Regulation of Plant Photoresponses by Protein Kinase Activity of Phytochrome A
Abstract
1. Introduction
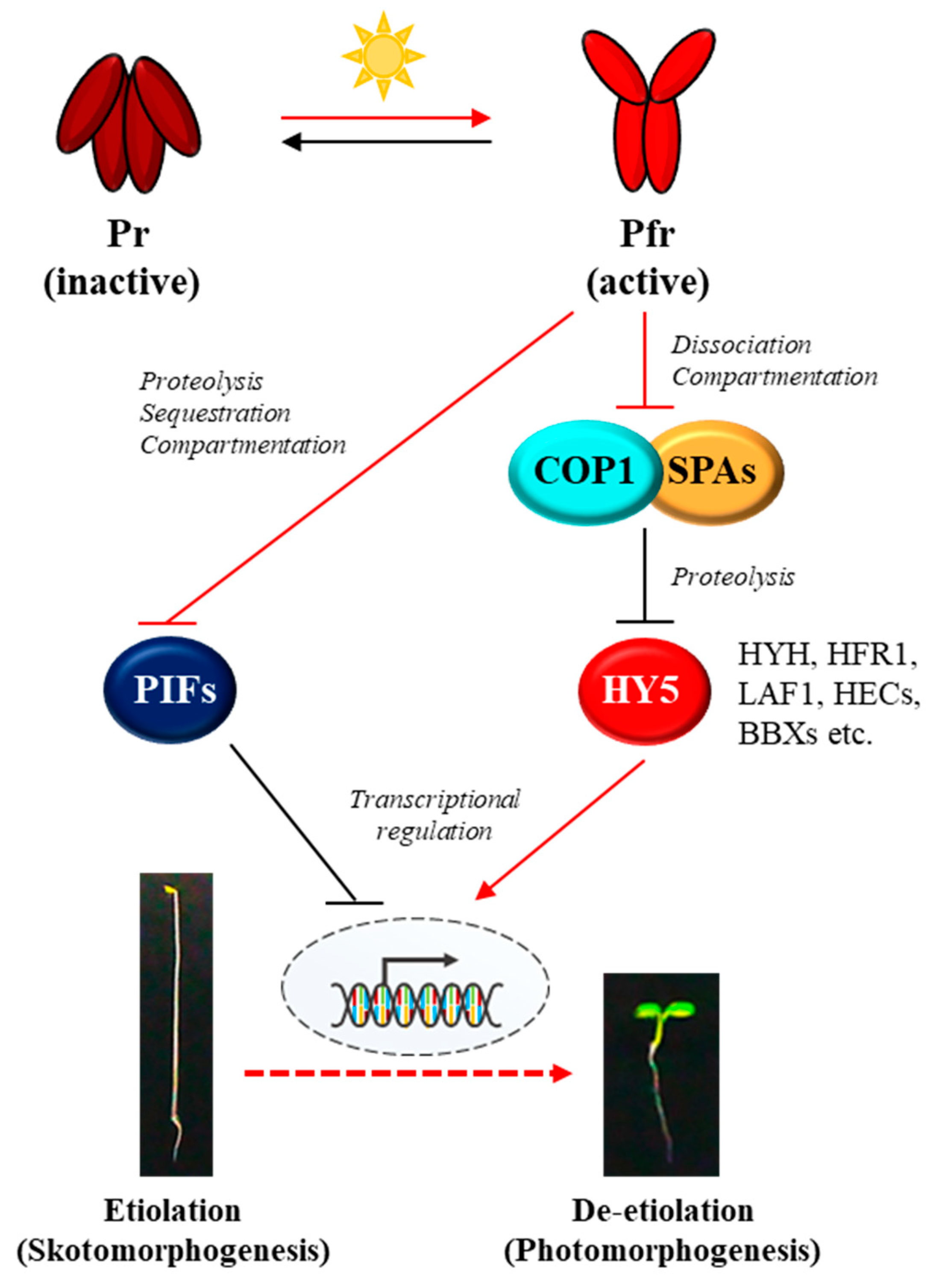
2. The Core Phytochrome Signaling Pathway

3. Regulation of phyA as Phosphoproteins
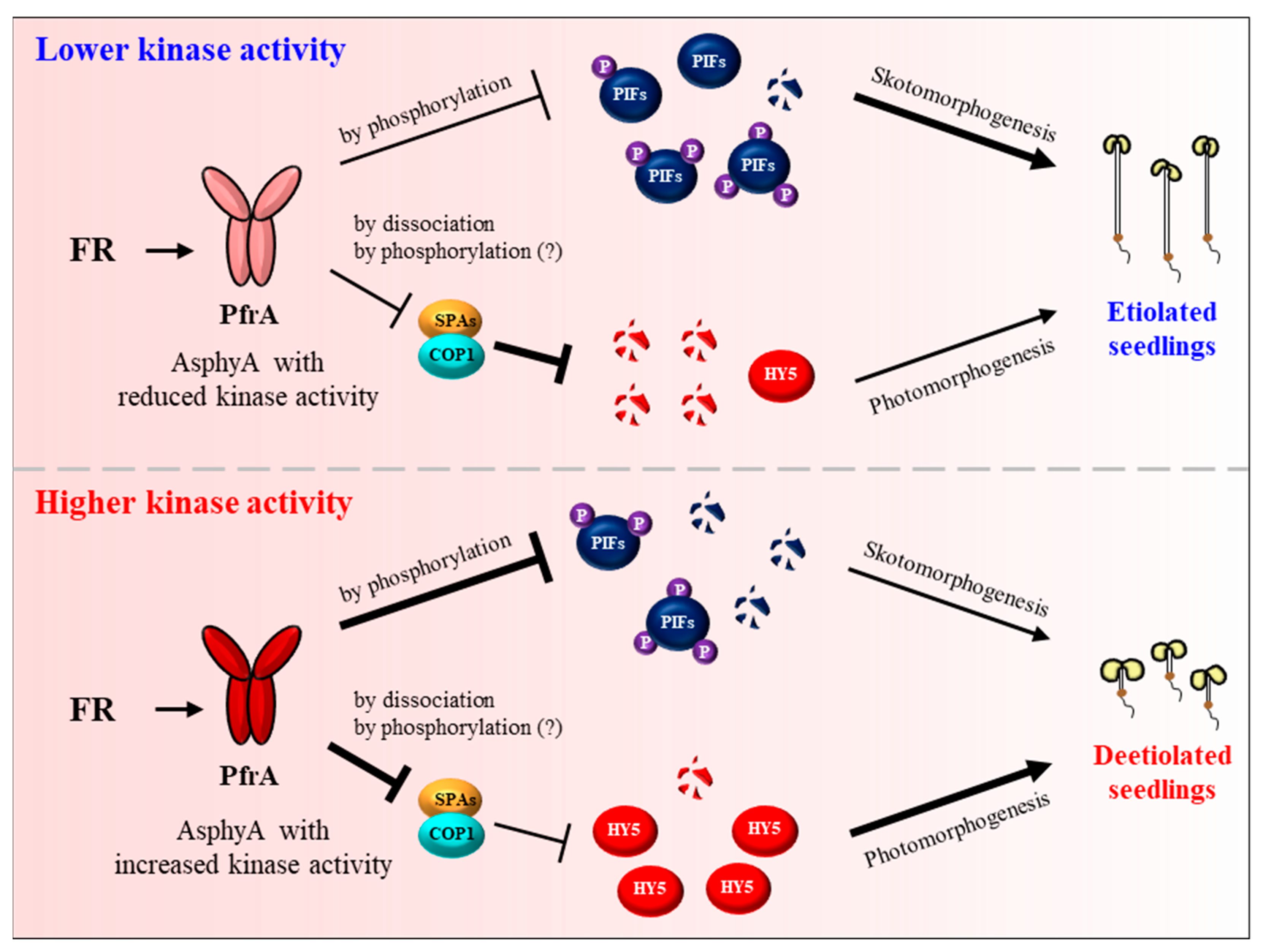
4. Function of phyA as Protein Kinases
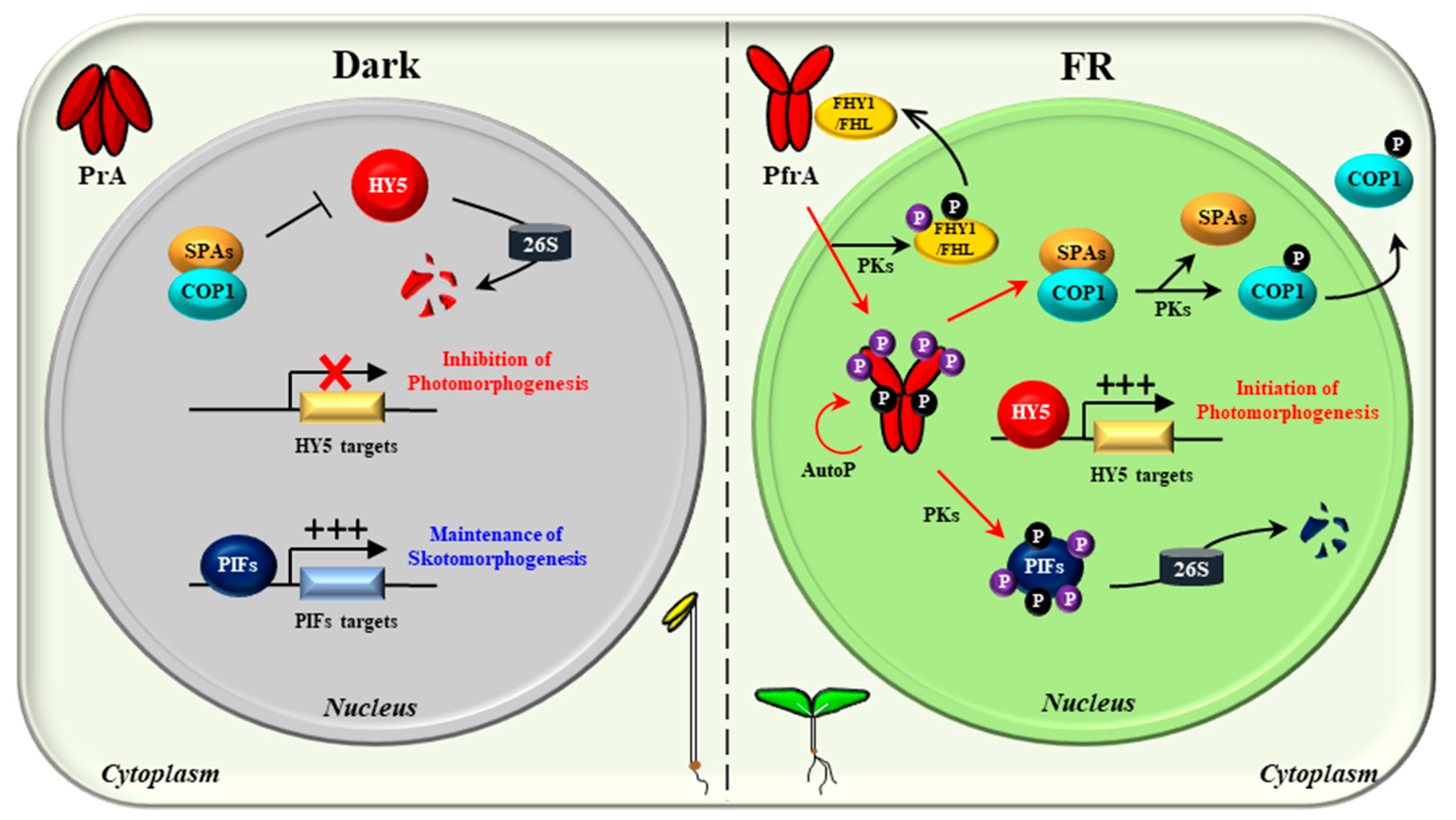
5. The Regulatory and Molecular Mechanisms of phyA in Plant Light Signaling
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huber, M.; Nieuwendijk, N.M.; Pantazopoulou, C.K.; Pierik, R. Light signalling shapes plant-plant interactions in dense canopies. Plant Cell Environ. 2021, 44, 1014–1029. [Google Scholar] [CrossRef] [PubMed]
- Paik, I.; Huq, E. Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol. 2019, 92, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Galvao, V.C.; Fankhauser, C. Sensing the light environment in plants: Photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 2015, 34, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.-C.; Kathare, P.K.; Paik, I.; Huq, E. Phytochrome signaling networks. Annu. Rev. Plant Biol. 2021, 72, 217–244. [Google Scholar] [CrossRef]
- Legris, M.; Ince, Y.C.; Fankhauser, C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 2019, 10, 5219. [Google Scholar] [CrossRef]
- Mathews, S. Evolutionary studies illuminate the structural-functional model of plant phytochromes. Plant Cell 2010, 22, 4–16. [Google Scholar] [CrossRef]
- Li, F.-W.; Melkonian, M.; Rothfels, C.J.; Villarreal, J.C.; Stevenson, D.W.; Graham, S.W.; Wong, G.K.; Pryer, K.M.; Mathews, S. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat. Commun. 2015, 6, 7852. [Google Scholar] [CrossRef]
- Sharrock, R.A.; Clack, T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002, 130, 442–456. [Google Scholar] [CrossRef]
- Sheerin, D.J.; Hiltbrunner, A. Molecular mechanisms and ecological function of far-red light signalling. Plant Cell Environ. 2017, 40, 2509–2529. [Google Scholar] [CrossRef]
- Burgie, E.S.; Gannam, Z.T.K.; McLoughlin, K.E.; Sherman, C.D.; Holehouse, A.S.; Stankey, R.J.; Vierstra, R.D. Differing biophysical properties underpin the unique signaling potentials within the plant phytochrome photoreceptor families. Proc. Natl. Acad. Sci. USA 2021, 118, e2105649118. [Google Scholar] [CrossRef]
- Seaton, D.D.; Toledo-Ortiz, G.; Ganpudi, A.; Kubota, A.; Imaizumi, T.; Halliday, K.J. Dawn and photoperiod sensing by phytochrome A. Proc. Natl. Acad. Sci. USA 2018, 115, 10523–10528. [Google Scholar] [CrossRef] [PubMed]
- Rausenberger, J.; Tscheuschler, A.; Nordmeier, W.; Wust, F.; Timmer, J.; Schafer, E.; Fleck, C.; Hiltbrunner, A. Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 2011, 146, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Klose, C.; Viczian, A.; Kircher, S.; Schafer, E.; Nagy, F. Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol. 2015, 206, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Menon, C.; Klose, C.; Hiltbrunner, A. Arabidopsis FHY1 and FHY1-LIKE are not required for phytochrome A signal transduction in the nucleus. Plant Commun. 2020, 1, 100007. [Google Scholar] [CrossRef] [PubMed]
- Possart, A.; Fleck, C.; Hiltbrunner, A. Shedding (far-red) light on phytochrome mechanisms and responses in land plants. Plant Sci. 2014, 217-218, 36–46. [Google Scholar] [CrossRef]
- Casal, J.J.; Candia, A.N.; Sellaro, R. Light perception and signalling by phytochrome A. J. Exp. Bot. 2014, 65, 2835–2845. [Google Scholar] [CrossRef]
- Tripathi, S.; Hoang, Q.T.N.; Han, Y.-J.; Kim, J.-I. Regulation of photomorphogenic development by plant phytochromes. Int. J. Mol. Sci. 2019, 20, 6165. [Google Scholar] [CrossRef]
- Cordeiro, A.M.; Andrade, L.; Monteiro, C.C.; Leitao, G.; Wigge, P.A.; Saibo, N.J.M. PHYTOCHROME-INTERACTING FACTORS: A promising tool to improve crop productivity. J. Exp. Bot. 2022, 73, 3881–3897. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Chu, L.; Zhang, Y.; Bian, Y.; Xiao, J.; Xu, D. HY5: A pivotal regulator of light-dependent development in higher plants. Front. Plant. Sci. 2021, 12, 800989. [Google Scholar] [CrossRef]
- Podolec, R.; Ulm, R. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 2018, 45, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Franklin, K.A.; Sharrock, R.A.; Jones, M.A.; Harmer, S.L.; Lagarias, J.C. Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc. Natl. Acad. Sci. USA 2013, 110, 1542–1547. [Google Scholar] [CrossRef]
- Strasser, B.; Sanchez-Lamas, M.; Yanovsky, M.J.; Casal, J.J.; Cerdan, P.D. Arabidopsis thaliana life without phytochromes. Proc. Natl. Acad. Sci. USA 2010, 107, 4776–4781. [Google Scholar] [CrossRef]
- Borthwick, H.A.; Hendricks, S.B.; Parker, M.W.; Toole, E.H.; Toole, V.K. A reversible photoreaction controlling seed germination. Proc. Natl. Acad. Sci. USA 1952, 38, 662–666. [Google Scholar] [CrossRef]
- Butler, W.L.; Norris, K.H.; Siegelman, H.W.; Hendricks, S.B. Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc. Natl. Acad. Sci. USA 1959, 45, 1703–1708. [Google Scholar] [CrossRef] [PubMed]
- Fankhauser, C.; Chen, M. Transposing phytochrome into the nucleus. Trends Plant Sci. 2008, 13, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Helizon, H.; Rosler-Dalton, J.; Gasch, P.; von Horsten, S.; Essen, L.-O.; Zeidler, M. Arabidopsis phytochrome A nuclear translocation is mediated by a far-red elongated hypocotyl 1-importin complex. Plant J. 2018, 96, 1255–1268. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Paik, I.; Zhu, L.; Huq, E. Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 2015, 20, 641–650. [Google Scholar] [CrossRef]
- Al-Sady, B.; Ni, W.; Kircher, S.; Schafer, E.; Quail, P.H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 2006, 23, 439–446. [Google Scholar] [CrossRef]
- Leivar, P.; Monte, E.; Al-Sady, B.; Carle, C.; Storer, A.; Alonso, J.M.; Ecker, J.R.; Quail, P.H. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 2008, 20, 337–352. [Google Scholar] [CrossRef]
- Park, E.; Kim, Y.; Choi, G. Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell 2018, 30, 1277–1292. [Google Scholar] [CrossRef]
- Favero, D.S. Mechanisms regulating PIF transcription factor activity at the protein level. Physiol. Plant. 2020, 169, 325–335. [Google Scholar] [CrossRef]
- Park, E.; Park, J.; Kim, J.; Nagatani, A.; Lagarias, J.C.; Choi, G. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 2012, 72, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Sheerin, D.J.; Menon, C.; zur Oven-Krockhaus, S.; Enderle, B.; Zhu, L.; Johnen, P.; Schleifenbaum, F.; Stierhof, Y.-D.; Huq, E.; Hiltbrunner, A. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 2015, 27, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.-D.; Zhou, C.-M.; Xu, P.-B.; Luo, Q.; Lian, H.-L.; Yang, H.-Q. Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 2015, 8, 467–478. [Google Scholar] [CrossRef]
- Pacin, M.; Legris, M.; Casal, J.J. Rapid decline in nuclear costitutive photomorphogenesis1 abundance anticipates the stabilization of its target elongated hypocotyl5 in the light. Plant Physiol. 2014, 164, 1134–1138. [Google Scholar] [CrossRef]
- Subramanian, C.; Kim, B.-H.; Lyssenko, N.N.; Xu, X.; Johnson, C.H.; von Arnim, A.G. The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: Mutational analysis by bioluminescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 2004, 101, 6798–6802. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Huang, X.; Deng, X.W. The photomorphogenic central repressor COP1: Conservation and functional diversification during evolution. Plant Commun. 2020, 1, 100044. [Google Scholar] [CrossRef]
- Seo, H.S.; Yang, J.-Y.; Ishikawa, M.; Bolle, C.; Ballesteros, M.L.; Chua, N.-H. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 2003, 423, 995–999. [Google Scholar] [CrossRef]
- Kathare, P.K.; Xu, X.; Nguyen, A.; Huq, E. A COP1-PIF-HEC regulatory module fine-tunes photomorphogenesis in Arabidopsis. Plant J. 2020, 104, 113–123. [Google Scholar] [CrossRef]
- Lin, F.; Jiang, Y.; Li, J.; Yan, T.; Fan, L.; Liang, J.; Chen, Z.J.; Xu, D.; Deng, X.W. B-BOX DOMAIN PROTEIN28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 2018, 30, 2006–2019. [Google Scholar] [CrossRef]
- Holm, M.; Ma, L.-G.; Qu, L.J.; Deng, X.-W. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002, 16, 1247–1259. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, Y.; Li, J.; Lin, F.; Holm, M.; Deng, X.W. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655–7660. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lin, R.; Sullivan, J.; Hoecker, U.; Liu, B.; Xu, L.; Deng, X.W.; Wang, H. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 2005, 17, 804–821. [Google Scholar] [CrossRef]
- Jang, I.-C.; Yang, J.-Y.; Seo, H.S.; Chua, N.-H. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 2005, 19, 593–602. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Bae, G.; Choi, G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Park, E.; Song, K.; Bae, G.; Choi, G. PHYTOCHROME INTERACTING FACTOR8 inhibits phytochrome A-mediated far-red light responses in Arabidopsis. Plant Cell 2020, 32, 186–205. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and phytochrome interacting factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef]
- Paik, I.; Kathare, P.K.; Kim, J.-I.; Huq, E. Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 2017, 10, 1035–1046. [Google Scholar] [CrossRef]
- Oh, E.; Kim, J.; Park, E.; Kim, J.-I.; Kang, C.; Choi, G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004, 16, 3045–3058. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, X.; Sun, J.; Mao, T. Coordinated regulation of hypocotyl cell elongation by light and ethylene through a microtubule destabilizing protein. Plant Physiol. 2018, 176, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yi, H.; Choi, G.; Shin, B.; Song, P.-S.; Choi, G. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 2003, 15, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Legris, M.; Klose, C.; Burgie, E.S.; Rojas, C.C.; Neme, M.; Hiltbrunner, A.; Wigge, P.A.; Schafer, E.; Vierstra, R.D.; Casal, J.J. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 2016, 354, 897–900. [Google Scholar] [CrossRef]
- Fiorucci, A.-S.; Galvao, V.C.; Ince, Y.C.; Boccaccini, A.; Goyal, A.; Allenbach Petrolati, L.; Trevisan, M.; Fankhauser, C. PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytol. 2020, 226, 50–58. [Google Scholar] [CrossRef]
- Delker, C.; Quint, M.; Wigge, P.A. Recent advances in understanding thermomorphogenesis signaling. Curr. Opin. Plant Biol. 2022, 68, 102231. [Google Scholar] [CrossRef]
- Lee, N.; Choi, G. Phytochrome-interacting factor from Arabidopsis to liverwort. Curr. Opin. Plant Biol. 2017, 35, 54–60. [Google Scholar] [CrossRef]
- Huq, E.; Quail, P.H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002, 21, 2441–2450. [Google Scholar] [CrossRef]
- Huq, E.; Al-Sady, B.; Hudson, M.; Kim, C.; Apel, K.; Quail, P.H. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 2004, 305, 1937–1941. [Google Scholar] [CrossRef]
- Kim, K.; Shin, J.; Lee, S.-H.; Kweon, H.-S.; Maloof, J.N.; Choi, G. Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2011, 108, 1729–1734. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Liu, D.; Abdul, W.; Upreti, S.; Liu, Y.; Song, G.; Wu, J.; Liu, B.; Gan, Y. PIL5 represses floral transition in Arabidopsis under long day conditions. Biochem. Biophys. Res. Commun. 2018, 499, 513–518. [Google Scholar] [CrossRef]
- Roig-Villanova, I.; Bou, J.; Sorin, C.; Devlin, P.F.; Martinez-Garcia, J.F. Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 2006, 141, 85–96. [Google Scholar] [CrossRef]
- Salter, M.G.; Franklin, K.A.; Whitelam, G.C. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 2003, 426, 680–683. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Lian, H.-L.; He, S.-B.; Li, L.; Jia, K.-P.; Yang, H.-Q. COP1 and phyB physically Interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 2014, 26, 2441–2456. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Pedmale, U.V.; Nito, K.; Fu, W.; Lin, L.; Hazen, S.P.; Chory, J. PIL1 participates in a negative feedback loop that regulates its own gene expression in response to shade. Mol. Plant 2014, 7, 1582–1585. [Google Scholar] [CrossRef]
- Liu, X.; Chen, C.Y.; Wang, K.-C.; Luo, M.; Tai, R.; Yuan, L.; Zhao, M.; Yang, S.; Tian, G.; Cui, Y.; et al. PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 2013, 25, 1258–1273. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, P.G.; Fankhauser, C.; Terry, M.J. PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 2009, 106, 7654–7659. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z.; et al. Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef]
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef]
- Lorrain, S.; Allen, T.; Duek, P.D.; Whitelam, G.C.; Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008, 53, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef]
- Xie, Y.; Liu, Y.; Wang, H.; Ma, X.; Wang, B.; Wu, G.; Wang, H. Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 2017, 8, 348. [Google Scholar] [CrossRef]
- Sun, J.; Qi, L.; Li, Y.; Zhai, Q.; Li, C. PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 2013, 25, 2102–2114. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.-Y.; Kim, J.; Paek, N.-C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef]
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787. [Google Scholar] [CrossRef]
- Li, N.; Bo, C.; Zhang, Y.; Wang, L. PHYTOCHROME INTERACTING FACTORS PIF4 and PIF5 promote heat stress induced leaf senescence in Arabidopsis. J. Exp. Bot. 2021, 72, 4577–4589. [Google Scholar] [CrossRef] [PubMed]
- Koini, M.A.; Alvey, L.; Allen, T.; Tilley, C.A.; Harberd, N.P.; Whitelam, G.C.; Franklin, K.A. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 2009, 19, 408–413. [Google Scholar] [CrossRef]
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alos, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liang, T.; Liu, H. How plants coordinate their development in response to light and temperature signals. Plant Cell 2022, 34, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Wang, J.; Li, P.; Zhao, C.; Chen, Y.; Bi, Y. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci. 2015, 238, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Z.; Chen, Y.; He, J.X.; Bi, Y. PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci. 2015, 237, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Fujimori, T.; Yamashino, T.; Kato, T.; Mizuno, T. Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Nozue, K.; Covington, M.F.; Duek, P.D.; Lorrain, S.; Fankhauser, C.; Harmer, S.L.; Maloof, J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007, 448, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Josse, E.-M.; Halliday, K.J. A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol. Biol. 2010, 73, 89–95. [Google Scholar] [CrossRef]
- Yang, C.; Huang, S.; Zeng, Y.; Liu, C.; Ma, Q.; Pruneda-Paz, J.; Kay, S.A.; Li, L. Two bHLH transcription factors, bHLH48 and bHLH60, associate with phytochrome interacting factor 7 to regulate hypocotyl elongation in Arabidopsis. Cell Rep. 2021, 35, 109054. [Google Scholar] [CrossRef] [PubMed]
- Kidokoro, S.; Maruyama, K.; Nakashima, K.; Imura, Y.; Narusaka, Y.; Shinwari, Z.K.; Osakabe, Y.; Fujita, Y.; Mizoi, J.; Shinozaki, K.; et al. The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 2009, 151, 2046–2057. [Google Scholar] [CrossRef]
- Lee, C.-M.; Thomashow, M.F. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 15054–15059. [Google Scholar] [CrossRef]
- Mizuno, T.; Oka, H.; Yoshimura, F.; Ishida, K.; Yamashino, T. Insight into the mechanism of end-of-day far-red light (EODFR)-induced shade avoidance responses in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2015, 79, 1987–1994. [Google Scholar] [CrossRef]
- Li, L.; Ljung, K.; Breton, G.; Schmitz, R.J.; Pruneda-Paz, J.; Cowing-Zitron, C.; Cole, B.J.; Ivans, L.J.; Pedmale, U.V.; Jung, H.S.; et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012, 26, 785–790. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, Q.; Jiang, Y.; Yang, C.; Wang, Q.; Li, L. Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. eLife 2018, 7, e31636. [Google Scholar] [CrossRef]
- Burko, Y.; Willige, B.C.; Seluzicki, A.; Novak, O.; Ljung, K.; Chory, J. PIF7 is a master regulator of thermomorphogenesis in shade. Nat. Commun. 2022, 13, 4942. [Google Scholar] [CrossRef]
- Hunt, R.E.; Pratt, L.H. Partial characterization of undegraded oat phytochrome. Biochemistry 1980, 19, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Burgie, E.S.; Gannam, Z.T.K.; Li, H.; Vierstra, R.D. Plant phytochrome B is an asymmetric dimer with unique signalling potential. Nature 2022, 604, 127–133. [Google Scholar] [CrossRef]
- von Horsten, S.; Strass, S.; Hellwig, N.; Gruth, V.; Klasen, R.; Mielcarek, A.; Linne, U.; Morgner, N.; Essen, L.-O. Mapping light-driven conformational changes within the photosensory module of plant phytochrome B. Sci. Rep. 2016, 6, 34366. [Google Scholar] [CrossRef] [PubMed]
- Burgie, E.S.; Bussell, A.N.; Walker, J.M.; Dubiel, K.; Vierstra, R.D. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. USA 2014, 111, 10179–10184. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Mroginski, M.A.; Lang, C.; Kopycki, J.; Gartner, W.; Matysik, J.; Hughes, J. 3D Structures of plant phytochrome A as Pr and Pfr from solid-state NMR: Implications for molecular function. Front. Plant Sci. 2018, 9, 498. [Google Scholar] [CrossRef]
- Cherry, J.R.; Hondred, D.; Walker, J.M.; Vierstra, R.D. Phytochrome requires the 6-kDa N-terminal domain for full biological activity. Proc. Natl. Acad. Sci. USA 1992, 89, 5039–5043. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Lagarias, J.C. Phytochrome evolution in 3D: Deletion, duplication, and diversification. New Phytol. 2020, 225, 2283–2300. [Google Scholar] [CrossRef]
- Qiu, Y.; Pasoreck, E.K.; Reddy, A.K.; Nagatani, A.; Ma, W.; Chory, J.; Chen, M. Mechanism of early light signaling by the carboxy-terminal output module of Arabidopsis phytochrome B. Nat. Commun. 2017, 8, 1905. [Google Scholar] [CrossRef]
- Wong, Y.S.; Cheng, H.C.; Walsh, D.A.; Lagarias, J.C. Phosphorylation of Avena phytochrome in vitro as a probe of light-induced conformational changes. J. Biol. Chem. 1986, 261, 12089–12097. [Google Scholar] [CrossRef]
- Biermann, B.J.; Pao, L.I.; Feldman, L.J. Pr-specific phytochrome phosphorylation in vitro by a protein kinase present in anti-phytochrome maize immunoprecipitates. Plant Physiol. 1994, 105, 243–251. [Google Scholar] [CrossRef]
- Lapko, V.N.; Jiang, X.Y.; Smith, D.L.; Song, P.S. Mass spectrometric characterization of oat phytochrome A: Isoforms and posttranslational modifications. Protein Sci. 1999, 8, 1032–1044. [Google Scholar] [CrossRef]
- Kim, J.-I.; Shen, Y.; Han, Y.-J.; Park, J.-E.; Kirchenbauer, D.; Soh, M.S.; Nagy, F.; Schafer, E.; Song, P.-S. Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 2004, 16, 2629–2640. [Google Scholar] [CrossRef]
- Choi, G.; Yi, H.; Lee, J.; Kwon, Y.K.; Soh, M.S.; Shin, B.; Luka, Z.; Hahn, T.-R.; Song, P.-S. Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 1999, 401, 610–613. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, L.; Duan, J.; Cheng, J.; Shen, Y.; Wang, X.; Han, R.; Li, H.; Li, Z.; Wang, L.; et al. Hinge region of Arabidopsis phyA plays an important role in regulating phyA function. Proc. Natl. Acad. Sci. USA 2018, 115, E11864–E11873. [Google Scholar] [CrossRef]
- Han, Y.-J.; Kim, H.-S.; Kim, Y.-M.; Shin, A.-Y.; Lee, S.-S.; Bhoo, S.H.; Song, P.-S.; Kim, J.-I. Functional characterization of phytochrome autophosphorylation in plant light signaling. Plant Cell Physiol. 2010, 51, 596–609. [Google Scholar] [CrossRef] [PubMed]
- Stockhaus, J.; Nagatani, A.; Halfter, U.; Kay, S.; Furuya, M.; Chua, N.H. Serine-to-alanine substitutions at the amino-terminal region of phytochrome A result in an increase in biological activity. Genes Dev. 1992, 6, 2364–2372. [Google Scholar] [CrossRef] [PubMed]
- Arshavsky, V.Y. Rhodopsin phosphorylation: From terminating single photon responses to photoreceptor dark adaptation. Trends Neurosci. 2002, 25, 124–126. [Google Scholar] [CrossRef]
- Eichenberg, K.; Kunkel, T.; Kretsch, T.; Speth, V.; Schafer, E. In vivo characterization of chimeric phytochromes in yeast. J. Biol. Chem. 1999, 274, 354–359. [Google Scholar] [CrossRef]
- Clough, R.C.; Jordan-Beebe, E.T.; Lohman, K.N.; Marita, J.M.; Walker, J.M.; Gatz, C.; Vierstra, R.D. Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J. 1999, 17, 155–167. [Google Scholar] [CrossRef]
- Debrieux, D.; Fankhauser, C. Light-induced degradation of phyA is promoted by transfer of the photoreceptor into the nucleus. Plant Mol. Biol. 2010, 73, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Allen, T.; Whitelam, G.C. Phytochrome A is an irradiance-dependent red light sensor. Plant J. 2007, 50, 108–117. [Google Scholar] [CrossRef]
- Seo, H.S.; Watanabe, E.; Tokutomi, S.; Nagatani, A.; Chua, N.-H. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004, 18, 617–622. [Google Scholar] [CrossRef]
- Saijo, Y.; Zhu, D.; Li, J.; Rubio, V.; Zhou, Z.; Shen, Y.; Hoecker, U.; Wang, H.; Deng, X.W. Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol. Cell 2008, 31, 607–613. [Google Scholar] [CrossRef]
- Medzihradszky, M.; Bindics, J.; Adam, E.; Viczian, A.; Klement, E.; Lorrain, S.; Gyula, P.; Merai, Z.; Fankhauser, C.; Medzihradszky, K.F.; et al. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 2013, 25, 535–544. [Google Scholar] [CrossRef]
- Nito, K.; Wong, C.C.; Yates, J.R., 3rd; Chory, J. Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Rep. 2013, 3, 1970–1979. [Google Scholar] [CrossRef] [PubMed]
- Viczian, A.; Adam, E.; Staudt, A.M.; Lambert, D.; Klement, E.; Romero Montepaone, S.; Hiltbrunner, A.; Casal, J.; Schafer, E.; Nagy, F.; et al. Differential phosphorylation of the N-terminal extension regulates phytochrome B signaling. New Phytol. 2020, 225, 1635–1650. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Kang, J.-G.; Yang, S.-S.; Chung, K.-S.; Song, P.-S.; Park, C.-M. A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 2002, 14, 3043–3056. [Google Scholar] [CrossRef]
- Phee, B.-K.; Kim, J.-I.; Shin, D.H.; Yoo, J.; Park, K.-J.; Han, Y.-J.; Kwon, Y.-K.; Cho, M.-H.; Jeon, J.-S.; Bhoo, S.H.; et al. A novel protein phosphatase indirectly regulates phytochrome-interacting factor 3 via phytochrome. Biochem. J. 2008, 415, 247–255. [Google Scholar] [CrossRef]
- Ryu, J.S.; Kim, J.-I.; Kunkel, T.; Kim, B.C.; Cho, D.S.; Hong, S.-H.; Kim, S.H.; Fernandez, A.P.; Kim, Y.; Alonso, J.M.; et al. Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 2005, 120, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Boylan, M.T.; Quail, P.H. Are the phytochromes protein kinases? Protoplasma 1996, 195, 12–17. [Google Scholar] [CrossRef]
- Thummler, F.; Algarra, P.; Fobo, G.M. Sequence similarities of phytochrome to protein-kinases - implication for the structure, function and evolution of the phytochrome gene family. FEBS. Lett. 1995, 357, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-I.; Park, J.E.; Zarate, X.; Song, P.-S. Phytochrome phosphorylation in plant light signaling. Photoch. Photobio. Sci. 2005, 4, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Yeh, K.C.; Wu, S.H.; Murphy, J.T.; Lagarias, J.C. A cyanobacterial phytochrome two-component light sensory system. Science 1997, 277, 1505–1508. [Google Scholar] [CrossRef]
- Yeh, K.C.; Lagarias, J.C. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 1998, 95, 13976–13981. [Google Scholar] [CrossRef]
- Shin, A.-Y.; Han, Y.-J.; Baek, A.; Ahn, T.; Kim, S.Y.; Nguyen, T.S.; Son, M.; Lee, K.W.; Shen, Y.; Song, P.-S.; et al. Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 2016, 7, 11545. [Google Scholar] [CrossRef]
- Wong, Y.S.; Lagarias, J.C. Affinity labeling of Avena phytochrome with ATP analogs. Proc. Natl. Acad. Sci. USA 1989, 86, 3469–3473. [Google Scholar] [CrossRef]
- Essen, L.-O.; Mailliet, J.; Hughes, J. The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. USA 2008, 105, 14709–14714. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Jarillo, J.A.; Smirnova, O.; Cashmore, A.R. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1998, 1, 939–948. [Google Scholar] [CrossRef]
- Colon-Carmona, A.; Chen, D.L.; Yeh, K.C.; Abel, S. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 2000, 124, 1728–1738. [Google Scholar] [CrossRef]
- Fankhauser, C.; Yeh, K.C.; Lagarias, J.C.; Zhang, H.; Elich, T.D.; Chory, J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 1999, 284, 1539–1541. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, Z.; Feng, S.; Li, J.; Tan-Wilson, A.; Qu, L.J.; Wang, H.; Deng, X.W. Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 2009, 21, 494–506. [Google Scholar] [CrossRef]
- Shen, H.; Zhu, L.; Castillon, A.; Majee, M.; Downie, B.; Huq, E. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 2008, 20, 1586–1602. [Google Scholar] [CrossRef]
- Shen, Y.; Khanna, R.; Carle, C.M.; Quail, P.H. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007, 145, 1043–1051. [Google Scholar] [CrossRef]
- Hoang, Q.T.N.; Cho, J.-Y.; Choi, D.-M.; Shin, A.-Y.; Kim, J.A.; Han, Y.-J.; Kim, J.-I. Protein kinase activity of phytochrome A positively correlates with photoresponses in Arabidopsis. Front. Plant Sci. 2021, 12, 706316. [Google Scholar] [CrossRef] [PubMed]
- Bernardo-Garcia, S.; de Lucas, M.; Martinez, C.; Espinosa-Ruiz, A.; Daviere, J.M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Zhu, L.; Dennis, M.D.; Yu, L.; Lu, S.X.; Person, M.D.; Tobin, E.M.; Browning, K.S.; Huq, E. Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. J. Biol. Chem. 2011, 286, 12066–12074. [Google Scholar] [CrossRef]
- Ni, W.; Xu, S.L.; Gonzalez-Grandio, E.; Chalkley, R.J.; Huhmer, A.F.R.; Burlingame, A.L.; Wang, Z.-Y.; Quail, P.H. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 2017, 8, 15236. [Google Scholar] [CrossRef] [PubMed]
- Paik, I.; Chen, F.; Ngoc Pham, V.; Zhu, L.; Kim, J.-I.; Huq, E. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun. 2019, 10, 4216. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.-J.; Li, J.; Zhu, D.; Deng, X.W. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. USA 2017, 114, 3539–3544. [Google Scholar] [CrossRef]
- Chen, F.; Shi, X.; Chen, L.; Dai, M.; Zhou, Z.; Shen, Y.; Li, J.; Li, G.; Wei, N.; Deng, X.W. Phosphorylation of FAR-RED ELONGATED HYPOCOTYL1 is a key mechanism defining signaling dynamics of phytochrome A under red and far-red light in Arabidopsis. Plant Cell 2012, 24, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Xu, D.; Jiang, Y.; Chen, H.; Fan, L.; Holm, M.; Deng, X.W. Phosphorylation and negative regulation of CONSTITUTIVELY PHOTOMORPHOGENIC 1 by PINOID in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 6617–6622. [Google Scholar] [CrossRef]
- Tepperman, J.M.; Hwang, Y.-S.; Quail, P.H. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 2006, 48, 728–742. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, B.S.; Li, G.; Charron, J.-B.; Dai, M.Q.; Shi, X.R.; Deng, X.W. Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell 2014, 26, 1949–1966. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Lin, R. Transcriptional regulatory network of the light signaling pathways. New Phytol. 2020, 227, 683–697. [Google Scholar] [CrossRef]
- Pfeiffer, A.; Shi, H.; Tepperman, J.M.; Zhang, Y.; Quail, P.H. Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 2014, 7, 1598–1618. [Google Scholar] [CrossRef]
- Burko, Y.; Seluzicki, A.; Zander, M.; Pedmale, U.V.; Ecker, J.R.; Chory, J. Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell 2020, 32, 967–983. [Google Scholar] [CrossRef]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.; Zhou, Y.; Wang, X.; Li, H.; Feng, Z.; Chen, H.; Qin, G.; Jin, D.; Terzaghi, W.; et al. TANDEM ZINC-FINGER/PLUS3 is a key component of phytochrome A signaling. Plant Cell 2018, 30, 835–852. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Qi, L.; Zhang, S.; Dong, X.; Jing, Y.; Cheng, J.; Feng, Z.; Peng, J.; Li, H.; Zhou, Y.; et al. Mutual upregulation of HY5 and TZP in mediating phytochrome A signaling. Plant Cell 2022, 34, 633–654. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, D.-M.; Kim, S.-H.; Han, Y.-J.; Kim, J.-I. Regulation of Plant Photoresponses by Protein Kinase Activity of Phytochrome A. Int. J. Mol. Sci. 2023, 24, 2110. https://doi.org/10.3390/ijms24032110
Choi D-M, Kim S-H, Han Y-J, Kim J-I. Regulation of Plant Photoresponses by Protein Kinase Activity of Phytochrome A. International Journal of Molecular Sciences. 2023; 24(3):2110. https://doi.org/10.3390/ijms24032110
Chicago/Turabian StyleChoi, Da-Min, Seong-Hyeon Kim, Yun-Jeong Han, and Jeong-Il Kim. 2023. "Regulation of Plant Photoresponses by Protein Kinase Activity of Phytochrome A" International Journal of Molecular Sciences 24, no. 3: 2110. https://doi.org/10.3390/ijms24032110
APA StyleChoi, D.-M., Kim, S.-H., Han, Y.-J., & Kim, J.-I. (2023). Regulation of Plant Photoresponses by Protein Kinase Activity of Phytochrome A. International Journal of Molecular Sciences, 24(3), 2110. https://doi.org/10.3390/ijms24032110






