Association of GILZ with MUC2, TLR2, and TLR4 in Inflammatory Bowel Disease
Abstract
:1. Introduction
2. Results
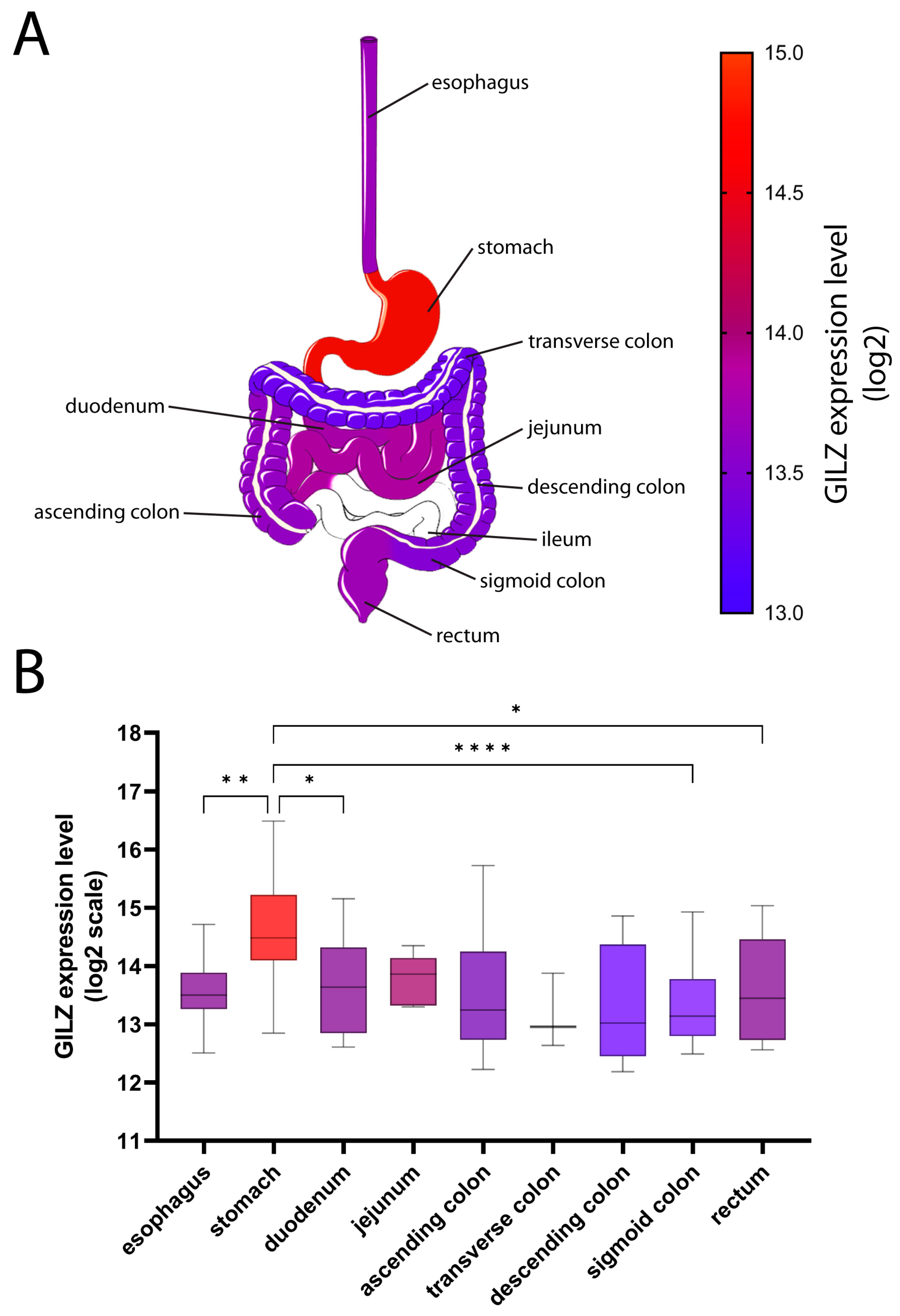
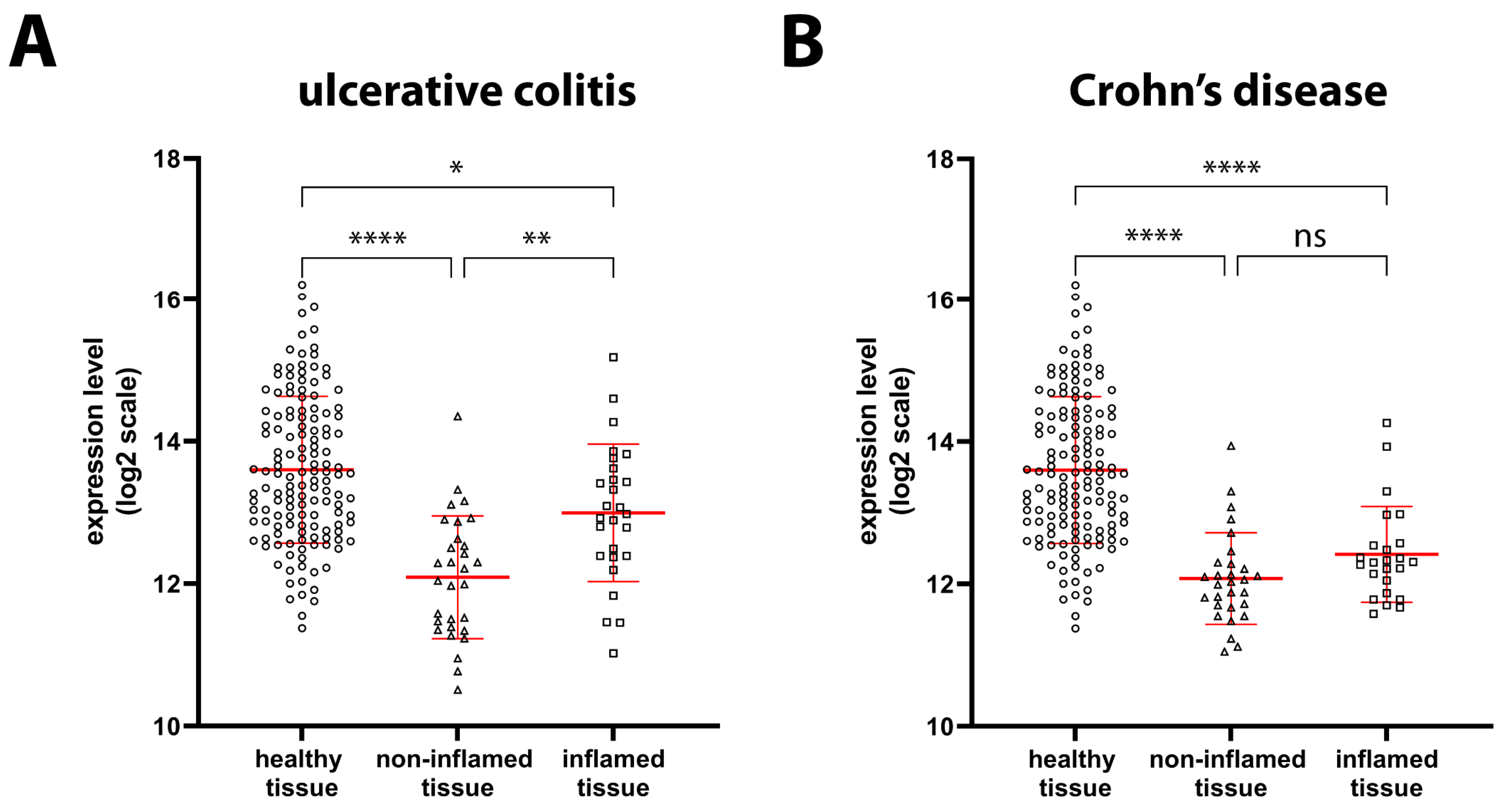
2.1. GILZ Expression in GI Tract Tissue from Healthy and IBD Patients
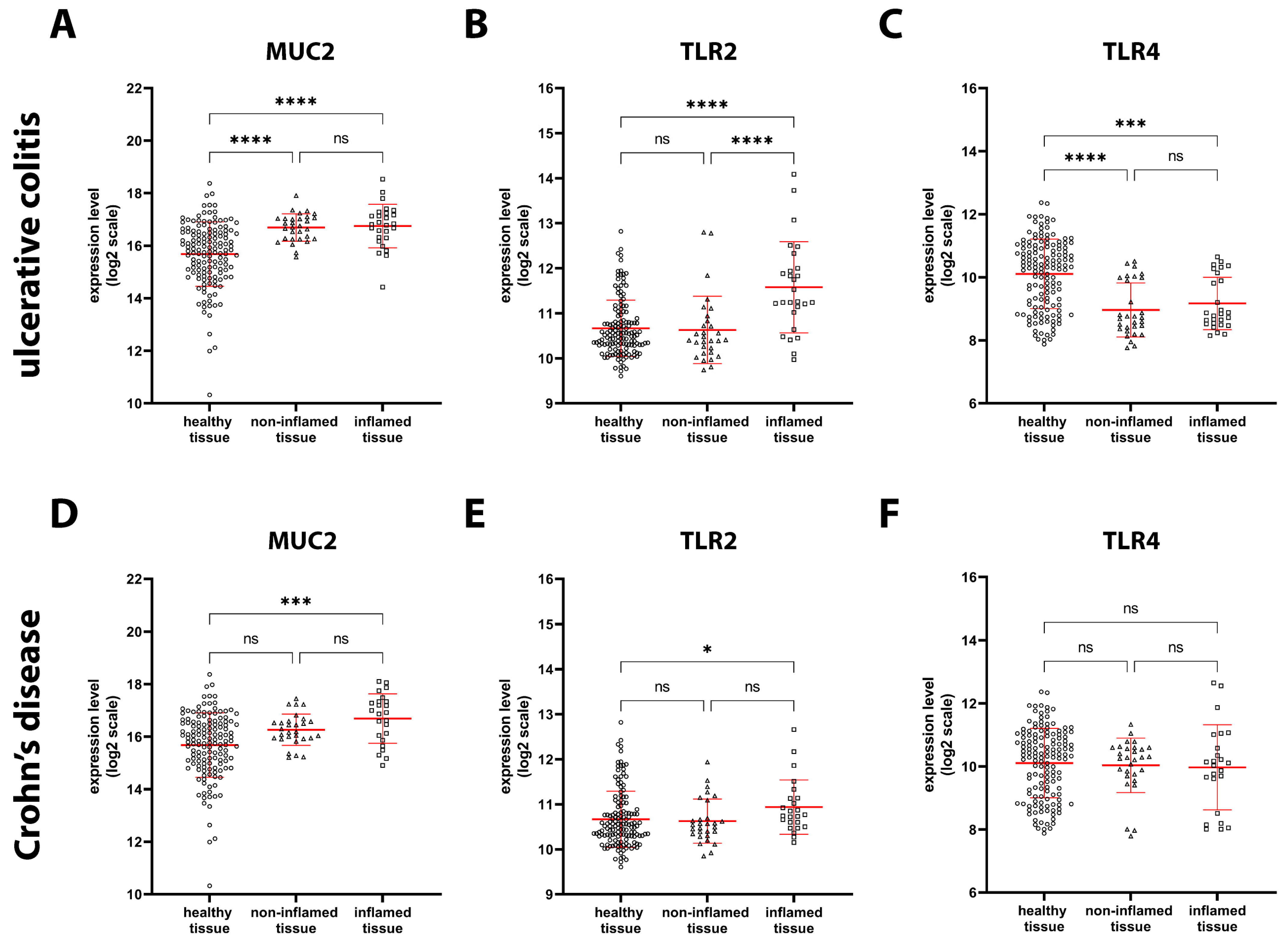
2.2. Expression of Genes Involved in Intestinal Barrier Function in Healthy and IBD Patients
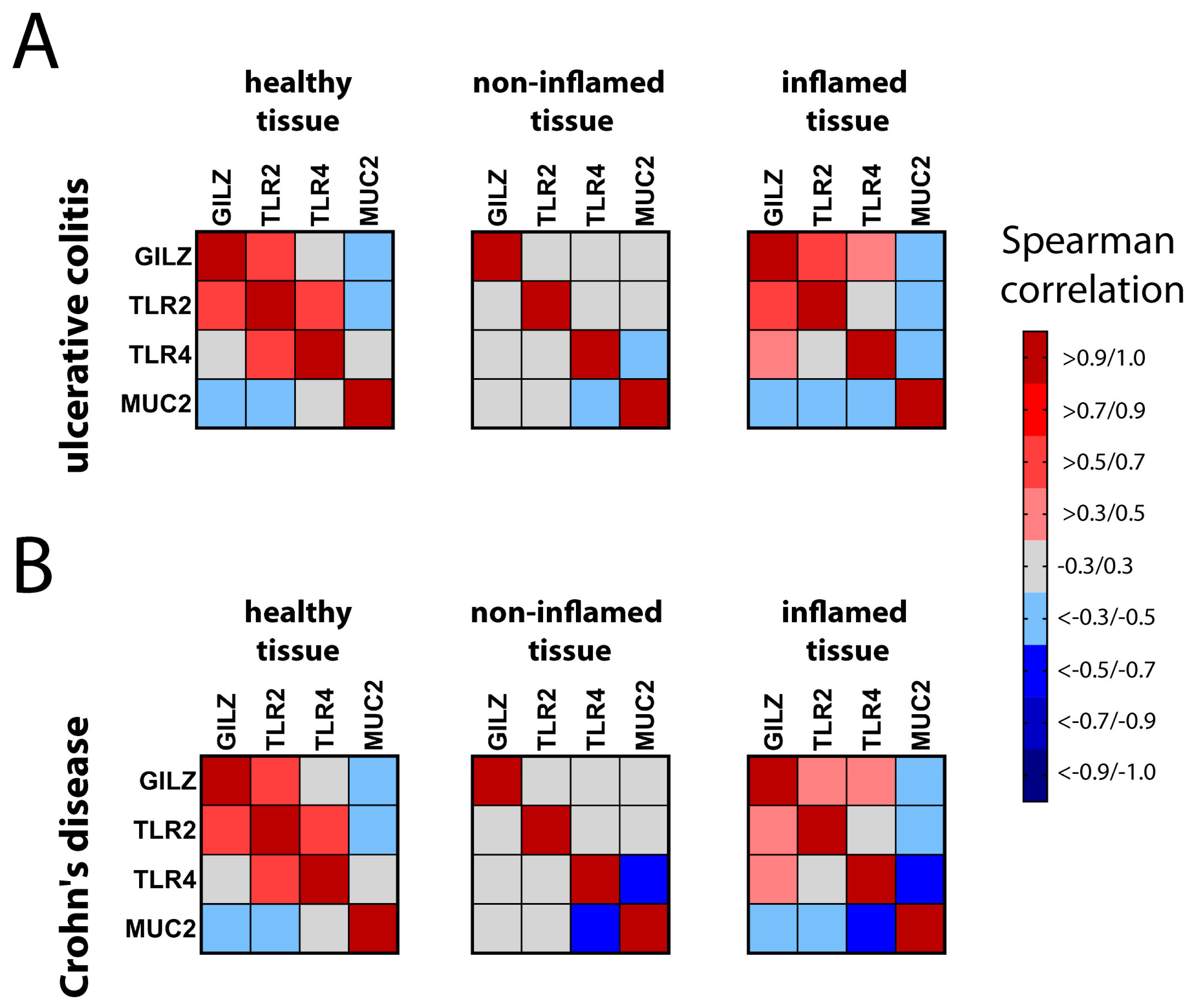
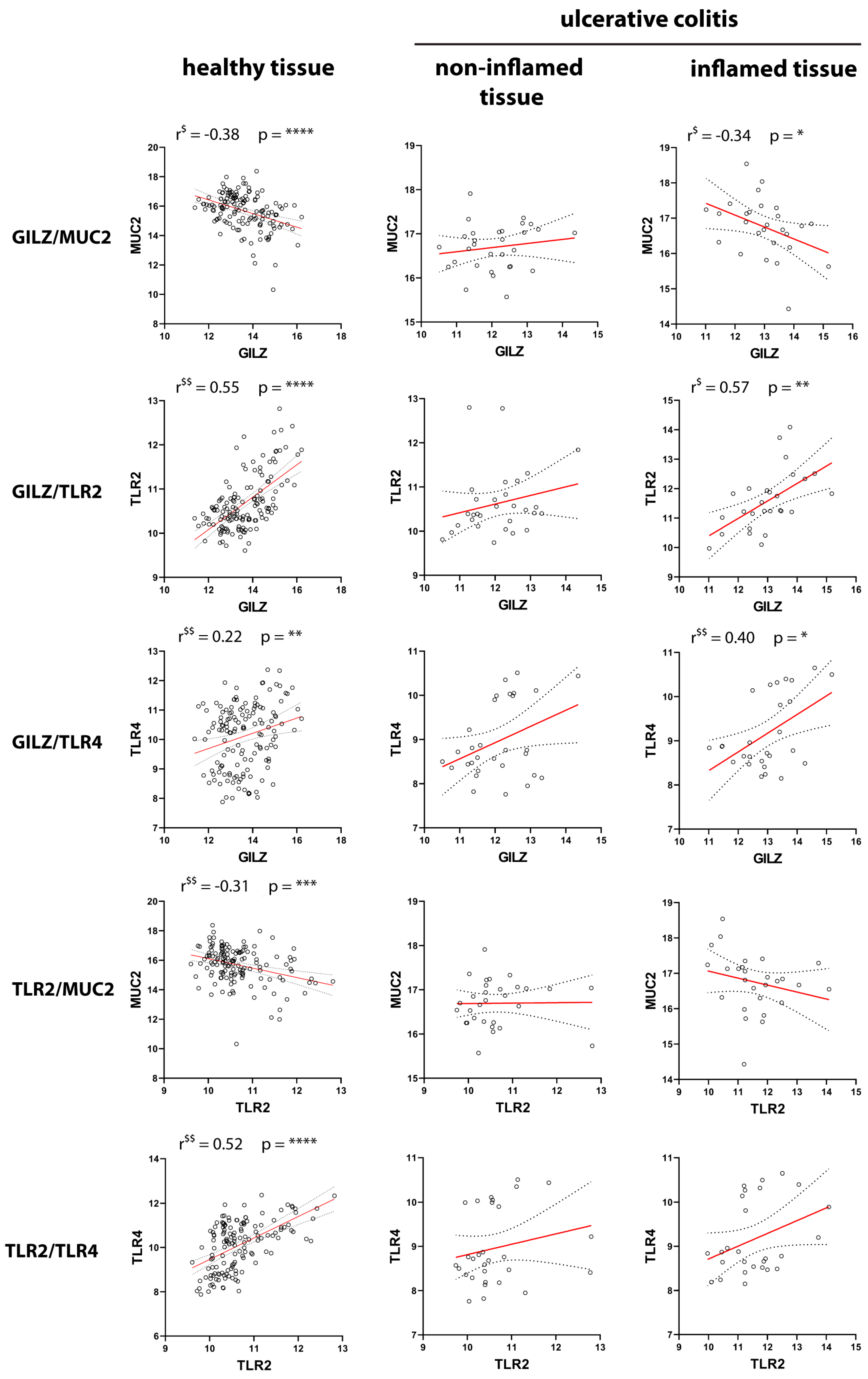
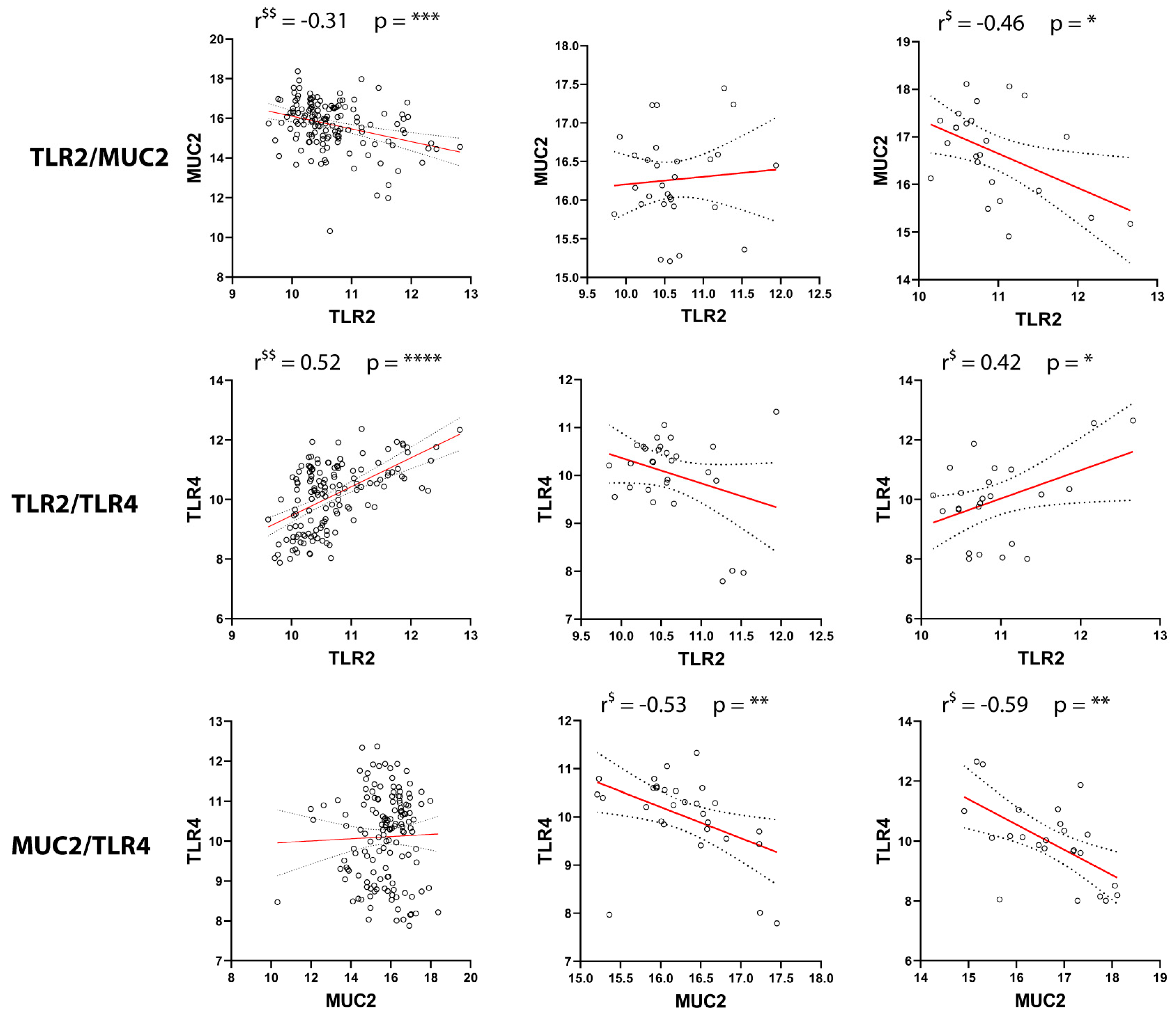
2.3. Correlation of GILZ Expression with MUC2, TLR2, and TLR4 Expression
2.4. Effect of LPS and Proinflammatory Cytokines on the Expression of Genes Involved in Gut Barrier Function
3. Discussion
4. Materials and Methods
4.1. Datasets
4.2. Gene Expression Analysis
4.3. Cell Lines and In Vitro Treatments
4.4. RNA Extraction and Reverse Transcription
4.5. Quantitative PCR
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Villanacci, V.; Reggiani-Bonetti, L.; Caprioli, F.; Saragoni, L.; Salviato, T.; Mescoli, C.; Canavese, G.; Manenti, S.; Spada, E.; Baron, L.; et al. Histopathology of inflammatory bowel disease—Position statement of the Pathologists of the Italian Group for the Study of Inflammatory Bowel Disease (IG-IBD) and Italian Group of Gastrointestinal Pathologists (GIPAD-SIAPEC). Dig. Liver Dis. 2020, 52, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.M.; Brummer, R.J.; Derrien, M.; MacDonald, T.T.; Troost, F.; Cani, P.D.; Theodorou, V.; Dekker, J.; Meheust, A.; de Vos, W.M.; et al. Homeostasis of the gut barrier and potential biomarkers. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G171–G193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aamann, L.; Vestergaard, E.M.; Gronbaek, H. Trefoil factors in inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 3223–3230. [Google Scholar] [CrossRef]
- Yao, D.; Dai, W.; Dong, M.; Dai, C.; Wu, S. MUC2 and related bacterial factors: Therapeutic targets for ulcerative colitis. EBioMedicine 2021, 74, 103751. [Google Scholar] [CrossRef]
- Burgueno, J.F.; Abreu, M.T. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 263–278. [Google Scholar] [CrossRef]
- Gibson, D.L.; Ma, C.; Bergstrom, K.S.; Huang, J.T.; Man, C.; Vallance, B.A. MyD88 signalling plays a critical role in host defence by controlling pathogen burden and promoting epithelial cell homeostasis during Citrobacter rodentium-induced colitis. Cell. Microbiol. 2008, 10, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Cario, E.; Gerken, G.; Podolsky, D.K. Toll-like receptor 2 enhances ZO-1-associated intestinal epithelial barrier integrity via protein kinase C. Gastroenterology 2004, 127, 224–238. [Google Scholar] [CrossRef]
- Cario, E.; Gerken, G.; Podolsky, D.K. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology 2007, 132, 1359–1374. [Google Scholar] [CrossRef] [PubMed]
- Birchenough, G.M.; Nystrom, E.E.; Johansson, M.E.; Hansson, G.C. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 2016, 352, 1535–1542. [Google Scholar] [CrossRef] [Green Version]
- Naito, T.; Mulet, C.; De Castro, C.; Molinaro, A.; Saffarian, A.; Nigro, G.; Berard, M.; Clerc, M.; Pedersen, A.B.; Sansonetti, P.J.; et al. Lipopolysaccharide from Crypt-Specific Core Microbiota Modulates the Colonic Epithelial Proliferation-to-Differentiation Balance. mBio 2017, 8, e01680-17. [Google Scholar] [CrossRef] [PubMed]
- Dheer, R.; Santaolalla, R.; Davies, J.M.; Lang, J.K.; Phillips, M.C.; Pastorini, C.; Vazquez-Pertejo, M.T.; Abreu, M.T. Intestinal Epithelial Toll-Like Receptor 4 Signaling Affects Epithelial Function and Colonic Microbiota and Promotes a Risk for Transmissible Colitis. Infect. Immun. 2016, 84, 798–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leoncini, G.; Gentili, M.; Lusenti, E.; Caruso, L.; Calafa, C.; Migliorati, G.; Riccardi, C.; Villanacci, V.; Ronchetti, S. The novel role of glucocorticoid-induced leucine zipper as a marker of mucosal healing in inflammatory bowel diseases. Pharmacol. Res. 2022, 182, 106353. [Google Scholar] [CrossRef]
- Ronchetti, S.; Migliorati, G.; Riccardi, C. GILZ as a Mediator of the Anti-Inflammatory Effects of Glucocorticoids. Front. Endocrinol. 2015, 6, 170. [Google Scholar] [CrossRef] [Green Version]
- Bruscoli, S.; Riccardi, C.; Ronchetti, S. GILZ as a Regulator of Cell Fate and Inflammation. Cells 2021, 11, 122. [Google Scholar] [CrossRef]
- Bereshchenko, O.; Coppo, M.; Bruscoli, S.; Biagioli, M.; Cimino, M.; Frammartino, T.; Sorcini, D.; Venanzi, A.; Di Sante, M.; Riccardi, C. GILZ promotes production of peripherally induced Treg cells and mediates the crosstalk between glucocorticoids and TGF-beta signaling. Cell Rep. 2014, 7, 464–475. [Google Scholar] [CrossRef] [Green Version]
- Bruscoli, S.; Sorcini, D.; Flamini, S.; Gagliardi, A.; Adamo, F.; Ronchetti, S.; Migliorati, G.; Bereshchenko, O.; Riccardi, C. Glucocorticoid-Induced Leucine Zipper Inhibits Interferon-Gamma Production in B Cells and Suppresses Colitis in Mice. Front. Immunol. 2018, 9, 1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, E.; Ronchetti, S.; Gabrielli, E.; Pericolini, E.; Gentili, M.; Roselletti, E.; Vecchiarelli, A.; Riccardi, C. GILZ restrains neutrophil activation by inhibiting the MAPK pathway. J. Leukoc. Biol. 2019, 105, 187–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentili, M.; Hidalgo-Garcia, L.; Vezza, T.; Ricci, E.; Migliorati, G.; Rodriguez-Nogales, A.; Riccardi, C.; Galvez, J.; Ronchetti, S. A recombinant glucocorticoid-induced leucine zipper protein ameliorates symptoms of dextran sulfate sodium-induced colitis by improving intestinal permeability. FASEB J. 2021, 35, e21950. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Kolesnikov, N.; Hastings, E.; Keays, M.; Melnichuk, O.; Tang, Y.A.; Williams, E.; Dylag, M.; Kurbatova, N.; Brandizi, M.; Burdett, T.; et al. ArrayExpress update--simplifying data submissions. Nucleic Acids Res. 2015, 43, D1113–D1116. [Google Scholar] [CrossRef]
- Thoo, L.; Noti, M.; Krebs, P. Keep calm: The intestinal barrier at the interface of peace and war. Cell Death Dis. 2019, 10, 849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artis, D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 2008, 8, 411–420. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Gersemann, M.; Becker, S.; Kubler, I.; Koslowski, M.; Wang, G.; Herrlinger, K.R.; Griger, J.; Fritz, P.; Fellermann, K.; Schwab, M.; et al. Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation 2009, 77, 84–94. [Google Scholar] [CrossRef]
- Hanski, C.; Born, M.; Foss, H.D.; Marowski, B.; Mansmann, U.; Arasteh, K.; Bachler, B.; Papenfuss, M.; Niedobitek, F. Defective post-transcriptional processing of MUC2 mucin in ulcerative colitis and in Crohn’s disease increases detectability of the MUC2 protein core. J. Pathol. 1999, 188, 304–311. [Google Scholar] [CrossRef]
- Bankole, E.; Read, E.; Curtis, M.A.; Neves, J.F.; Garnett, J.A. The Relationship between Mucins and Ulcerative Colitis: A Systematic Review. J. Clin. Med. 2021, 10, 1935. [Google Scholar] [CrossRef] [PubMed]
- Tawiah, A.; Cornick, S.; Moreau, F.; Gorman, H.; Kumar, M.; Tiwari, S.; Chadee, K. High MUC2 Mucin Expression and Misfolding Induce Cellular Stress, Reactive Oxygen Production, and Apoptosis in Goblet Cells. Am. J. Pathol. 2018, 188, 1354–1373. [Google Scholar] [CrossRef] [Green Version]
- Tytgat, K.M.; van der Wal, J.W.; Einerhand, A.W.; Buller, H.A.; Dekker, J. Quantitative analysis of MUC2 synthesis in ulcerative colitis. Biochem. Biophys. Res. Commun. 1996, 224, 397–405. [Google Scholar] [CrossRef]
- Price, A.E.; Shamardani, K.; Lugo, K.A.; Deguine, J.; Roberts, A.W.; Lee, B.L.; Barton, G.M. A Map of Toll-like Receptor Expression in the Intestinal Epithelium Reveals Distinct Spatial, Cell Type-Specific, and Temporal Patterns. Immunity 2018, 49, 560–575.e6. [Google Scholar] [CrossRef] [Green Version]
- Abreu, M.T. Toll-like receptor signalling in the intestinal epithelium: How bacterial recognition shapes intestinal function. Nat Rev. Immunol. 2010, 10, 131–144. [Google Scholar] [CrossRef]
- Ricci, E.; Roselletti, E.; Gentili, M.; Sabbatini, S.; Perito, S.; Riccardi, C.; Migliorati, G.; Monari, C.; Ronchetti, S. Glucocorticoid-Induced Leucine Zipper-Mediated TLR2 Downregulation Accounts for Reduced Neutrophil Activity Following Acute DEX Treatment. Cells 2021, 10, 2228. [Google Scholar] [CrossRef] [PubMed]
- Cario, E. Barrier-protective function of intestinal epithelial Toll-like receptor 2. Mucosal Immunol. 2008, 1 (Suppl. S1), S62–S66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehli, M.; Poltorak, A.; Schwarzfischer, L.; Krause, S.W.; Andreesen, R.; Beutler, B. PU.1 and interferon consensus sequence-binding protein regulate the myeloid expression of the human Toll-like receptor 4 gene. J. Biol. Chem. 2000, 275, 9773–9781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukata, M.; Chen, A.; Klepper, A.; Krishnareddy, S.; Vamadevan, A.S.; Thomas, L.S.; Xu, R.; Inoue, H.; Arditi, M.; Dannenberg, A.J.; et al. Cox-2 is regulated by Toll-like receptor-4 (TLR4) signaling: Role in proliferation and apoptosis in the intestine. Gastroenterology 2006, 131, 862–877. [Google Scholar] [CrossRef] [Green Version]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.W.; Kim, S.; Son, M.; Cheon, J.H.; Park, Y.S. Melatonin controls microbiota in colitis by goblet cell differentiation and antimicrobial peptide production through Toll-like receptor 4 signalling. Sci. Rep. 2020, 10, 2232. [Google Scholar] [CrossRef] [Green Version]
- Sodhi, C.P.; Neal, M.D.; Siggers, R.; Sho, S.; Ma, C.; Branca, M.F.; Prindle, T., Jr.; Russo, A.M.; Afrazi, A.; Good, M.; et al. Intestinal epithelial Toll-like receptor 4 regulates goblet cell development and is required for necrotizing enterocolitis in mice. Gastroenterology 2012, 143, 708–718.e5. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.D.; Guk, S.M.; Chai, J.Y. Toll-like receptor 2 and Muc2 expression on human intestinal epithelial cells by Gymnophalloides seoi adult antigen. J. Parasitol. 2010, 96, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef] [Green Version]
- Bjerrum, J.T.; Hansen, M.; Olsen, J.; Nielsen, O.H. Genome-wide gene expression analysis of mucosal colonic biopsies and isolated colonocytes suggests a continuous inflammatory state in the lamina propria of patients with quiescent ulcerative colitis. Inflamm. Bowel. Dis. 2010, 16, 999–1007. [Google Scholar] [CrossRef]
- Olsen, J.; Gerds, T.A.; Seidelin, J.B.; Csillag, C.; Bjerrum, J.T.; Troelsen, J.T.; Nielsen, O.H. Diagnosis of ulcerative colitis before onset of inflammation by multivariate modeling of genome-wide gene expression data. Inflamm. Bowel. Dis. 2009, 15, 1032–1038. [Google Scholar] [CrossRef]
- Nielsen, O.H.; Bjerrum, J.T.; Csillag, C.; Nielsen, F.C.; Olsen, J. Influence of smoking on colonic gene expression profile in Crohn’s disease. PLoS ONE 2009, 4, e6210. [Google Scholar] [CrossRef] [Green Version]
- Funke, B.; Lasitschka, F.; Roth, W.; Penzel, R.; Meuer, S.; Saile, M.; Gretz, N.; Sido, B.; Schirmacher, P.; Autschbach, F. Selective downregulation of retinoic acid-inducible gene I within the intestinal epithelial compartment in Crohn’s disease. Inflamm. Bowel. Dis. 2011, 17, 1943–1954. [Google Scholar] [CrossRef]
- Funke, B.; Autschbach, F.; Kim, S.; Lasitschka, F.; Strauch, U.; Rogler, G.; Gdynia, G.; Li, L.; Gretz, N.; Macher-Goeppinger, S.; et al. Functional characterisation of decoy receptor 3 in Crohn’s disease. Gut 2009, 58, 483–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwashita, J.; Sato, Y.; Sugaya, H.; Takahashi, N.; Sasaki, H.; Abe, T. mRNA of MUC2 is stimulated by IL-4, IL-13 or TNF-alpha through a mitogen-activated protein kinase pathway in human colon cancer cells. Immunol. Cell Biol. 2003, 81, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Cari, L.; Ricci, E.; Gentili, M.; Petrillo, M.G.; Ayroldi, E.; Ronchetti, S.; Nocentini, G.; Riccardi, C. A focused Real Time PCR strategy to determine GILZ expression in mouse tissues. Res. Immunol. 2015, 5, 37–42. [Google Scholar] [CrossRef] [PubMed]



| Gene | Forward | Reverse |
|---|---|---|
| MUC2 | ACTCTCCACACCCAGCATCATC | GTGTCTCCGTATGTGCCGTTGT |
| TLR4 | CTGCCACATGTCAGGCCTTAT | AATGCCCACCTGGAAGACTCT |
| GAPDH | GCTCCTCCTGTTCGACAGTCA | GCAACAATATCCACTTTACCAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cari, L.; Rosati, L.; Leoncini, G.; Lusenti, E.; Gentili, M.; Nocentini, G.; Riccardi, C.; Migliorati, G.; Ronchetti, S. Association of GILZ with MUC2, TLR2, and TLR4 in Inflammatory Bowel Disease. Int. J. Mol. Sci. 2023, 24, 2235. https://doi.org/10.3390/ijms24032235
Cari L, Rosati L, Leoncini G, Lusenti E, Gentili M, Nocentini G, Riccardi C, Migliorati G, Ronchetti S. Association of GILZ with MUC2, TLR2, and TLR4 in Inflammatory Bowel Disease. International Journal of Molecular Sciences. 2023; 24(3):2235. https://doi.org/10.3390/ijms24032235
Chicago/Turabian StyleCari, Luigi, Lucrezia Rosati, Giuseppe Leoncini, Eleonora Lusenti, Marco Gentili, Giuseppe Nocentini, Carlo Riccardi, Graziella Migliorati, and Simona Ronchetti. 2023. "Association of GILZ with MUC2, TLR2, and TLR4 in Inflammatory Bowel Disease" International Journal of Molecular Sciences 24, no. 3: 2235. https://doi.org/10.3390/ijms24032235






