Associated Diseases. Front. Pharmacol. 2016, 7, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanda, T.; Matsuoka, S.; Yamazaki, M.; Shibata, T.; Nirei, K.; Takahashi, H.; Kaneko, T.; Fujisawa, M.; Higuchi, T.; Nakamura, H. Apoptosis and non-alcoholic fatty liver diseases. World J. Gastroenterol. 2018, 24, 2661. [Google Scholar] [CrossRef]
- Ma, J.; Yin, H.; Li, M.; Deng, Y.; Ahmad, O.; Qin, G.; He, Q.; Li, J.; Gao, K.; Zhu, J. A comprehensive study of high cholesterol diet-induced larval zebrafish model: A short-time in vivo screening method for non-alcoholic fatty liver disease drugs. Int. J. Biol. Sci. 2019, 15, 973. [Google Scholar] [CrossRef]
- Kobatake, E.; Nakagawa, H.; Seki, T.; Miyazaki, T. Protective effects and functional mechanisms of Lactobacillus gasseri SBT2055 against oxidative stress. PLoS ONE 2017, 12, e0177106. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Gu, S.; Liu, D.; Zhao, L.; Xia, S.; He, X.; Chen, H.; Ge, J. Lactobacillus brevis 23017 Relieves Mercury Toxicity in the Colon by Modulation of Oxidative Stress and Inflammation Through the Interplay of MAPK and NF-κB Signaling Cascades. Front. Microbiol. 2018, 9, 2425. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, Y.; Cheng, Y.; Yan, Q.; Zhou, C.; He, Z.; Zeng, J.; He, J.; Tan, Z. Supplementation of Lactobacillus plantarum or Macleaya cordata Extract Alleviates Oxidative Damage Induced by Weaning in the Lower Gut of Young Goats. Animals 2020, 10, 548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amaretti, A.; di Nunzio, M.; Pompei, A.; Raimondi, S.; Rossi, M.; Bordoni, A. Antioxidant properties of potentially probiotic bacteria: In vitro and in vivo activities. Appl. Microbiol. Biotechnol. 2013, 97, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Y.; Wang, Y.; Xu, H.; Mei, X.; Yu, D.; Wang, Y.; Li, W. Antioxidant Properties of Probiotic Bacteria. Nutrients 2017, 9, 521. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Cui, S.; Song, W.; Jiang, Y.; Chen, H.; Liao, D.; Lu, X.; Li, J.; Chen, X.; Peng, L. Interleukin-17 Weakens the NAFLD/NASH Process by Facilitating Intestinal Barrier Restoration Depending on the Gut Microbiota. Mbio 2022, 13, e03688-21. [Google Scholar] [CrossRef] [PubMed]
- Harley, I.T.W.; Stankiewicz, T.E.; Giles, D.A.; Softic, S.; Flick, L.M.; Cappelletti, M.; Sheridan, R.; Xanthakos, S.A.; Steinbrecher, K.A.; Sartor, R.B.; et al. IL-17 signaling accelerates the progression of nonalcoholic fatty liver disease in mice. Hepatology 2014, 59, 1830–1839. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Wang, K.; Aoyama, T.; Grivennikov, S.I.; Paik, Y.; Scholten, D.; Cong, M.; Iwaisako, K.; Liu, X.; Zhang, M. Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology 2012, 143, 765–776.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garidou, L.; Pomié, C.; Klopp, P.; Waget, A.; Charpentier, J.; Aloulou, M.; Giry, A.; Serino, M.; Stenman, L.; Lahtinen, S. The gut microbiota regulates intestinal CD4 T cells expressing RORγt and controls metabolic disease. Cell Metab. 2015, 22, 100–112. [Google Scholar] [CrossRef] [Green Version]
- Gomes, A.L.; Teijeiro, A.; Burén, S.; Tummala, K.S.; Yilmaz, M.; Waisman, A.; Theurillat, J.-P.; Perna, C.; Djouder, N. Metabolic inflammation-associated IL-17A causes non-alcoholic steatohepatitis and hepatocellular carcinoma. Cancer Cell 2016, 30, 161–175. [Google Scholar] [CrossRef]
- Yu, J.; Ip, E.; dela Peña, A.; Hou, J.Y.; Sesha, J.; Pera, N.; Hall, P.; Kirsch, R.; Leclercq, I.; Farrell, G.C. COX-2 induction in mice with experimental nutritional steatohepatitis: Role as pro-inflammatory mediator. Hepatology 2006, 43, 826–836. [Google Scholar] [CrossRef]
- Lee, J.S.; Tato, C.M.; Joyce-Shaikh, B.; Gulen, M.F.; Cayatte, C.; Chen, Y.; Blumenschein, W.M.; Judo, M.; Ayanoglu, G.; McClanahan, T.K. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 2015, 43, 727–738. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Zhou, H.; Jin, H.; Liu, K. Role of Inflammatory Factors in Mediating the Effect of Lipids on Nonalcoholic Fatty Liver Disease: A Two-Step, Multivariable Mendelian Randomization Study. Nutrients 2022, 14, 4434. [Google Scholar] [CrossRef]
- Bi, Y.; Li, C.; Liu, L.; Zhou, J.; Li, Z.; Deng, H.; Wu, C.; Han, Y.; Song, Y.; Tan, Y. IL-17A-dependent gut microbiota is essential for regulating diet-induced disorders in mice. Sci. Bull. 2017, 62, 1052–1063. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lin, Y.; Lin, S.; Li, C.; Gao, J.; Feng, Z.; Wang, J.; Zhang, J.; Zhang, H.; Zhang, Y.; et al. Silencing of functional p53 attenuates NAFLD by promoting HMGB1-related autophagy induction. Hepatol. Int. 2020, 14, 828–841. [Google Scholar] [CrossRef]
- Yan, Z.; Miao, X.; Zhang, B.; Xie, J. p53 as a double-edged sword in the progression of non-alcoholic fatty liver disease. Life Sci. 2018, 215, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Prokesch, A.; Graef, F.A.; Madl, T.; Kahlhofer, J.; Heidenreich, S.; Schumann, A.; Moyschewitz, E.; Pristoynik, P.; Blaschitz, A.; Knauer, M. Liver p53 is stabilized upon starvation and required for amino acid catabolism and gluconeogenesis. FASEB J. 2017, 31, 732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yahagi, N.; Shimano, H.; Matsuzaka, T.; Sekiya, M.; Najima, Y.; Okazaki, S.; Okazaki, H.; Tamura, Y.; Iizuka, Y.; Inoue, N. p53 involvement in the pathogenesis of fatty liver disease. J. Biol. Chem. 2004, 279, 20571–20575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillen-Sacoto, M.J.; Martinez, A.F.; Abe, Y.; Kruszka, P.; Weiss, K.; Everson, J.L.; Bataller, R.; Kleiner, D.E.; Ward, J.M.; Sulik, K.K. Human germline hedgehog pathway mutations predispose to fatty liver. J. Hepatol. 2017, 67, 809–817. [Google Scholar] [CrossRef]
- Deisenroth, C.; Itahana, Y.; Tollini, L.; Jin, A.; Zhang, Y. p53-Inducible DHRS3 is an endoplasmic reticulum protein associated with lipid droplet accumulation. J. Biol. Chem. 2011, 286, 28343–28356. [Google Scholar] [CrossRef]
- Goldstein, I.; Rotter, V. Regulation of lipid metabolism by p53–fighting two villains with one sword. Trends Endocrinol. Metab. 2012, 23, 567–575. [Google Scholar] [CrossRef]
- Kung, C.-P.; Basu, S.; Murphy, M.E. A link between TP53 polymorphisms and metabolism. Mol. Cell. Oncol. 2016, 3, e1173769. [Google Scholar] [CrossRef] [Green Version]
- Parrales, A.; Iwakuma, T. p53 as a regulator of lipid metabolism in cancer. Int. J. Mol. Sci. 2016, 17, 2074. [Google Scholar] [CrossRef] [Green Version]
- Molchadsky, A.; Ezra, O.; Amendola, P.; Krantz, D.; Kogan-Sakin, I.; Buganim, Y.; Rivlin, N.; Goldfinger, N.; Folgiero, V.; Falcioni, R. p53 is required for brown adipogenic differentiation and has a protective role against diet-induced obesity. Cell Death Differ. 2013, 20, 774–783. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease—Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Derdak, Z.; Villegas, K.A.; Harb, R.; Wu, A.M.; Sousa, A.; Wands, J.R. Inhibition of p53 attenuates steatosis and liver injury in a mouse model of non-alcoholic fatty liver disease. J. Hepatol. 2013, 58, 785–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dibra, D.; Xia, X.; Mitra, A.; Cutrera, J.J.; Lozano, G.; Li, S. Mutant p53 in concert with an interleukin-27 receptor alpha deficiency causes spontaneous liver inflammation, fibrosis, and steatosis in mice. Hepatology 2016, 63, 1000–1012. [Google Scholar] [CrossRef] [Green Version]
- Porteiro, B.; Fondevila, M.F.; Delgado, T.C.; Iglesias, C.; Imbernon, M.; Iruzubieta, P.; Crespo, J.; Zabala-Letona, A.; Fernø, J.; González-Terán, B. Hepatic p63 regulates steatosis via IKKβ/ER stress. Nat. Commun. 2017, 8, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Teratani, T.; Suzuki, T.; Oshikawa, T.; Yokoyama, H.; Shimamura, K.; Nishiyama, K.; Mataki, N.; Irie, R.; Minamino, T. p53/p66Shc-mediated signaling contributes to the progression of non-alcoholic steatohepatitis in humans and mice. J. Hepatol. 2012, 57, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Jia, L.; Lin, M.; Shi, Y.; Yin, J.; Liu, Y.; Chen, D.; Meng, Q. ASPP 2 attenuates triglycerides to protect against hepatocyte injury by reducing autophagy in a cell and mouse model of non-alcoholic fatty liver disease. J. Cell. Mol. Med. 2015, 19, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Derdak, Z.; Lang, C.H.; Villegas, K.A.; Tong, M.; Mark, N.M.; Suzanne, M.; Wands, J.R. Activation of p53 enhances apoptosis and insulin resistance in a rat model of alcoholic liver disease. J. Hepatol. 2011, 54, 164–172. [Google Scholar] [CrossRef] [Green Version]
- Farrell, G.C.; Larter, C.Z.; Hou, J.Y.; Zhang, R.H.; Yeh, M.M.; Williams, J.; Dela Peňa, A.; Francisco, R.; Osvath, S.R.; Brooling, J. Apoptosis in experimental NASH is associated with p53 activation and TRAIL receptor expression. J. Gastroenterol. Hepatol. 2009, 24, 443–452. [Google Scholar] [CrossRef]
- Yokoyama, M.; Okada, S.; Nakagomi, A.; Moriya, J.; Shimizu, I.; Nojima, A.; Yoshida, Y.; Ichimiya, H.; Kamimura, N.; Kobayashi, Y. Inhibition of endothelial p53 improves metabolic abnormalities related to dietary obesity. Cell Rep. 2014, 7, 1691–1703. [Google Scholar] [CrossRef] [Green Version]
- Panasiuk, A.; Dzieciol, J.; Panasiuk, B.; Prokopowicz, D. Expression of p53, Bax and Bcl-2 proteins in hepatocytes in non-alcoholic fatty liver disease. World J. Gastroenterol. WJG 2006, 12, 6198. [Google Scholar] [CrossRef]
- Palmisano, B.T.; Zhu, L.; Stafford, J.M. Role of estrogens in the regulation of liver lipid metabolism. In Sex and Gender Factors Affecting Metabolic Homeostasis, Diabetes and Obesity; Springer: Cham, Switzerland, 2017; pp. 227–256. [Google Scholar]
- Ciana, P.; Raviscioni, M.; Mussi, P.; Vegeto, E.; Que, I.; Parker, M.G.; Lowik, C.; Maggi, A. In vivo imaging of transcriptionally active estrogen receptors. Nat. Med. 2003, 9, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Ji, X.; Zhu, Y.; Zhao, D.; Han, K.; Gu, S.; Ma, L.; Jin, S.; Chen, J.; Li, T. Effects of Sheep Bone Collagen Peptide on Liver Lipid Deposition in Ovariectomized Rats. J. Nutr. Sci. Vitaminol. 2022, 68, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L.; Toft, D.O. A novel chaperone complex for steroid receptors involving heat shock proteins, immunophilins, and p23. J. Biol. Chem. 1994, 269, 24989–24993. [Google Scholar] [CrossRef]
- Roe, S.M.; Prodromou, C.; O’Brien, R.; Ladbury, J.E.; Piper, P.W.; Pearl, L.H. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J. Med. Chem. 1999, 42, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Maloney, A.; Workman, P. HSP90 as a new therapeutic target for cancer therapy: The story unfolds. Expert Opin. Biol. Ther. 2002, 2, 3–24. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, T.; Chen, Z.; Ren, H.; Song, P.; Zhu, Y.; Liang, S.; Tzeng, C. Development and Evaluation of a Thermosensitive In Situ Gel Formulation for Intravaginal Delivery of Lactobacillus gasseri. Pharmaceutics 2022, 14, 1934. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Y.; Sun, J.; Sha, H.; Yang, Y.; Ye, Q.; Yang, Q.; Huang, B.; Yu, Y.; Huang, H. Acute toxic responses of embryo-larval zebrafish to zinc pyrithione (ZPT) reveal embryological and developmental toxicity. Chemosphere 2018, 205, 62–70. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, H.-L.; Li, T.-T.; Yang, F.; Tzeng, C.-M. Baicalin ameliorates dexamethasone-induced osteoporosis by regulation of the RANK/RANKL/OPG signaling pathway. Drug Des. Dev. Ther. 2020, 14, 195. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Zheng, Y.-M.; Zhang, J.-P. Comparative study of different diets-induced NAFLD models of zebrafish. Front. Endocrinol. 2018, 9, 366. [Google Scholar] [CrossRef]
- Henderson, J.M.; Polak, N.; Chen, J.; Roediger, B.; Weninger, W.; Kench, J.G.; McCaughan, G.W.; Zhang, H.E.; Gorrell, M.D. Multiple liver insults synergize to accelerate experimental hepatocellular carcinoma. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Bu, Y.; Wu, Q.; Wang, X.; Chang, N.; Lei, L.; Chen, S.; Liu, D.; Zhu, X.; Hu, K. Mecp2 regulates neural cell differentiation by suppressing the Id1 to Her2 axis in zebrafish. J. Cell Sci. 2015, 128, 2340–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, S.H.; Wu, Y.L.; Vega, V.B.; Miller, L.D.; Spitsbergen, J.; Tong, Y.; Zhan, H.; Govindarajan, K.R.; Lee, S.; Mathavan, S. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat. Biotechnol. 2006, 24, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Huang, X.; Ding, T.W.; Gong, Z. Enhanced angiogenesis, hypoxia and neutrophil recruitment during Myc-induced liver tumorigenesis in zebrafish. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]







| Gene | Gene ID | Forward Sequence | Reverse Sequence |
|---|---|---|---|
| papss2a | 777719 | GCAAGTTGTAGGCACCAGAG | CCATGACGGATGTTGTCACC |
| hsd17b7 | 768185 | TAGCTTAGCCTTGAACCGCC | TGGTATTTGGCCATCGGGTC |
| cdk21 | 569515 | CTGAAGCCTGACAATGTGCT | GCAAGCCAATTACCTCAAAGA |
| myl9b | 406493 | CTGCTTCGATGAGGAGGGAT | GGCTCCGTGTTTGAGGATTC |
| iras2a | 393285 | CTCCGAGGTGGCATCAGTTAC | CCCCTTCACTTGCAGTCCGTATT |
| ptgs2b | 559020 | CCCCAGAGTACTGGAAACCA | ACATGGCCCGTTGACATTAT |
| hsp90aa1.1 | 30591 | AGGCCTTTGTGCCGGATTTA | TTGCAGTGGGGTGTTTATGC |
| kyat3 | 393315 | TGGAATGTGGGACAGAACCA | GCCAACAGCCTCCTGTAATG |
| gapdh | 317743 | CCAACTGCCTGGCTCCTT | CCCATCAACGGTCTTCTGTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, T.; Lykov, N.; Luo, X.; Wang, H.; Du, Z.; Chen, Z.; Chen, S.; Zhu, L.; Zhao, Y.; Tzeng, C. Protective Effects of Lactobacillus gasseri against High-Cholesterol Diet-Induced Fatty Liver and Regulation of Host Gene Expression Profiles. Int. J. Mol. Sci. 2023, 24, 2053. https://doi.org/10.3390/ijms24032053
He T, Lykov N, Luo X, Wang H, Du Z, Chen Z, Chen S, Zhu L, Zhao Y, Tzeng C. Protective Effects of Lactobacillus gasseri against High-Cholesterol Diet-Induced Fatty Liver and Regulation of Host Gene Expression Profiles. International Journal of Molecular Sciences. 2023; 24(3):2053. https://doi.org/10.3390/ijms24032053
Chicago/Turabian StyleHe, Tianhua, Nikita Lykov, Xu Luo, Huiling Wang, Zhanxiang Du, Ziyi Chen, Shitian Chen, Lin Zhu, Ye Zhao, and Chimeng Tzeng. 2023. "Protective Effects of Lactobacillus gasseri against High-Cholesterol Diet-Induced Fatty Liver and Regulation of Host Gene Expression Profiles" International Journal of Molecular Sciences 24, no. 3: 2053. https://doi.org/10.3390/ijms24032053






