Gut Microbiome Proteomics in Food Allergies
Abstract
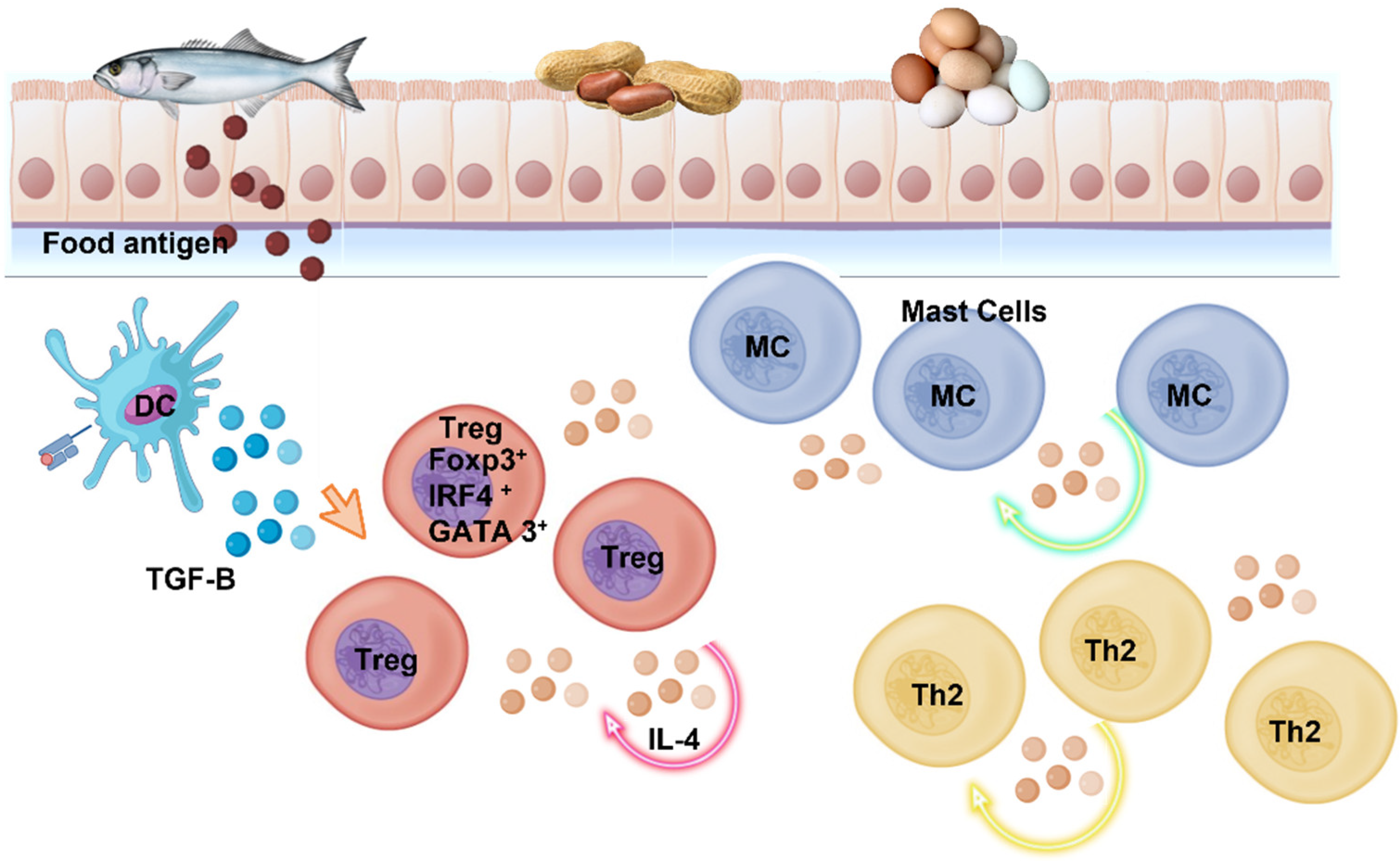
:1. Imbalances in the Human GI Tract Microbial Ecosystem and Their Association with Food Allergies
2. Allergic Reactions in the GI
3. Proteomics to Study Food Allergies
3.1. Allergy Proteomics and Microorganism
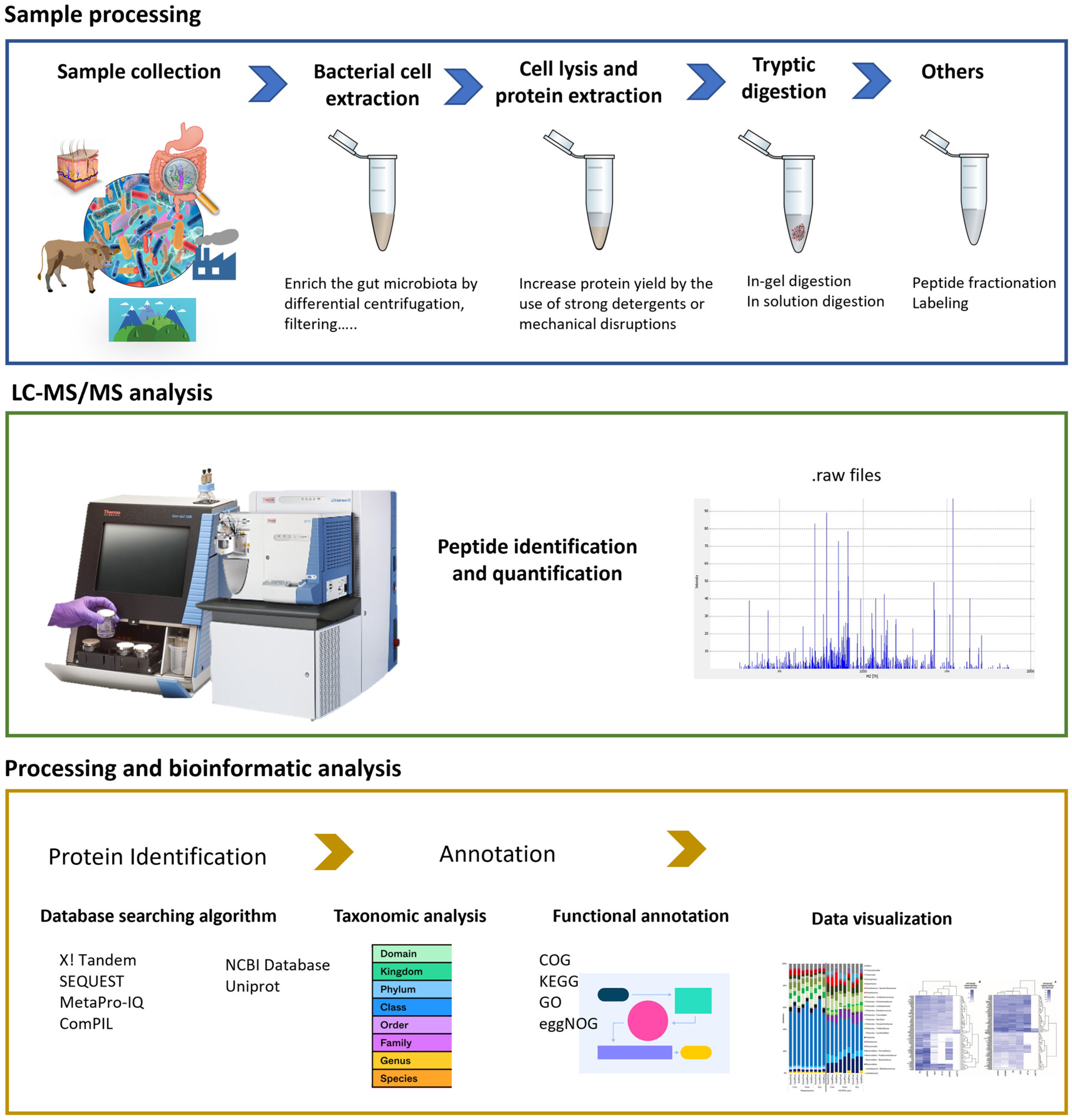
Metaproteomics
4. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stephen-Victor, E.; Chatila, T.A. Regulation of oral immune tolerance by the microbiome in food allergy. Curr. Opin. Immunol. 2019, 60, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Salameh, M.; Burney, Z.; Mhaimeed, N.; Laswi, I.; Yousri, N.A.; Bendriss, G.; Zakaria, D. The role of gut microbiota in atopic asthma and allergy, implications in the understanding of disease pathogenesis. Scand. J. Immunol. 2020, 91, e12855. [Google Scholar] [CrossRef] [PubMed]
- Di Costanzo, M.; Paparo, L.; Cosenza, L.; Di Scala, C.; Nocerino, R.; Aitoro, R.; Canani, R.B. Food allergies: Novel mechanisms and therapeutic perspectives. Methods Mol. Biol. 2016, 1371, 215–221. [Google Scholar] [PubMed]
- Zubeldia-Varela, E.; Barker-Tejeda, T.C.; Obeso, D.; Villaseñor, A.; Barber, D.; Pérez-Gordo, M. Microbiome and Allergy: New Insights and Perspectives. J. Investig. Allergol. Clin. Immunol. 2022, 32, 327–344. [Google Scholar] [CrossRef] [PubMed]
- Bunyavanich, S.; Berin, M.C. Food allergy and the microbiome: Current understandings and future directions. J. Allergy Clin. Immunol. 2019, 144, 1468–1477. [Google Scholar] [CrossRef]
- Burks, A.W.; Tang, M.; Sicherer, S.; Muraro, A.; Eigenmann, P.A.; Ebisawa, M.; Sampson, H.A. ICON: Food allergy. J. Allergy Clin. Immunol. 2012, 129, 906–920. [Google Scholar] [CrossRef]
- Suzuki, T.A.; Fitzstevens, J.L.; Schmidt, V.T.; Enav, H.; Huus, K.E.; Mbong Ngwese, M.; Ley, R.E. Codiversification of gut microbiota with humans. Science 2022, 377, 1328–1332. [Google Scholar] [CrossRef]
- Larsen, O.F.A.; Claassen, E.; Brummer, R.J. On the importance of intraindividual variation in nutritional research. Benef. Microbes 2020, 11, 511–517. [Google Scholar] [CrossRef]
- Rachid, R.; Stephen-Victor, E.; Chatila, T.A. The microbial origins of food allergy. J. Allergy Clin. Immunol. 2021, 147, 808–813. [Google Scholar] [CrossRef]
- Goguyer-Deschaumes, R.; Waeckel, L.; Killian, M.; Rochereau, N.; Paul, S. Metabolites and secretory immunoglobulins: Messengers and effectors of the host–microbiota intestinal equilibrium. Trends Immunol. 2022, 43, 63–77. [Google Scholar] [CrossRef]
- Berni Canani, R.; De Filippis, F.; Nocerino, R.; Paparo, L.; Di Scala, C.; Cosenza, L.; Della Gatta, G.; Calignano, A.; De Caro, C.; Laiola, M.; et al. Gut microbiota composition and butyrate production in children affected by non-IgE-mediated cow’s milk allergy. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Gadir, A.; Stephen-Victor, E.; Gerber, G.K.; Noval Rivas, M.; Wang, S.; Harb, H.; Wang, L.; Li, N.; Crestani, E.; Spielman, S.; et al. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat. Med. 2019, 25, 1164–1174. [Google Scholar] [CrossRef]
- Bao, R.; Hesser, L.A.; He, Z.; Zhou, X.; Nadeau, K.C.; Nagler, C.R. Fecal microbiome and metabolome differ in healthy and food-allergic twins. J. Clin. Invest. 2021, 131, e141935. [Google Scholar] [CrossRef]
- Rodriguez, B.; Prioult, G.; Hacini-Rachinel, F.; Moine, D.; Bruttin, A.; Ngom-Bru, C.; Labellie, C.; Nicolis, I.; Berger, B.; Mercenier, A.; et al. Infant gut microbiota is protective against cow’s milk allergy in mice despite immature ileal T-cell response. FEMS Microbiol. Ecol. 2012, 79, 192–202. [Google Scholar] [CrossRef]
- Bunyavanich, S.; Shen, N.; Grishin, A.; Wood, R.; Burks, W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.; Sicherer, S.; et al. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 2016, 138, 1122–1130. [Google Scholar] [CrossRef] [Green Version]
- Savage, J.H.; Lee-Sarwar, K.A.; Sordillo, J.; Bunyavanich, S.; Zhou, Y.; O’Connor, G.; Sandel, M.; Bacharier, L.B.; Zeiger, R.; Sodergren, E.; et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2018, 73, 145–152. [Google Scholar] [CrossRef]
- Azad, M.B.; Konya, T.; Guttman, D.S.; Field, C.J.; Sears, M.R.; Hayglass, K.T.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Becker, A.B.; et al. Infant gut microbiota and food sensitization: Associations in the first year of life. Clin. Exp. Allergy 2015, 45, 632–643. [Google Scholar] [CrossRef]
- Hua, X.; Goedert, J.J.; Pu, A.; Yu, G.; Shi, J. Allergy associations with the adult fecal microbiota: Analysis of the American Gut Project. EBioMedicine 2016, 3, 172–179. [Google Scholar] [CrossRef] [Green Version]
- Inoue, R.; Sawai, T.; Sawai, C.; Nakatani, M.; Romero-Pérez, G.A.; Ozeki, M.; Nonomura, K.; Tsukahara, T. A preliminary study of gut dysbiosis in children with food allergy. Biosci. Biotechnol. Biochem. 2017, 81, 2396–2399. [Google Scholar] [CrossRef] [Green Version]
- Mills, R.H.; Vázquez-Baeza, Y.; Zhu, Q.; Jiang, L.; Gaffney, J.; Humphrey, G.; Smarr, L.; Knight, R.; Gonzalez, D.J. Evaluating Metagenomic Prediction of the Metaproteome in a 4.5-Year Study of a Patient with Crohn’s Disease. mSystems 2019, 4, e00337-18. [Google Scholar] [CrossRef]
- Fazlollahi, M.; Chun, Y.; Grishin, A.; Wood, R.A.; Burks, A.W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.A.; Sicherer, S.H.; et al. Early-life gut microbiome and egg allergy. Allergy 2018, 73, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Feng, J.; Yan, D.Y.; Lyu, Y.J.; Xu, X. Early-life gut microbiome and cow’s milk allergy—A prospective case—Control 6-month follow-up study. Saudi J. Biol. Sci. 2018, 25, 875–880. [Google Scholar] [CrossRef] [PubMed]
- Díaz, M.; Guadamuro, L.; Espinosa-Martos, I.; Mancabelli, L.; Jiménez, S.; Molinos-Norniella, C.; Pérez-Solis, D.; Milani, C.; Rodríguez, J.M.; Ventura, M.; et al. Microbiota and Derived Parameters in Fecal Samples of Infants with Non-IgE Cow’s Milk Protein Allergy under a Restricted Diet. Nutrients 2018, 10, 1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kourosh, A.; Luna, R.A.; Balderas, M.; Nance, C.; Anagnostou, A.; Devaraj, S.; Davis, C.M. Fecal microbiome signatures are different in food-allergic children compared to siblings and healthy children. Pediatr. Allergy Immunol. 2018, 29, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Ho, H.; Bunyavanich, S. The gut microbiome in food allergy. Ann. Allergy Asthma Immunol. 2019, 122, 276–282. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Charbonnier, L.M.; Noval Rivas, M.; Georgiev, P.; Li, N.; Gerber, G.; Bry, L.; Chatila, T.A. MyD88 Adaptor-Dependent Microbial Sensing by Regulatory T Cells Promotes Mucosal Tolerance and Enforces Commensalism. Immunity 2015, 43, 289–303. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic T reg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.H.; Wu, C.C.; Tyan, Y.C.; Yu, W.T.; Huang, E.S.; Yu, H.S. Identification of pyruvate kinase as a novel allergen in whiteleg shrimp (Litopenaeus vannamei) by specific-IgE present in patients with shrimp allergy. Food Chem. 2018, 258, 359–365. [Google Scholar] [CrossRef]
- Georgiev, P.; Charbonnier, L.M.; Chatila, T.A. Regulatory T Cells: The Many Faces of Foxp3. J. Clin. Immunol. 2019, 39, 623–640. [Google Scholar] [CrossRef]
- Sharma, A.; Rudra, D. Emerging functions of regulatory T cells in tissue homeostasis. Front. Immunol. 2018, 9, 883. [Google Scholar] [CrossRef]
- Mayorga, C.; Palomares, F.; Cañas, J.A.; Pérez-Sánchez, N.; Núñez, R.; Torres, M.J.; Gómez, F. New Insights in Therapy for Food Allergy. Foods 2021, 10, 1037. [Google Scholar] [CrossRef]
- NovalRivas, M.; Burton, O.T.; Wise, P.; Charbonnier, L.M.; Georgiev, P.; Oettgen, H.C.; Rachid, R.; Chatila, T.A. Regulatory T Cell Reprogramming toward a Th2-Cell-like Lineage Impairs Oral Tolerance and Promotes Food Allergy. Immunity 2015, 42, 512–523. [Google Scholar] [CrossRef] [Green Version]
- Marrs, T.; Sim, K. Demystifying Dysbiosis: Can the Gut Microbiome Promote Oral Tolerance Over IgE-mediated Food Allergy? Curr. Pediatr. Rev. 2018, 14, 156–163. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [Green Version]
- Holmstrøm, K.; Collins, M.D.; Møller, T.; Falsen, E.; Lawson, P.A. Subdoligranulum variabile gen. nov., sp. nov. from human feces. Anaerobe 2004, 10, 197–203. [Google Scholar] [CrossRef]
- Noval Rivas, M.; Burton, O.T.; Wise, P.; Zhang, Y.Q.; Hobson, S.A.; Garcia Lloret, M.; Chehoud, C.; Kuczynski, J.; Desantis, T.; Warrington, J.; et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immunol. 2013, 131, 201–212. [Google Scholar] [CrossRef]
- Chatila, T.A.; Abdel-Gadir, A.; Massoud, A.H. Antigen-specific Treg cells in immunological tolerance: Implications for allergic diseases. F1000Research 2018, 7, 38. [Google Scholar]
- Albuhairi, S.; Rachid, R. Biologics and Novel Therapies for Food Allergy. Immunol. Allergy Clin. N. Am. 2021, 41, 271–283. [Google Scholar] [CrossRef]
- Khodadoust, M.S.; Olsson, N.; Wagar, L.E.; Haabeth, O.A.W.; Chen, B.; Swaminathan, K.; Rawson, K.; Liu, C.L.; Steiner, D.; Lund, P.; et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 2017, 543, 723–727. [Google Scholar] [CrossRef] [Green Version]
- Kalliomäki, M.; Salminen, S.; Arvilommi, H.; Kero, P.; Koskinen, P.; Isolauri, E. Probiotics in primary prevention of atopic disease: A randomised placebo-controlled trial. Lancet 2001, 357, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Candy, D.C.A.; Van Ampting, M.T.J.; Oude Nijhuis, M.M.; Wopereis, H.; Butt, A.M.; Peroni, D.G.; Vandenplas, Y.; Fox, A.T.; Shah, N.; West, C.E.; et al. A synbiotic-containing amino-acid-based formula improves gut microbiota in non-IgE-mediated allergic infants. Pediatr. Res. 2017, 83, 677–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virkud, Y.V.; Kelly, R.S.; Wood, C.; Lasky-Su, J.A. The nuts and bolts of omics for the clinical allergist. Ann. Allergy Asthma Immunol. 2019, 123, 558–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; Donovan, D.M.; García, P. Enhanced staphylolytic activity of the Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88 HydH5 virion-associated peptidoglycan hydrolase: Fusions, deletions, and synergy with LysH5. Appl. Environ. Microbiol. 2012, 78, 2241–2248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diamant, Z.; Boot, J.D.; Mantzouranis, E.; Flohr, R.; Sterk, P.J.; Gerth van Wijk, R. Biomarkers in asthma and allergic rhinitis. Pulm. Pharmacol. Ther. 2010, 23, 468–481. [Google Scholar] [CrossRef] [Green Version]
- Dhondalay, G.K.; Rael, E.; Acharya, S.; Zhang, W.; Sampath, V.; Galli, S.J.; Tibshirani, R.; Boyd, S.D.; Maecker, H.; Nadeau, K.C.; et al. Food allergy and omics. J. Allergy Clin. Immunol. 2018, 141, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Carrera, M.; Cañas, B.; Gallardo, J.M. Advanced proteomics and systems biology applied to study food allergy. Curr. Opin. Food Sci. 2018, 22, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Piras, C.; Roncada, P.; Rodrigues, P.M.; Bonizzi, L.; Soggiu, A. Proteomics in food: Quality, safety, microbes, and allergens. Proteomics 2016, 16, 799–815. [Google Scholar] [CrossRef]
- Yagami, T.; Haishima, Y.; Tsuchiya, T.; Tomitaka-Yagami, A.; Kano, H.; Matsunaga, K. Proteomic Analysis of Putative Latex Allergens. Int. Arch. Allergy Immunol. 2004, 135, 3–11. [Google Scholar] [CrossRef]
- Nony, E.; Le Mignon, M.; Brier, S.; Martelet, A.; Moingeon, P. Proteomics for Allergy: From Proteins to the Patients. Curr. Allergy Asthma Rep. 2016, 16, 1–9. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; Vázquez, J.; Gallardo, J.M. Extensive de Novo sequencing of new parvalbumin isoforms using a novel combination of bottom-up proteomics, accurate molecular mass measurement by FTICR-MS, and selected MS/MS ion monitoring. J. Proteome Res. 2010, 9, 4393–4406. [Google Scholar] [CrossRef]
- Goliáš, J.; Humlová, Z.; Halada, P.; Hábová, V.; Janatková, I.; Tučková, L. Identification of rice proteins recognized by the IgE antibodies of patients with food allergies. J. Agric. Food Chem. 2013, 61, 8851–8860. [Google Scholar] [CrossRef]
- Hirano, K.; Hino, S.; Oshima, K.; Nadano, D.; Urisu, A.; Takaiwa, F.; Matsuda, T. Evaluation of allergenic potential for rice seed protein components utilizing a rice proteome database and an allergen database in combination with IgE-binding of recombinant proteins. Biosci. Biotechnol. Biochem. 2016, 80, 564–573. [Google Scholar] [CrossRef]
- Bouakkadia, H.; Boutebba, A.; Haddad, I.; Vinh, J.; Guilloux, L.; Sutra, J.P.; Sénéchal, H.; Poncet, P. Analyse immunoprotéomique des allergènes non-hydrosolubles de farines de quatre légumineuses: Arachide, soja, sésame et lentille. Ann. Biol. Clin. 2015, 73, 690–704. [Google Scholar]
- Akagawa, M.; Handoyo, T.; Ishii, T.; Kumazawa, S.; Morita, N.; Suyama, K. Proteomic Analysis of Wheat Flour Allergens. J. Agric. Food Chem. 2007, 55, 6863–6870. [Google Scholar] [CrossRef]
- Marzban, G.; Herndl, A.; Maghuly, F.; Katinger, H.; Laimer, M. Mapping of fruit allergens by 2D electrophoresis and immunodetection. Expert Rev. Proteom. 2014, 5, 61–75. [Google Scholar] [CrossRef]
- Grishina, G.; Bardina, L.; Grishin, A. 2D-electrophoresis and immunoblotting in food allergy. Methods Mol. Biol. 2017, 1592, 59–69. [Google Scholar]
- Odedra, K.M. Milk allergy in adults and children. Nurs. Stand. 2015, 29, 43–48. [Google Scholar] [CrossRef]
- Hettinga, K.A.; Reina, F.M.; Boeren, S.; Zhang, L.; Koppelman, G.H.; Postma, D.S.; Vervoort, J.J.M.; Wijga, A.H. Difference in the Breast Milk Proteome between Allergic and Non-Allergic Mothers. PLoS ONE 2015, 10, e0122234. [Google Scholar] [CrossRef] [Green Version]
- Betzen, C.; Alhamdani, M.S.S.; Lueong, S.; Schröder, C.; Stang, A.; Hoheisel, J.D. Clinical proteomics: Promises, challenges and limitations of affinity arrays. Proteom. Clin. Appl. 2015, 9, 342–347. [Google Scholar] [CrossRef] [Green Version]
- Bavaro, S.L.; De Angelis, E.; Barni, S.; Pilolli, R.; Mori, F.; Novembre, E.M.; Monaci, L. Modulation of Milk Allergenicity by Baking Milk in Foods: A Proteomic Investigation. Nutrients 2019, 11, 1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apostolovic, D.; Tran, T.A.T.; Hamsten, C.; Starkhammar, M.; Cirkovic Velickovic, T.; Van Hage, M. Immunoproteomics of processed beef proteins reveal novel galactose-α-1,3-galactose-containing allergens. Allergy 2014, 69, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- Picariello, G.; Mamone, G.; Cutignano, A.; Fontana, A.; Zurlo, L.; Addeo, F.; Ferranti, P. Proteomics, Peptidomics, and Immunogenic Potential of Wheat Beer (Weissbier). J. Agric. Food Chem. 2015, 63, 3579–3586. [Google Scholar] [CrossRef] [PubMed]
- Tomm, J.M.; Van Do, T.; Jende, C.; Simon, J.C.; Treudler, R.; Von Bergen, M.; Averbeck, M. Identi cation of New Allergens From L niloticus and G morhua Identi cation of New Potential Allergens From Nile Perch (Lates niloticus) and Cod (Gadus morhua). J. Investig. Allergol. Clin. Immunol. 2013, 23, 159–167. [Google Scholar] [PubMed]
- Prodic, I.; Stanic-Vucinic, D.; Apostolovic, D.; Mihailovic, J.; Radibratovic, M.; Radosavljevic, J.; Burazer, L.; Milcic, M.; Smiljanic, K.; van Hage, M.; et al. Influence of peanut matrix on stability of allergens in gastric-simulated digesta: 2S albumins are main contributors to the IgE reactivity of short digestion-resistant peptides. Clin. Exp. Allergy 2018, 48, 731–740. [Google Scholar] [CrossRef] [Green Version]
- Carrera, M.; González-Fernández, Á.; Magadán, S.; Mateos, J.; Pedrós, L.; Medina, I.; Gallardo, J.M. Molecular characterization of B-cell epitopes for the major fish allergen, parvalbumin, by shotgun proteomics, protein-based bioinformatics and IgE-reactive approaches. J. Proteom. 2019, 200, 123–133. [Google Scholar] [CrossRef]
- Ortea, I.; Cañas, B.; Gallardo, J.M. Mass spectrometry characterization of species-specific peptides from arginine kinase for the identification of commercially relevant shrimp species. J. Proteome Res. 2009, 8, 5356–5362. [Google Scholar] [CrossRef]
- Korte, R.; Brockmeyer, J. MRM3-based LC-MS multi-method for the detection and quantification of nut allergens. Anal. Bioanal. Chem. 2016, 408, 7845–7855. [Google Scholar] [CrossRef]
- Martínez-Esteso, M.J.; Nørgaard, J.; Brohée, M.; Haraszi, R.; Maquet, A.; O’Connor, G. Defining the wheat gluten peptide fingerprint via a discovery and targeted proteomics approach. J. Proteom. 2016, 147, 156–168. [Google Scholar] [CrossRef]
- Weber, D.; Cléroux, C.; Godefroy, S.B. Emerging analytical methods to determine gluten markers in processed foods-method development in support of standard setting. Anal. Bioanal. Chem. 2009, 395, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Kim, C.J. Determination of Allergenic Egg Proteins in Food by Protein-, Mass Spectrometry-, and DNA-Based Methods. J. AOAC Int. 2010, 93, 462–477. [Google Scholar] [CrossRef]
- Monaci, L.; Losito, I.; De Angelis, E.; Pilolli, R.; Visconti, A. Multi-allergen quantification of fining-related egg and milk proteins in white wines by high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 2009–2018. [Google Scholar] [CrossRef]
- De Angelis, E.; Pilolli, R.; Monaci, L. Coupling SPE on-line pre-enrichment with HPLC and MS/MS for the sensitive detection of multiple allergens in wine. Food Control. 2017, 73, 814–820. [Google Scholar] [CrossRef]
- Cereda, A.; Kravchuk, A.V.; D’Amato, A.; Bachi, A.; Righetti, P.G. Proteomics of wine additives: Mining for the invisible via combinatorial peptide ligand libraries. J. Proteom. 2010, 73, 1732–1739. [Google Scholar] [CrossRef]
- D’Amato, A.; Kravchuk, A.V.; Bachi, A.; Righetti, P.G. Noah’s nectar: The proteome content of a glass of red wine. J. Proteom. 2010, 73, 2370–2377. [Google Scholar] [CrossRef]
- Ortea, I.; Cañas, B.; Gallardo, J.M. Selected tandem mass spectrometry ion monitoring for the fast identification of seafood species. J. Chromatogr. A 2011, 1218, 4445–4451. [Google Scholar] [CrossRef] [Green Version]
- Pilolli, R.; De Angelis, E.; Monaci, L. Streamlining the analytical workflow for multiplex MS/MS allergen detection in processed foods. Food Chem. 2017, 221, 1747–1753. [Google Scholar] [CrossRef]
- Mattarozzi, M.; Bignardi, C.; Elviri, L.; Careri, M. Rapid shotgun proteomic liquid chromatography-electrospray ionization-tandem mass spectrometry-based method for the lupin (Lupinus albus L.) multi-allergen determination in foods. J. Agric. Food Chem. 2012, 60, 5841–5846. [Google Scholar] [CrossRef]
- Careri, M.; Costa, A.; Elviri, L.; Lagos, J.B.; Mangia, A.; Terenghi, M.; Cereti, A.; Garoffo, L.P. Use of specific peptide biomarkers for quantitative confirmation of hidden allergenic peanut proteins Ara h 2 and Ara h 3/4 for food control by liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2007, 389, 1901–1907. [Google Scholar] [CrossRef]
- Hebling, C.M.; McFarland, M.A.; Callahan, J.H.; Ross, M.M. Global proteomic screening of protein allergens and advanced glycation endproducts in thermally processed peanuts. J. Agric. Food Chem. 2013, 61, 5638–5648. [Google Scholar] [CrossRef]
- Monaci, L.; De Angelis, E.; Bavaro, S.L.; Pilolli, R. High-resolution OrbitrapTM-based mass spectrometry for rapid detection of peanuts in nuts. Food Addit. Contam. Part A 2015, 32, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Chassaigne, H.; Nørgaard, J.V.; Van Hengel, A.J. Proteomics-based approach to detect and identify major allergens in processed peanuts by capillary LC-Q-TOF (MS/MS). J. Agric. Food Chem. 2007, 55, 4461–4473. [Google Scholar] [CrossRef] [PubMed]
- Pedreschi, R.; Nørgaard, J.; Maquet, A. Current Challenges in Detecting Food Allergens by Shotgun and Targeted Proteomic Approaches: A Case Study on Traces of Peanut Allergens in Baked Cookies. Nutrients 2012, 4, 132–150. [Google Scholar] [CrossRef] [PubMed]
- Careri, M.; Elviri, L.; Maffini, M.; Mangia, A.; Mucchino, C.; Terenghi, M. Determination of peanut allergens in cereal-chocolate-based snacks: Metal-tag inductively coupled plasma mass spectrometry immunoassay versus liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2008, 22, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Careri, M.; Elviri, L.; Lagos, J.B.; Mangia, A.; Speroni, F.; Terenghi, M. Selective and rapid immunomagnetic bead-based sample treatment for the liquid chromatography–electrospray ion-trap mass spectrometry detection of Ara h3/4 peanut protein in foods. J. Chromatogr. A 2008, 1206, 89–94. [Google Scholar] [CrossRef]
- Johnson, P.E.; Sayers, R.L.; Gethings, L.A.; Balasundaram, A.; Marsh, J.T.; Langridge, J.I.; Mills, E.N.C. Quantitative Proteomic Profiling of Peanut Allergens in Food Ingredients Used for Oral Food Challenges. Anal. Chem. 2016, 88, 5689–5695. [Google Scholar] [CrossRef] [Green Version]
- Lutter, P.; Parisod, V.; Weymuth, H. Development and Validation of a Method for the Quantification of Milk Proteins in Food Products Based on Liquid Chromatography with Mass Spectrometric Detection. J. AOAC Int. 2011, 94, 1043–1059. [Google Scholar] [CrossRef] [Green Version]
- Monaci, L.; Losito, I.; Palmisano, F.; Visconti, A. Identification of allergenic milk proteins markers in fined white wines by capillary liquid chromatography–electrospray ionization-tandem mass spectrometry. J. Chromatogr. A 2010, 1217, 4300–4305. [Google Scholar] [CrossRef]
- Hong, C.; Jiang, H.; Lü, E.; Wu, Y.; Guo, L.; Xie, Y.; Wang, C.; Yang, Y. Identification of Milk Component in Ancient Food Residue by Proteomics. PLoS ONE 2012, 7, e37053. [Google Scholar] [CrossRef]
- Monaci, L.; Losito, I.; Palmisano, F.; Visconti, A. Reliable Detection of Milk Allergens in Food Using a High-Resolution, Stand-Alone Mass Spectrometer. J. AOAC Int. 2011, 94, 1034–1042. [Google Scholar] [CrossRef] [Green Version]
- Czerwenka, C.; Maier, I.; Potocnik, N.; Pittner, F.; Lindner, W. Absolute Quantitation of β-Lactoglobulin by Protein Liquid Chromatography−Mass Spectrometry and Its Application to Different Milk Products. Anal. Chem. 2007, 79, 5165–5172. [Google Scholar] [CrossRef]
- Houston, N.L.; Lee, D.G.; Stevenson, S.E.; Ladies, G.S.; Bannon, G.A.; McClain, S.; Privalle, L.; Stagg, N.; Herouet-Guieheney, C.; MacIntosh, S.C.; et al. Quantitation of soybean allergens using tandem mass spectrometry. J. Proteome Res. 2011, 10, 763–773. [Google Scholar] [CrossRef]
- Koeberl, M.; Clarke, D.; Lopata, A.L. Next generation of food allergen quantification using mass spectrometric systems. J. Proteome Res. 2014, 13, 3499–3509. [Google Scholar] [CrossRef]
- Monaci, L.; van Hengel, A.J. Development of a method for the quantification of whey allergen traces in mixed-fruit juices based on liquid chromatography with mass spectrometric detection. J. Chromatogr. A 2008, 1192, 113–120. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; Gallardo, J.M. Rapid direct detection of the major fish allergen, parvalbumin, by selected MS/MS ion monitoring mass spectrometry. J. Proteom. 2012, 75, 3211–3220. [Google Scholar] [CrossRef] [Green Version]
- Carrera, M.; Cañas, B.; López-Ferrer, D.; Piñeiro, C.; Vázquez, J.; Gallardo, J.M. Fast monitoring of species-specific peptide biomarkers using high-intensity-focused-ultrasound-assisted tryptic digestion and selected MS/MS ion monitoring. Anal. Chem. 2011, 83, 5688–5695. [Google Scholar] [CrossRef] [Green Version]
- Saelens, G.; Planckaert, S.; Martínez-Sernández, V.; Ubeira, F.M.; Devreese, B.; Gabriël, S. Targeted proteomics and specific immunoassays reveal the presence of shared allergens between the zoonotic nematodes Anisakis simplex and Pseudoterranova decipiens. Sci. Rep. 2022, 12, 1–11. [Google Scholar] [CrossRef]
- Carrera, M.; Gallardo, J.M.; Pascual, S.; González, Á.F.; Medina, I. Protein biomarker discovery and fast monitoring for the identification and detection of Anisakids by parallel reaction monitoring (PRM) mass spectrometry. J. Proteom. 2016, 142, 130–137. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.S.; Han, D.; Kim, J.Y.; Kim, D.W.; Kim, Y.M.; Mo, J.H.; Choi, H.G.; Park, J.W.; Shin, H.W. In-Depth, Proteomic Analysis of Nasal Secretions from Patients With Chronic Rhinosinusitis and Nasal Polyps. Allergy Asthma Immunol. Res. 2019, 11, 691–708. [Google Scholar] [CrossRef]
- Yan, B.; Lou, H.; Wang, Y.; Li, Y.; Meng, Y.; Qi, S.; Wang, M.; Xiao, L.; Wang, C.; Zhang, L. Epithelium-derived cystatin SN enhances eosinophil activation and infiltration through IL-5 in patients with chronic rhinosinusitis with nasal polyps. J. Allergy Clin. Immunol. 2019, 144, 455–469. [Google Scholar] [CrossRef] [Green Version]
- Schofield, J.P.R.; Burg, D.; Nicholas, B.; Strazzeri, F.; Brandsma, J.; Staykova, D.; Folisi, C.; Bansal, A.T.; Xian, Y.; Guo, Y.; et al. Stratification of asthma phenotypes by airway proteomic signatures. J. Allergy Clin. Immunol. 2019, 144, 70–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candreva, Á.M.; Ferrer-Navarro, M.; Bronsoms, S.; Quiroga, A.; Curciarello, R.; Cauerhff, A.; Petruccelli, S.; Docena, G.H.; Trejo, S.A. Identification of cross-reactive B-cell epitopes between Bos d 9.0101(Bos Taurus) and Gly m 5.0101 (Glycine max) by epitope mapping MALDI-TOF MS. Proteomics 2017, 17, 1700069. [Google Scholar] [CrossRef] [PubMed]
- Orrù, S.; Di Nicola, P.; Giuliani, F.; Fabris, C.; Conti, A.; Coscia, A.; Bertino, E. Detection of Bovine Alpha-S1-Casein in Term and Preterm Human Colostrum with Proteomic Techniques. Int. J. Immunopathol. Pharmacol. 2013, 26, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Fisher, D.A.C.; Malkova, O.; Oh, S.T. Analysis of signaling networks at the single-cell level using mass cytometry. Methods Mol. Biol. 2017, 1636, 371–392. [Google Scholar] [PubMed]
- Bendall, S.C.; Simonds, E.F.; Qiu, P.; Amir, E.A.D.; Krutzik, P.O.; Finck, R.; Bruggner, R.V.; Melamed, R.; Trejo, A.; Ornatsky, O.I.; et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011, 332, 687–696. [Google Scholar] [CrossRef] [Green Version]
- Bandura, D.R.; Baranov, V.I.; Ornatsky, O.I.; Antonov, A.; Kinach, R.; Lou, X.; Pavlov, S.; Vorobiev, S.; Dick, J.E.; Tanner, S.D. Mass Cytometry: Technique for Real Time Single Cell Multitarget Immunoassay Based on Inductively Coupled Plasma Time-of-Flight Mass Spectrometry. Anal. Chem. 2009, 81, 6813–6822. [Google Scholar] [CrossRef]
- Hoh, R.A.; Joshi, S.A.; Liu, Y.; Wang, C.; Roskin, K.M.; Lee, J.Y.; Pham, T.; Looney, T.J.; Jackson, K.J.L.; Dixit, V.P.; et al. Single B-cell deconvolution of peanut-specific antibody responses in allergic patients. J. Allergy Clin. Immunol. 2016, 137, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Martino, D.; Allen, K. Meeting the challenges of measuring human immune regulation. J. Immunol. Methods 2015, 424, 1–6. [Google Scholar] [CrossRef]
- Goswami, R.; Blazquez, A.B.; Kosoy, R.; Rahman, A.; Nowak-Węgrzyn, A.; Berin, M.C. Systemic innate immune activation in food protein–induced enterocolitis syndrome. J. Allergy Clin. Immunol. 2017, 139, 1885–1896.e9. [Google Scholar] [CrossRef] [Green Version]
- Frazier, A.; Schulten, V.; Hinz, D.; Oseroff, C.; Sidney, J.; Peters, B.; Sette, A. Allergy-associated T cell epitope repertoires are surprisingly diverse and include non-IgE reactive antigens. World Allergy Organ. J. 2014, 7, 26. [Google Scholar] [CrossRef] [Green Version]
- Li Xu, L.; Wei Zhang, H.; Lin, H.; Mei Zhang, X.; Qi Wen, Y.; Long Zhao, J.; Xing Li, Z.; Gasset, M. SWATH-MS-based proteomics reveals functional biomarkers of Th1/Th2 responses of tropomyosin allergy in mouse models. Food Chem. 2022, 383, 132474. [Google Scholar] [CrossRef]
- Cong, Y.; Motamedchaboki, K.; Misal, S.A.; Liang, Y.; Guise, A.J.; Truong, T.; Huguet, R.; Plowey, E.D.; Zhu, Y.; Lopez-Ferrer, D.; et al. Ultrasensitive single-cell proteomics workflow identifies >1000 protein groups per mammalian cell. Chem. Sci. 2021, 12, 1001–1006. [Google Scholar] [CrossRef]
- Alves, T.O.; D’Almeida, C.T.S.; Victorio, V.C.M.; Souza, G.H.M.F.; Cameron, L.C.; Ferreira, M.S.L. Immunogenic and allergenic profile of wheat flours from different technological qualities revealed by ion mobility mass spectrometry. J. Food Compos. Anal. 2018, 73, 67–75. [Google Scholar] [CrossRef]
- Miron, R.; Hulea, M.; Folea, S. Food Allergens Monitoring System Backed-up by Blockchain Technology. In Proceedings of the 2020 IEEE International Conference on Automation, Quality and Testing, Robotics (AQTR), Cluj-Napoca, Romania, 21–23 May 2020. [Google Scholar]
- Hong, L.; Pan, M.; Xie, X.; Liu, K.; Yang, J.; Wang, S.; Wang, S. Aptamer-Based Fluorescent Biosensor for the Rapid and Sensitive Detection of Allergens in Food Matrices. Foods 2021, 10, 2598. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Q.; Sun, M.; Zhang, J.; Mo, S.; Wang, J.; Wei, X.; Bai, J. Magnetic-assisted aptamer-based fluorescent assay for allergen detection in food matrix. Sens. Actuators B Chem. 2018, 263, 43–49. [Google Scholar] [CrossRef]
- Chinnappan, R.; Rahamn, A.A.; AlZabn, R.; Kamath, S.; Lopata, A.L.; Abu-Salah, K.M.; Zourob, M. Aptameric biosensor for the sensitive detection of major shrimp allergen, tropomyosin. Food Chem. 2020, 314, 126133. [Google Scholar] [CrossRef]
- Zhou, J.; Ai, R.; Weng, J.; Li, L.; Zhou, C.; Ma, A.; Fu, L.; Wang, Y. A “on-off-on” fluorescence aptasensor using carbon quantum dots and graphene oxide for ultrasensitive detection of the major shellfish allergen Arginine kinase. Microchem. J. 2020, 158, 105171. [Google Scholar] [CrossRef]
- Shi, Y.; Pan, Y.; Zhang, H.; Zhang, Z.; Li, M.J.; Yi, C.; Yang, M. A dual-mode nanosensor based on carbon quantum dots and gold nanoparticles for discriminative detection of glutathione in human plasma. Biosens. Bioelectron. 2014, 56, 39–45. [Google Scholar] [CrossRef]
- Phadke, C.; Tada, S.; Kono, I.; Hiyama, A.; Takase, Y.; Gayama, S.; Aigaki, T.; Ito, Y.; Uzawa, T. Instantaneous detection of αs-casein in cow’s milk using fluorogenic peptide aptamers. Anal. Methods 2020, 12, 1368–1373. [Google Scholar] [CrossRef]
- Shi, M.; Cen, Y.; Sohail, M.; Xu, G.; Wei, F.; Ma, Y.; Xu, X.; Ma, Y.; Song, Y.; Hu, Q. Aptamer based fluorometric β-lactoglobulin assay based on the use of magnetic nanoparticles and carbon dots. Microchim. Acta 2018, 185, 1–8. [Google Scholar] [CrossRef]
- Qi, S.; Duan, N.; Sun, Y.; Zhou, Y.; Ma, P.; Wu, S.; Wang, Z. High-affinity aptamer of allergen β-lactoglobulin: Selection, recognition mechanism and application. Sens. Actuators B Chem. 2021, 340, 129956. [Google Scholar] [CrossRef]
- Jimenez-Lopez, J.C.; Foley, R.C.; Brear, E.; Clarke, V.C.; Lima-Cabello, E.; Florido, J.F.; Singh, K.B.; Alché, J.D.; Smith, P.M.C. Characterization of narrow-leaf lupin (Lupinus angustifolius L.) recombinant major allergen IgE-binding proteins and the natural β-conglutin counterparts in sweet lupin seed species. Food Chem. 2018, 244, 60–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, C.K.; Mairal, T.; Jauset-Rubio, M.; Svobodova, M.; Skouridou, V.; Esposito, V.; Virgilio, A.; Galeone, A. Aptamers Against the β-Conglutin Allergen: Insights into the Behavior of the Shortest Multimeric(Intra)Molecular DNA G-Quadruplex. Int. J. Mol. Sci. 2021, 22, 1150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Q.; Wang, C.; Chen, J.; Cheng, Z.; Luo, L.; Sun, J.; Fu, L. Variations in oral microbiota and salivary proteomics reveal distinct patterns in polysensitized individuals. Allergy 2022, 77, 1899–1902. [Google Scholar] [CrossRef] [PubMed]
- Erban, T.; Klimov, P.B.; Harant, K.; Talacko, P.; Nesvorna, M.; Hubert, J. Label-free proteomic analysis reveals differentially expressed Wolbachia proteins in Tyrophagus putrescentiae: Mite allergens and markers reflecting population-related proteome differences. J. Proteom. 2021, 249, 104356. [Google Scholar] [CrossRef]
- Xiao, M.; Yang, J.; Feng, Y.; Zhu, Y.; Chai, X.; Wang, Y. Metaproteomic strategies and applications for gut microbial research. Appl. Microbiol. Biotechnol. 2017, 101, 3077–3088. [Google Scholar] [CrossRef]
- Wilmes, P.; Bond, P.L. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ. Microbiol. 2004, 6, 911–920. [Google Scholar] [CrossRef]
- Carrasco-Navarro, U.; Vera-Estrella, R.; Barkla, B.J.; Zúñiga-León, E.; Reyes-Vivas, H.; Fernández, F.J.; Fierro, F. Proteomic analysis of the signaling pathway mediated by the heterotrimeric Ga protein Pga1 of Penicillium chrysogenum. Microb. Cell Fact. 2016, 15, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Verberkmoes, N.C.; Russell, A.L.; Shah, M.; Godzik, A.; Rosenquist, M.; Halfvarson, J.; Lefsrud, M.G.; Apajalahti, J.; Tysk, C.; Hettich, R.L.; et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2008, 3, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Mayne, J.; Ning, Z.; Zhang, X.; Starr, A.E.; Chen, R.; Deeke, S.; Chiang, C.K.; Xu, B.; Wen, M.; Cheng, K.; et al. Bottom-Up Proteomics (2013-2015): Keeping up in the Era of Systems Biology. Anal. Chem. 2016, 88, 95–121. [Google Scholar] [CrossRef]
- Fu, B.C.; Randolph, T.W.; Lim, U.; Monroe, K.R.; Cheng, I.; Wilkens, L.R.; Le Marchand, L.; Hullar, M.A.J.; Lampe, J.W. Characterization of the gut microbiome in epidemiologic studies: The multiethnic cohort experience. Ann. Epidemiol. 2016, 26, 373–379. [Google Scholar] [CrossRef] [Green Version]
- Curtis, H.; Dirk, G.; Rob, K.; Sahar, A.; Jonathan, H.B.; Asif, T.C. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Xu, X.; Shi, L.; Wang, M. Comparative quantitative proteomics unveils putative mechanisms involved into mercury toxicity and tolerance in Tigriopus japonicus under multigenerational exposure scenario. Environ. Pollut. 2016, 218, 1287–1297. [Google Scholar] [CrossRef]
- Wang, H.B.; Zhang, Z.X.; Li, H.; He, H.B.; Fang, C.X.; Zhang, A.J.; Li, Q.S.; Chen, R.S.; Guo, X.K.; Lin, H.F.; et al. Characterization of metaproteomics in crop rhizospheric soil. J. Proteome Res. 2011, 10, 932–940. [Google Scholar] [CrossRef]
- López, J.L.; Marina, A.; Álvarez, G.; Vázquez, J. Application of proteomics for fast identification of species-specific peptides from marine species—López—2002—PROTEOMICS—Wiley Online Library. Proteom. Syst. Biol. 2002, 2, 1658–1665. [Google Scholar]
- Tanca, A.; Palomba, A.; Pisanu, S.; Addis, M.F.; Uzzau, S. Enrichment or depletion? The impact of stool pretreatment on metaproteomic characterization of the human gut microbiota. Proteomics 2015, 15, 3474–3485. [Google Scholar] [CrossRef]
- Rose, C.; Parker, A.; Jefferson, B.; Cartmell, E. The Characterization of Feces and Urine: A Review of the Literature to Inform Advanced Treatment Technology. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1827–1879. [Google Scholar] [CrossRef] [Green Version]
- Stephen, A.M.; Cummings, J.H. The microbial contribution to human faecal mass. J. Med. Microbiol. 1980, 13, 45–56. [Google Scholar] [CrossRef]
- Van der Post, S.; Arike, L. Metaproteomics Analysis of Host–Microbiota Interfaces. Methods Mol. Biol. 2021, 2259, 167–179. [Google Scholar]
- Zhang, X.; Deeke, S.A.; Ning, Z.; Starr, A.E.; Butcher, J.; Li, J.; Mayne, J.; Cheng, K.; Liao, B.; Li, L.; et al. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Blackburn, J.M.; Martens, L. The challenge of metaproteomic analysis in human samples. Expert Rev. Proteom. 2016, 13, 135–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalski, A.; Damoc, E.; Hauschild, J.P.; Lange, O.; Wieghaus, A.; Makarov, A.; Nagaraj, N.; Cox, J.; Mann, M.; Horning, S. Mass spectrometry-based proteomics using Q exactive, a high-performance benchtop quadrupole orbitrap mass spectrometer. Mol. Cell. Proteom. 2011, 10, M111.011015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleiner, M. Metaproteomics: Much More than Measuring Gene Expression in Microbial Communities. mSystems 2019, 4, e00115-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, A.; Nayfach, S.; Boland, M.; Strozzi, F.; Beracochea, M.; Shi, Z.J.; Pollard, K.S.; Sakharova, E.; Parks, D.H.; Hugenholtz, P.; et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 2020, 39, 105–114. [Google Scholar] [CrossRef]
- Tanca, A.; Palomba, A.; Deligios, M.; Cubeddu, T.; Fraumene, C.; Biosa, G.; Pagnozzi, D.; Addis, M.F.; Uzzau, S. Evaluating the Impact of Different Sequence Databases on Metaproteome Analysis: Insights from a Lab-Assembled Microbial Mixture. PLoS ONE 2013, 8, e82981. [Google Scholar] [CrossRef]
- De Souza, G.A.; Wiker, H.G. The Impact of Proteomic Advances on Bacterial Gene Annotation. Curr. Proteom. 2012, 6, 84–92. [Google Scholar] [CrossRef]
- Stamboulian, M.; Li, S.; Ye, Y. Using high-abundance proteins as guides for fast and effective peptide/protein identification from human gut metaproteomic data. Microbiome 2021, 9, 148. [Google Scholar] [CrossRef]
- Stamboulian, M.; Canderan, J.; Ye, Y. Metaproteomics as a tool for studying the protein landscape of human-gut bacterial species. PLOS Comput. Biol. 2022, 18, e1009397. [Google Scholar] [CrossRef]
- Presley, L.L.; Ye, J.; Li, X.; Leblanc, J.; Zhang, Z.; Ruegger, P.M.; Allard, J.; McGovern, D.; Ippoliti, A.; Roth, B.; et al. Host–Microbe Relationships in Inflammatory Bowel Disease Detected by Bacterial and Metaproteomic Analysis of the Mucosal–Luminal Interface. Inflamm. Bowel Dis. 2012, 18, 409–417. [Google Scholar] [CrossRef] [Green Version]
- Juste, C.; Kreil, D.P.; Beauvallet, C.; Guillot, A.; Vaca, S.; Carapito, C.; Mondot, S.; Sykacek, P.; Sokol, H.; Blon, F.; et al. Bacterial protein signals are associated with Crohn’s disease. Gut 2014, 63, 1566–1577. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, T.; Schallert, K.; Vilchez-Vargas, R.; Benndorf, D.; Püttker, S.; Sydor, S.; Schulz, C.; Bechmann, L.; Canbay, A.; Heidrich, B.; et al. Metaproteomics of fecal samples of Crohn’s disease and Ulcerative Colitis. J. Proteom. 2019, 201, 93–103. [Google Scholar] [CrossRef]
- Kolmeder, C.A.; Salojärvi, J.; Ritari, J.; De Been, M.; Raes, J.; Falony, G.; Vieira-Silva, S.; Kekkonen, R.A.; Corthals, G.L.; Palva, A.; et al. Faecal Metaproteomic Analysis Reveals a Personalized and Stable Functional Microbiome and Limited Effects of a Probiotic Intervention in Adults. PLoS ONE 2016, 11, e0153294. [Google Scholar] [CrossRef] [Green Version]
- Kingkaw, A.; Nakphaichit, M.; Suratannon, N.; Nitisinprasert, S.; Wongoutong, C.; Chatchatee, P.; Krobthong, S.; Charoenlappanit, S.; Roytrakul, S.; Vongsangnak, W. Analysis of the infant gut microbiome reveals metabolic functional roles associated with healthy infants and infants with atopic dermatitis using metaproteomics. PeerJ 2020, 8, e9988. [Google Scholar] [CrossRef]
- Mok, K.; Suratanon, N.; Roytrakul, S.; Charoenlappanit, S.; Patumcharoenpol, P.; Chatchatee, P.; Vongsangnak, W.; Nakphaichit, M. ITS2 Sequencing and Targeted Meta-Proteomics of Infant Gut Mycobiome Reveal the Functional Role of Rhodotorula sp. during Atopic Dermatitis Manifestation. J. Fungi 2021, 7, 748. [Google Scholar] [CrossRef]
- Maier, T.V.; Lucio, M.; Lee, L.H.; Verberkmoes, N.C.; Brislawn, C.J.; Bernhardt, J.; Lamendella, R.; McDermott, J.E.; Bergeron, N.; Heinzmann, S.S.; et al. Impact of dietary resistant starch on the human gut Microbiome, Metaproteome, and Metabolome. MBio 2017, 8, e01343-17. [Google Scholar] [CrossRef] [Green Version]
- Ke, X.; Walker, A.; Haange, S.B.; Lagkouvardos, I.; Liu, Y.; Schmitt-Kopplin, P.; von Bergen, M.; Jehmlich, N.; He, X.; Clavel, T.; et al. Synbiotic-driven improvement of metabolic disturbances is associated with changes in the gut microbiome in diet-induced obese mice. Mol. Metab. 2019, 22, 96–109. [Google Scholar] [CrossRef]
- Pan, S.; Hullar, M.A.J.; Lai, L.A.; Peng, H.; May, D.H.; Noble, W.S.; Raftery, D.; Navarro, S.L.; Neuhouser, M.L.; Lampe, P.D.; et al. Gut Microbial Protein Expression in Response to Dietary Patterns in a Controlled Feeding Study: A Metaproteomic Approach. Microorganisms 2020, 8, 379. [Google Scholar] [CrossRef] [Green Version]
- Kolmeder, C.A.; Ritari, J.; Verdam, F.J.; Muth, T.; Keskitalo, S.; Varjosalo, M.; Fuentes, S.; Greve, J.W.; Buurman, W.A.; Reichl, U.; et al. Colonic metaproteomic signatures of active bacteria and the host in obesity. Proteomics 2015, 15, 3544–3552. [Google Scholar] [CrossRef]
- Pettersen, V.K.; Dufour, A.; Arrieta, M.-C. Metaproteomic profiling of fungal gut colonization in gnotobiotic mice. Anim. Microbiome 2022, 4, 1–17. [Google Scholar] [CrossRef]
- Choi, H.; Song, W.M.; Zhang, B. Linking childhood allergic asthma phenotypes with endotype through integrated systems biology: Current evidence and research needs. Rev. Environ. Health 2017, 32, 55–63. [Google Scholar] [CrossRef]
- Bräcker, J.; Brockmeyer, J. Characterization and Detection of Food Allergens Using High-Resolution Mass Spectrometry: Current Status and Future Perspective. J. Agric. Food Chem. 2018, 66, 8935–8940. [Google Scholar] [CrossRef] [PubMed]
| Types of Food Allergy | Association with Food Allergy | Reference |
|---|---|---|
| Cow’s milk | ↓Clostridia, Firmicutes | [15] |
| Cow’s milk, egg, wheat, soy, nuts | ↓Citrobacter, Oscillospira, Lactococcus, Dorea | [16] |
| Cow’s milk, egg, peanut | ↑Enterobacteriaceae ↓Bacteroidaceae | [17] |
| Peanut | ↓Clostridiales ↑Bacteroidales | [18] |
| Egg, wheat, soybean, sesame, cow’s milk, peanut, shrimp, crab | ↓Dorea, Akkermansia ↑Veillonella | [19] |
| Cow’s milk, egg, wheat, nut, peanuts, fish, shrimp, soybeans | ↓Bacteroidetes, Proteobacteria, Actinobacteria ↑Firmicutes | [20] |
| Egg | ↑Lachnospiraceae, Streptococcaceae, Leuconostocaceae | [21] |
| Cow’s milk | ↑Lactobacillaceae ↓Bifdobacteriaceae, Ruminococcaceae | [22] |
| Cow’s milk | ↓Coriobacteriaceae | [23] |
| Cow’s milk | ↑Bacteroides, Alistipes | [11] |
| Tree nuts, fish, milk, egg, sesame, soy | ↑Oscillibacter valericigenes, Lachnoclostridium bolteae, Faecalibacterium sp. | [24] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abril, A.G.; Carrera, M.; Sánchez-Pérez, Á.; Villa, T.G. Gut Microbiome Proteomics in Food Allergies. Int. J. Mol. Sci. 2023, 24, 2234. https://doi.org/10.3390/ijms24032234
Abril AG, Carrera M, Sánchez-Pérez Á, Villa TG. Gut Microbiome Proteomics in Food Allergies. International Journal of Molecular Sciences. 2023; 24(3):2234. https://doi.org/10.3390/ijms24032234
Chicago/Turabian StyleAbril, Ana G., Mónica Carrera, Ángeles Sánchez-Pérez, and Tomás G. Villa. 2023. "Gut Microbiome Proteomics in Food Allergies" International Journal of Molecular Sciences 24, no. 3: 2234. https://doi.org/10.3390/ijms24032234







