Identification and Analysis of the CBF Gene Family in Three Species of Acer under Cold Stress
Abstract
:1. Introduction
2. Results
2.1. Identification of CBF Genes in Three Acer Species
2.2. Construction of Phylogenetic Tree
2.3. Gene Structure and Conserved Motif of CBF Genes
2.4. Cis-Acting Elements of CBF Genes
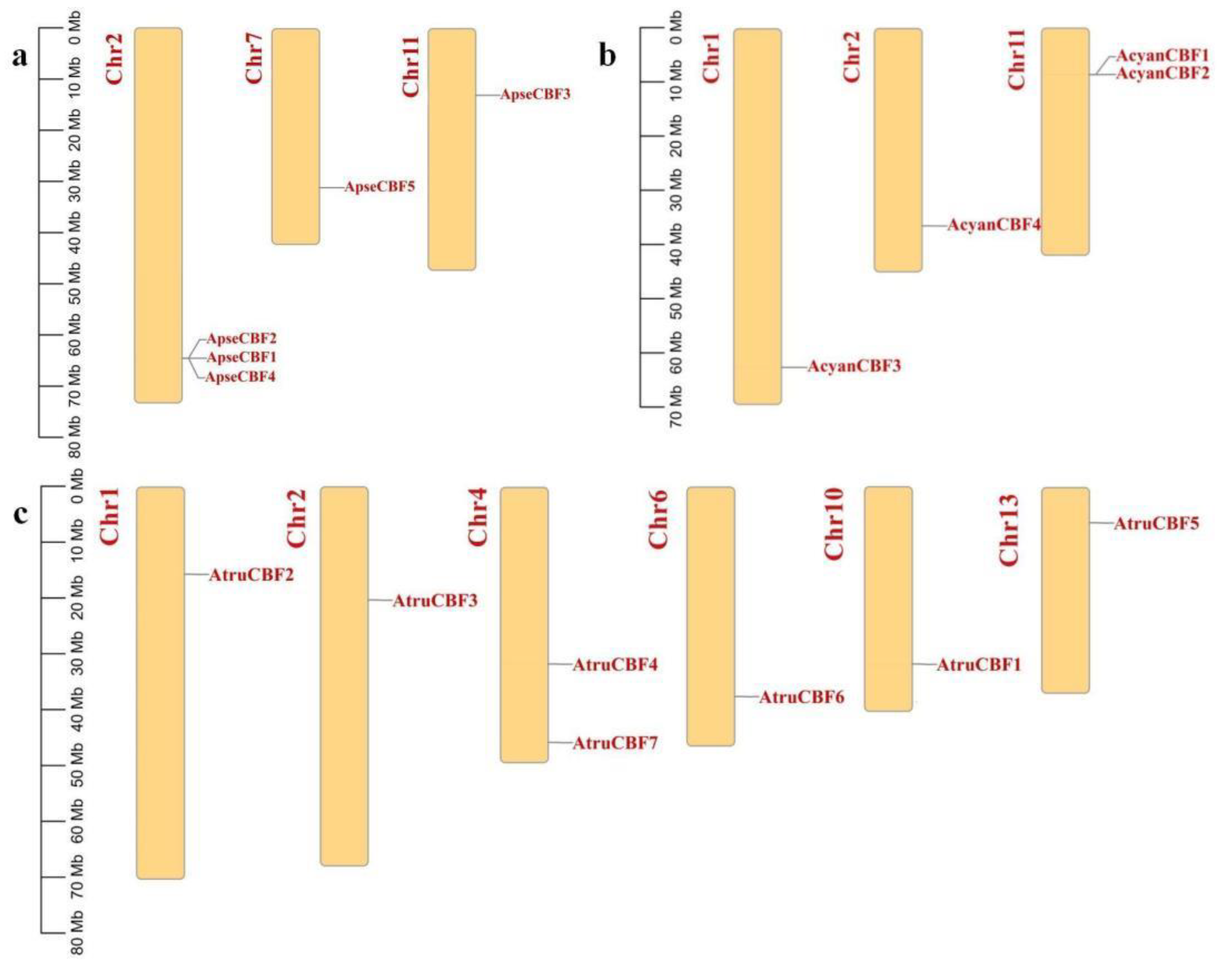
2.5. Chromosome Location of CBF Genes
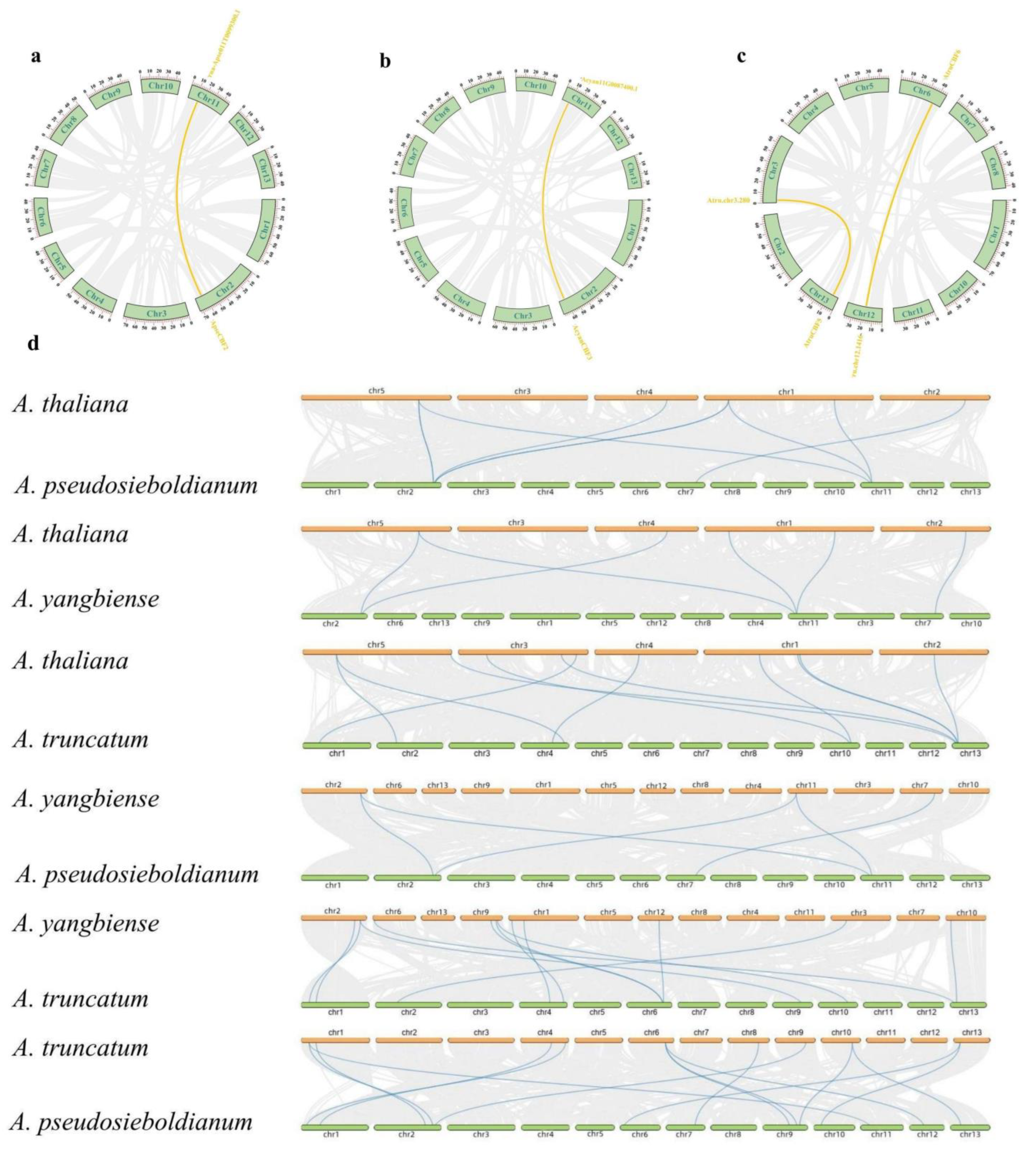
2.6. Synteny Analysis of CBF Genes
2.7. The Expression of CBF Genes in Different Tissues
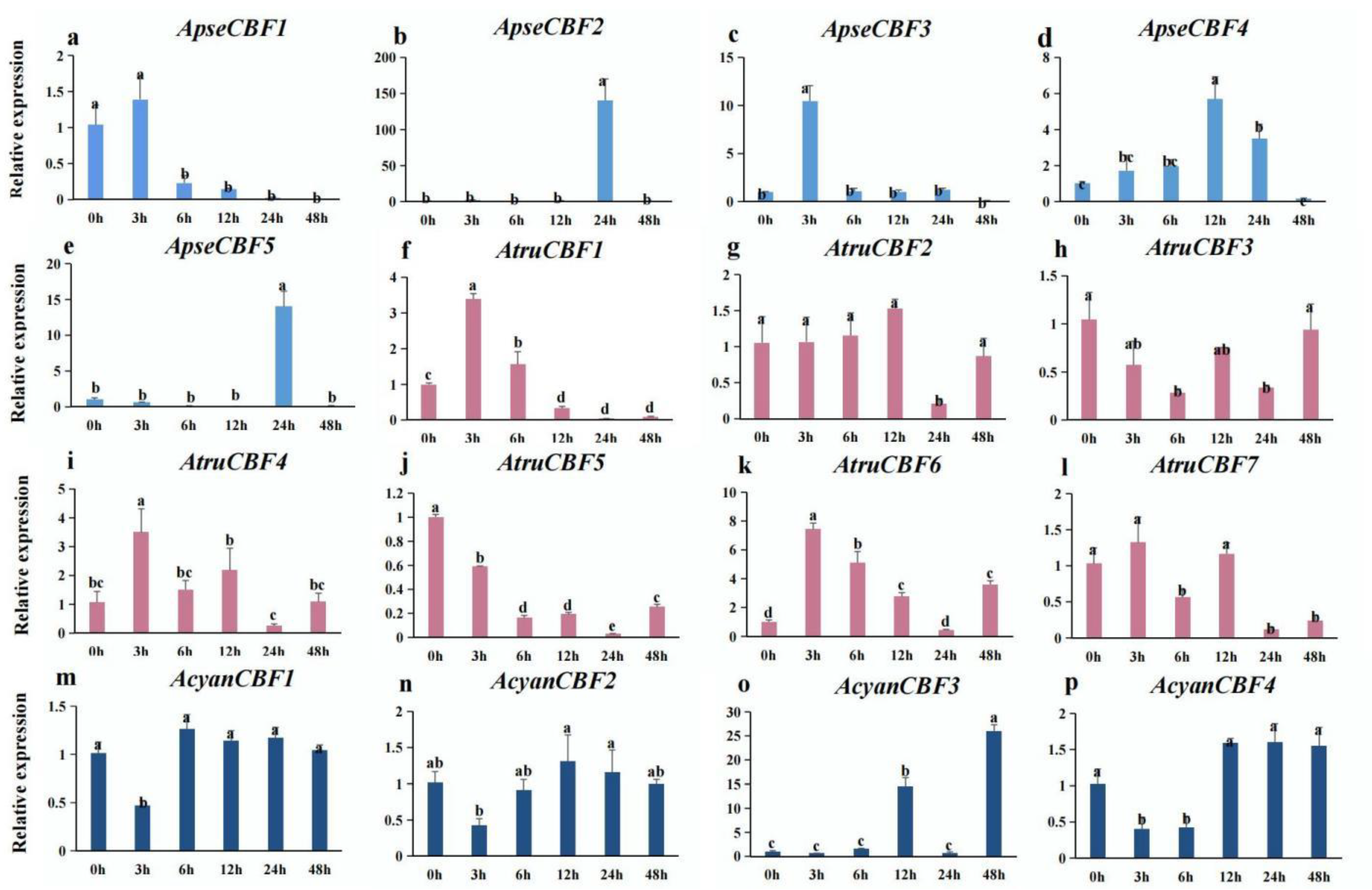
2.8. Expression of CBF Genes in Three Acer Species under Low-Temperature Conditions
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Retrieving the CBF Gene Family Sequences
4.3. Analyses of Gene Structure and Protein Motif Composition
4.4. The Analyses of Chromosomal Location and Collinearity
4.5. The Analysis of Phylogenetic Tree
4.6. The Analysis of Cis-Acting Elements
4.7. The Analysis of Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, H.; Gu, B.; Zhang, J. The transcription factor VaMYC2 from Chinese wild Vitis amurensis enhances cold tolerance of grape (V. vinifera) by up-regulating VaCBF1 and VaP5CS. Plant Physiol. Biochem. 2022, 192, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Jankauskienė, J.; Mockevičiūtė, R.; Gavelienė, V.; Jurkonienė, S.; Anisimovienė, N. The application of auxin-like compounds promotes cold acclimation in the oilseed rape plant. Life 2022, 12, 1283. [Google Scholar] [CrossRef]
- Sun, B.; Liang, M.; Yang, Y.; Xu, H. A primary analysis on cold resistance of Acer pseudosieboldianum (pax) kom in Heilongjiang Province. Territ. Nat. Resour. Study 2013, 3, 88–89. [Google Scholar]
- Liu, C.; Tsuda, Y.; Shen, H.; Hu, L.; Saito, Y.; Ide, Y. Genetic structure and hierarchical population divergence history of Acer mono var. mono in South and Northeast China. PLoS ONE 2014, 9, e87187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renner, S.S.; Beenken, L.; Grimm, G.W.; Kocyan, A.; Ricklefs, R.E. The evolution of dioecy, heterodichogamy, and labile sex expression in Acer. Evolution 2007, 61, 2701–2719. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.S.; Grimm, G.W.; Schneeweiss, G.M.; Stuessy, T.F.; Ricklefs, R.E. Rooting and dating maples (Acer) with an uncorrelated-rates molecular clock: Implications for North American/Asian Disjunctions. Syst. Biol. 2008, 7, 795–808. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Sun, T.; Li, S.; Wen, J.; Zhu, L.; Yin, T.; Yan, K.; Xu, X.; Li, S.; Mao, J.; et al. The Acer truncatum genome provides insights into nervonic acid biosynthesis. Plant J. 2020, 104, 662–678. [Google Scholar] [CrossRef]
- Gao, Y.F.; Zhao, D.H.; Zhang, J.Q.; Chen, J.S.; Li, J.L.; Weng, Z.; Rong, L.P. De novo transcriptome sequencing and anthocyanin metabolite analysis reveals leaf color of Acer pseudosieboldianum in autumn. BMC Genom. 2021, 22, 383. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, W.; Yang, J. Development and characterization of microsatellite markers in the critically endangered species Acer yangbiense (Aceraceae). Am. J. Bot. 2011, 98, e247–e249. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Zhang, R.; Chi, B.; Li, H. Analysis of growth and physiological characteristics of different provenances of Acer truncatum. Liaoning For. Sci. Technol. 2020, 1, 17–20, 31, 72. [Google Scholar]
- Xiao, L.; Ren, J.Z.; Li, Q.; Yang, B.; Liu, Z.J.; Chen, R.B.; Zhang, L. Genome-wide analysis of AP2/ERF superfamily in Isatis indigotica. J. Integr. Med. 2022, 21, 77–78. [Google Scholar] [CrossRef]
- Dietz, K.J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Cui, B.; Guo, H.; Liu, Y.; Nie, S. Genome-wide identification and expression analysis of the CBF transcription factor family in Lolium perenne under abiotic stress. Plant Signal Behav. 2022, 17, 2086733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gong, Y.; Sun, P.; Chen, S.; Ma, C. Genome-wide identification of CBF genes and their responses to cold acclimation in Taraxacum koksaghyz. PeerJ 2022, 10, e13429. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ban, Q.Y.; Hao, J.; Zhu, X.X.; Cheng, Y.H.; Mao, J.L.; Lin, M.L.; Xia, E.H.; Li, Y.Y. Genome-wide characterization of the C-repeat Binding Factor (CBF) gene family involved in the response to abiotic stresses in tea plant (Camellia sinensis). Front. Plant Sci. 2020, 11, 921. [Google Scholar] [CrossRef]
- Cao, P.B.; Azar, S.; SanClemente, H.; Mounet, F.; Dunand, C.; Marque, G.; Marque, C.; Teulières, C. Genome-wide analysis of the AP2/ERF family in Eucalyptus grandis: An intriguing over-representation of stress-responsive DREB1/CBF genes. PLoS ONE 2015, 10, e0121041. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Lim, M.H.; Yu, J.G.; Park, B.S.; Yang, T.J. Genome-wide characterization of the CBF/DREB1 gene family in Brassica rapa. Plant Physiol. Biochem. 2012, 61, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Fowler, S.G.; Cheng, H.; Lou, Y.; Thomashow, M.F.; Stockinger, E.J.; Thomashow, M.F. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 2010, 39, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Maurya, N.K.; Goswami, A.K.; Singh, S.K.; Prakash, J.; Kumari, A.; Chinnusamy, V.; Talukdar, A.; Pradhan, S.; Kumari, A. Studies on expression of CBF1 and CBF2 genes and anti-oxidant enzyme activities in papaya genotypes exposed to low temperature stress. Sci. Hortic. 2019, 261, 108914. [Google Scholar] [CrossRef]
- Akhtar, M.; Jaiswal, A.; Taj, G.; Jaiswal, J.P.; Qureshi, M.I.; Singh, N.K. DREB1/CBF transcription factors: Their structure, function and role in abiotic stress tolerance in plants. J. Genet. 2012, 91, 385–395. [Google Scholar] [CrossRef]
- Haake, V.; Cook, D.; Riechmann, J.L.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and Grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Liang, X.; Li, W.; Yao, A.; Liu, W.; Wang, Y.; Yang, G.; Han, D. Isolation and functional analysis of MbCBF2, a Malus baccata (L.) Borkh CBF transcription factor gene, with functions in tolerance to cold and salt stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 9827. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wu, H.; He, F.; Qu, J.; Wang, Y.; Li, C.; Liu, J.H. Citrus sinensis CBF1 functions in cold tolerance by modulating putrescine biosynthesis through regulation of arginine decarboxylase. Plant Cell Physiol. 2022, 3, 19–29. [Google Scholar] [CrossRef]
- Maibam, P.; Nawkar, G.M.; Park, J.H.; Sahi, V.P.; Lee, S.Y.; Kang, C.H. The influence of light quality, Circadian rhythm, and photoperiod on the CBF-mediated freezing tolerance. Int. J. Mol. Sci. 2013, 14, 11527–11543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Wariss, H.M.; Tao, L.; Zhang, R.; Yun, Q.; Hollingsworth, P.; Dao, Z.; Luo, G.; Guo, H.; Ma, Y.; et al. De novo genome assembly of the endangered Acer yangbiense, a plant species with extremely small populations endemic to Yunnan Province, China. Gigascience 2019, 8, giz085. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Cai, K.W.; Han, Z.M.; Zhang, S.K.; Sun, A.R.; Xie, Y.; Han, R.; Guo, R.X.; Tigabu, M.; Sederoff, R.; et al. Chromosome-level genome assembly for Acer pseudosieboldianum and highlights to mechanisms for leaf color and shape change. Front. Plant Sci. 2022, 13, 850054. [Google Scholar] [CrossRef]
- Navarro, M.; Ayax, C.; Martinez, Y.; Laur, J.; El Kayal, W.; Marque, C.; Teulières, C. Two EguCBF1 genes overexpressed in Eucalyptus display a different impact on stress tolerance and plant development. Plant Biotechnol. J. 2011, 9, 50–63. [Google Scholar] [CrossRef]
- Wisniewski, M.; Norelli, J.; Artlip, T. Overexpression of a peach CBF gene in apple: A model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 2015, 6, 85. [Google Scholar] [CrossRef] [Green Version]
- Welling, A.; Palva, E.T. Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol. 2008, 147, 1199–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Cai, K.; Pei, X.; Li, Y.; Hu, Y.; Meng, F.; Song, X.; Tigabu, M.; Ding, C.; Zhao, X. Genome-wide identification of NAC transcription factor family in Juglans mandshurica and their expression analysis during the fruit development and ripening. Int. J. Mol. Sci. 2021, 22, 12414. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Fei, S.Z. Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 2006, 224, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar] [CrossRef]
- Shu, Y.; Liu, Y.; Zhang, J.; Song, L.; Guo, C. Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant Sci. 2016, 1247. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.J.; Cao, Y.; Wang, J.; Xiong, Z. Transcriptome sequencing of Pinus kesiya var. langbianensis and comparative analysis in the Pinus phylogeny. BMC Genom. 2018, 19, 725. [Google Scholar] [CrossRef] [Green Version]
- Doelling, J.H.; Pikaard, C.S. The minimal ribosomal RNA gene promoter of Arabidopsis thaliana includes a critical element at the transcription initiation site. Plant J. 1995, 8, 683–692. [Google Scholar] [CrossRef]
- McKhann, H.I.; Gery, C.; Bérard, A.; Lévêque, S.; Zuther, E.; Hincha, D.K.; De Mita, S.; Brunel, D.; Téoulé, E. Natural variation in CBF gene sequence, gene expression and freezing tolerance in the versailles core collection of Arabidopsis thaliana. BMC Plant Biol. 2008, 8, 105–118. [Google Scholar] [CrossRef]
- Kang, J.; Zhang, H.; Sun, T.; Shi, Y.; Wang, J.; Zhang, B.; Wang, Z.; Zhou, Y.; Gu, H. Natural variation of C-Repeat-Binding Factor (CBFs) genes is a major cause of divergence in freezing tolerance among a group of Arabidopsis thaliana populations along the Yangtze River in China. New Phytol. 2013, 199, 1069–1080. [Google Scholar] [CrossRef]
- Gehan, M.A.; Park, S.; Gilmour, S.J.; An, C.; Lee, C.M.; Thomashow, M.F. Natural variation in the C-Repeat Binding Factor cold response pathway correlates with local adaptation of Arabidopsis ecotypes. Plant J. 2015, 84, 682–693. [Google Scholar] [CrossRef] [Green Version]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.D.; Zhuang, K.Y.; Liu, Z.M.; Yang, D.Y.; Ma, N.N.; Meng, Q.W. Overexpression of a novel NAC-type tomato transcription factor, SlNAM1, enhances the chilling stress tolerance of transgenic tobacco. J. Plant Physiol. 2016, 204, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.J.; Seo, Y.W. Identification of novel C-repeat binding factor (CBF) genes in rye (Secale cereale L.) and expression studies. Gene 2019, 684, 82–94. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, S.; Zheng, J. Identification and expression analysis of CBF (C-repeat Binding Factor) gene family in Pomegranate (Punica granatum). Acta Agric. Boreali-Occident. Sin. 2022, 31, 1154–1167. [Google Scholar]
- Li, X.; Guo, C.; Ahmad, S.; Wang, Q.; Yu, J.; Liu, C.; Guo, Y. Systematic analysis of MYB family genes in potato and their multiple roles in development and stress responses. Biomolecules 2019, 9, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Cao, S.; Zhang, J.; Cheng, H.; Aslam, M.; Lv, H.; Dong, W.; Hu, A.; Guo, M.; Liu, Q.; Qin, Y. Identification and evolutionary analysis of FAD2 gene family in green plants. Trop. Plant Biol. 2021, 14, 239–250. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Gene Name | Gene ID | AA 1 | MW 2 (kDa) | PI 3 | SL 4 | CDS (bp) |
|---|---|---|---|---|---|---|
| ApseCBF1 | Apse002T0243400.1 | 253 | 27,693.81 | 4.9 | Nucleus | 759 |
| ApseCBF2 | Apse002T0243300.1 | 253 | 27,666.73 | 4.77 | Nucleus | 759 |
| ApseCBF3 | Apse011T0099600.1 | 246 | 27,542.91 | 6.68 | Cytoplasm, Nucleus | 738 |
| ApseCBF4 | Apse002T0243600.1 | 235 | 24,831.5 | 5.04 | Cytoplasm, Nucleus | 705 |
| ApseCBF5 | Apse007T0094500.1 | 212 | 22,876.3 | 5.46 | Nucleus | 636 |
| AcyanCBF1 | Acyan11G0087800.1 | 301 | 33,234.08 | 6.03 | Cytoplasm, Nucleus | 903 |
| AcyanCBF2 | Acyan11G0087900.1 | 382 | 42,556.73 | 8.9 | Cytoplasm | 1146 |
| AcyanCBF3 | Acyan02G0289000.1 | 249 | 26,360.25 | 5.42 | Cytoplasm, Nucleus | 747 |
| AcyanCBF4 | Acyan07G0093600.1 | 223 | 23,930.49 | 5.61 | Nucleus | 669 |
| AtruCBF1 | Atru.chr10.1810 | 258 | 27,824.85 | 9.35 | Nucleus | 774 |
| AtruCBF2 | Atru.chr1.1042 | 238 | 27,303.82 | 9.49 | Nucleus | 714 |
| AtruCBF3 | Atru.chr2.1230 | 190 | 20,944.53 | 9.73 | Nucleus | 570 |
| AtruCBF4 | Atru.chr4.1701 | 216 | 24,514.44 | 9.79 | Nucleus | 648 |
| AtruCBF5 | Atru.chr13.731 | 165 | 19,008.12 | 9.37 | Cytoplasm, Nucleus | 495 |
| AtruCBF6 | Atru.chr6.2697 | 291 | 31,912.79 | 7.37 | Nucleus | 873 |
| AtruCBF7 | Atru.chr4.2790 | 395 | 43,582.88 | 6.38 | Nucleus | 1185 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Q.; Han, R.; Cai, K.; Yan, H.; Li, Y.; Qu, G.; Liu, L.; Zhao, X. Identification and Analysis of the CBF Gene Family in Three Species of Acer under Cold Stress. Int. J. Mol. Sci. 2023, 24, 2088. https://doi.org/10.3390/ijms24032088
Zhao Q, Han R, Cai K, Yan H, Li Y, Qu G, Liu L, Zhao X. Identification and Analysis of the CBF Gene Family in Three Species of Acer under Cold Stress. International Journal of Molecular Sciences. 2023; 24(3):2088. https://doi.org/10.3390/ijms24032088
Chicago/Turabian StyleZhao, Qiushuang, Rui Han, Kewei Cai, Huiling Yan, Yan Li, Guanzheng Qu, Lin Liu, and Xiyang Zhao. 2023. "Identification and Analysis of the CBF Gene Family in Three Species of Acer under Cold Stress" International Journal of Molecular Sciences 24, no. 3: 2088. https://doi.org/10.3390/ijms24032088
APA StyleZhao, Q., Han, R., Cai, K., Yan, H., Li, Y., Qu, G., Liu, L., & Zhao, X. (2023). Identification and Analysis of the CBF Gene Family in Three Species of Acer under Cold Stress. International Journal of Molecular Sciences, 24(3), 2088. https://doi.org/10.3390/ijms24032088






